Abstract
WRKY transcription factors have been extensively characterized in the past 20 years, but in wheat, studies on WRKY genes and their function are lagging behind many other species. To explore the function of wheat WRKY genes, we identified a TaWRKY68 gene from a common wheat cultivar. It encodes a protein comprising 313 amino acids which harbors 19 conserved motifs or active sites. Gene expression patterns were determined by analyzing microarray data of TaWRKY68 in wheat and of orthologous genes from maize, rice and barley using Genevestigator. TaWRKY68 orthologs were identified and clustered using DELTA-BLAST and COBALT programs available at NCBI. The results showed that these genes, which are expressed in all tissues tested, had relatively higher levels in the roots and were up-regulated in response to biotic stresses. Bioinformatics results were confirmed by RT-PCR experiments using wheat plants infected by Agrobacterium tumefaciens and Blumeria graminis, or treated with Deoxynivalenol, a Fusarium graminearum-induced mycotoxin in wheat or barley. In summary, TaWRKY68 functions differ during plant developmental stages and might be representing a hub gene function in wheat responses to various biotic stresses. It was also found that including data from major cereal genes in the bioinformatics analysis gave more accurate and comprehensive predictions of wheat gene functions.
Keywords: wheat, TaWRKY68, Orthologous genes, genevestigator, gene functions
Introduction
WRKY transcription factors play important roles in various biological processes, such as plant growth and development and responses to stress. WRKY proteins are defined by one or two WRKY domains, which consist of about 60 highly conserved amino acids with WRKYGQK in the N-terminus and a zinc-finger pattern C-Xn-C-Xn-HX-H/C in the C-terminus (Rushton et al., 1996; Eulgem et al., 2000). WRKY proteins regulate gene expression through binding to the W-BOX motif TTGACC/T in gene promoters (Eulgem et al., 2000; Rushton et al., 2010).
The first WRKY gene was isolated from Ipomoea batatas and was named SWEET POTATO FACTOR1 (SPF1) with a potential role in regulating gene expression by sucrose (Ishiguro and Nakamura, 1994). In another study, two WRKY proteins from wild oat, ABF1 and ABF2, were found to bind to the W-BOX region of the α-amylase gene promoter. Rushton et al. (1996) coined the name WRKY proteins and identified WRKY1, WRKY2 and WRKY3 genes from parsley, involved in plant responses to pathogens. Since then, many WRKY members have been identified in different plant species, including wheat. Wu et al. (2008) cloned 15 wheat WRKY genes based on WRKY sequences from rice and analyzed gene expression patterns under environmental stresses. Other studies showed that TaWRKY78, WRKY2 and WRKY19 can enhance plant tolerance to biotic and abiotic stresses (Proietti et al., 2011; Niu et al., 2012).
Bioinformatics tools have been successfully used for the classification and evolutionary analysis of WRKY genes in plants (Eulgem et al., 2000; Ross et al., 2007). As for the prediction of gene function, many bioinformatics studies have linked genes of interest with published homologous genes, or have analyzed gene expression profiles from microarray samples under different conditions within the same species (Persson et al., 2005). But these methods are not applicable when homologous genes have not been identified or microarray samples are lacking, especially for genes from wheat, for which little genetic and genomic resources are available.
Genevestigator (Zimmermann et al., 2004; Hruz et al., 2008) provides an online platform to access large and well-annotated datasets of curated microarray samples testing thousands of genes. Expression patterns of individual genes under different conditions can be retrieved to provide deep insight into gene function. The expression signal of genes of interest is the average of gene expression levels across many samples sharing the same biological context. Therefore, the Genevestigator platform is a powerful bioinformatics tool for functional analysis of any gene that has corresponding probes in the Genevestigator database.
Here we cloned the wheat TaWRKY68 gene and used the sequence for BLAST queries in the NCBI database to obtain homologous genes from monocot plant species included in the Genevestigator database. Next, the expression profiles of TaWRKY68 and its homologs were analyzed under a much broader range of treatments and developmental stages using the Genevestigator tools. RT-PCR was then carried out to validate the results from the bioinformatic analysis. This study provides key clues in understanding the roles of TaWRKY68 gene in wheat.
Material and Methods
Cloning of the TaWRKY68 gene
Total RNA was extracted from fresh leaves of wheat cultivar Yangmai 158 and then reverse transcribed to cDNA using the Invitrogen Superscript III transcriptase kit (Invitrogen). PCR reaction mixtures of 20 ul reaction volume contained: 1 μL of cDNA, 1x Phusion HF Buffer, 200 μM dNTP mix, 0.5 μM gene specific primers, 1 U/50 μL Phusion DNA polymerase, 3% DMSO, and water. PCR parameters for TaWRKY68 amplification were: 98 °C for 1 min, 35 cycles of 98 °C for 20 s, 61 °C for 20 s and 72 °C for 1min 40s, and a final extension step of 72 °C for 10 min. TaWRKY68 primers, designed based on the coding sequence of the TaWRKY68 gene using DNAMAN software, were: Forward primer: 5’AGC GAG CCA AGA TCT GCA GAG T, Reverse primer: 5’AAC TAA GTC AGA CGT GCC CGT TG (Wu et al., 2008). The cDNA fragment of TaWRKY68 was then cloned into pGEM-T Easy vector (Promega) for confirmation by sequencing.
Homologous genes from main crops
The CDD (Conserved Domains Database) and Prosite databases were used to search for conserved motifs. The protein sequence of TaWRKY68 was submitted to a CD search in the CDD database (NCBI), and to a Motif Scan procedure in Myhits, with Prosite databases selected.
The TaWRKY68 sequence was used in BLASTP queries against the NCBI database and was aligned with the NR (Non-redundancy protein sequences) database using the newly developed algorithm DELTA-BLAST (Domain Enhanced Lookup Time Accelerated BLAST). Homologous genes from wheat and some well sequenced monocot plants (barley, Brachypodium, maize, sorghum and rice) were selected for further analysis.
Phylogenetic analysis
Sequences of homologous genes were submitted to the COBALT (Constraint-based Multiple Protein Alignment Tool)program for multiple alignment. Alignment results were then imported to Geneious software to construct a phylogenetic tree using the Neighbour-Joining method.
Expression profiles
Affymetrix probe IDs corresponding to the homologous genes were identified from wheat, barley, rice and maize using the BLAST tool NetAffx in the Affymetrix website. Their expression profiles were analyzed by submitting probe IDs to the corresponding organism databases in the Genevestigator online search engine. Gene function under different conditions (anatomy, development and treatments) could be revealed through this online bioinformatics tool. In order to maximize comparability, all experimental data from users were normalized using Affymetrix MAS 5.0 software. Signal intensities and P values were collected for each hybridized Affymetrix GeneChip array (Zimmermann et al., 2004) .
Experimental validation of bioinformatics analyses
Roots, leaves and spikelets in the booting and seedling stages and stems in booting stage were sampled from wheat cultivar Yangmai 158 for spatio-temporal expression analysis. Inflorescences of wheat cultivar Yangmai 158 susceptible to Fusarium Head Blight fungus were dipped into Deoxynivalenol solution (0.2μg/μL) (Gardiner et al., 2010) and sampled at 2, 6 and 10 h. Seedlings at the six-leaf stage were treated with Agrobaterium tumefaciens strain EHA105 for 1.5 h following a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana (Clough and Bent, 1998). The leaves of the seedlings were cut into pieces at 1, 6 and 12 h, respectively, and were collected randomly. Seedlings from the two-leaf stage of wheat cultivar Jingdong 8, which is susceptible to powdery mildew fungus, were inoculated with Blumeria graminis f. sp. tritici (Bgt) isolate E09, and leaves were sampled at 12 h after the inoculation. Mock-treated materials of wheat plants were collected, and water control for DNA contamination was used in this work.
All samples were subjected to total RNA extraction and reverse transcription for cDNA synthesis. Amplification of TaWRKY 68 was carried out in a 10 μL of reaction mix consisting of: 1x Phusion HF Buffer, 200 μM dNTP mix, 0.5 μM gene specific primers, 1 U/50 μL Phusion DNA polymerase (New England Biolabs, Inc), 3% DMSO, cDNA and water. The amplification of TaWRKY68 was done using the same primers described for cloning the gene. The reaction mix for the endogenous control gene β-Actin (TaACTIN) was: 1x reaction Buffer, 200 μM dNTP mix, 0.5 μM gene specific primers, 5 U/μL Taq DNA polymerase, cDNA and water. The PCR protocol for TaACTIN was: 94 °C for 10 min, 25 cycles of 95 °C for 30 s, 58 °C for 30 s and 72 °C for 40 s, and a final extension step of 72 °C for 10min. The TaACTIN primer sequences were as follows: Forward primer:GGAATCCATGAGACCACCTAC, Reverse Primer: GACCCAGACAACTCGCAAC. Each experiment was conducted with two biological repeats and at least two technical repeats, and apmplification products were visualized by ethidium bromide staining in gels.
Results
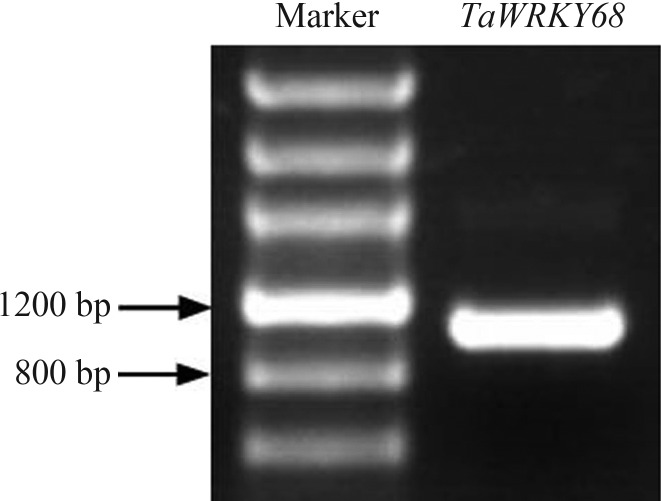
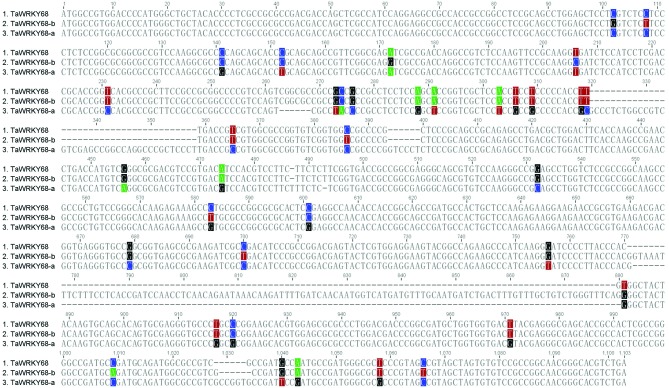
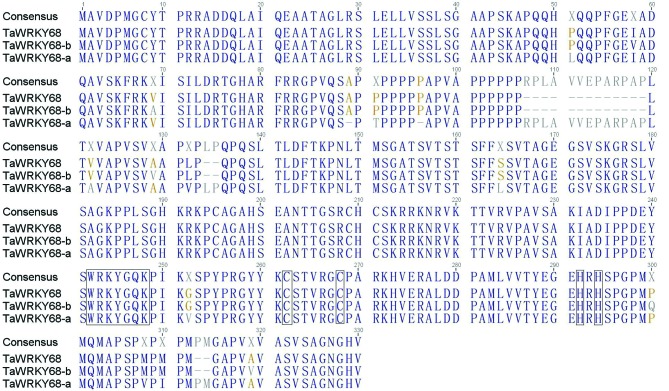
TaWRKY68 was cloned from fresh leaves of wheat cultivar Yangmai158, and the size of its cDNA fragment was 996 bp (Figure 1). BLAST analysis indicated that TaWRKY68 shares 91% and 99% similarity (Figure 2) in DNA sequence with TaWRKY68-a and TaWRKY68-b, respectively. The similarity values in protein sequence were 91% and 98% respectively (Figure 3). This newly cloned gene is, thus, an allelic variant of TaWRKY68-a and TaWRKY68-b.
Figure 1.
Isolation of TaWRKY68 by RT-PCR. The first lane and the second lane indicate the DNA marker and the coding sequence of the TaWRKY68 gene, respectively. Arrows indicate the size of DNA fragments in the marker.
Figure 2.
Coding sequence alignment of TaWRKY68 genes. Polymorphic sites are colored and gap sequences are hyphened in the diagram.
Figure 3.
Protein sequence alignment of TaWRKY68 transcription factors. Conserved WRKYGQK and zinc-finger patterns C-X5-C-X23-H-X1-H are boxed in the diagram.
Sequence features are the foundation of biological functions of genes. Therefore, we identified conserved domains, motifs and active sites by searching the Prosite and Pfam databases using the TaWRKY68 sequence as a query. This analysis predicted 19 sequence features for TaWRKY68. Six conserved domains or motifs were identified including the WRKY domain, a Plant Zinc Clust domain, a Pro_Rich region, a Met_Rich region, a Nuclear Localization Signal (NLS) and a Filamin domain (Figure 4). Thirteen active sites were detected, including two CK2_Phospho_Sites, six PKC_Phospho_Sites, two Asn_Glycosylation sites, one Microbodies_Cter signal and two Myristyle sites. These features are related to subcellular localization, signal transduction, transcriptional regulation and protein interaction, and build a basis for TaWRKY68 function.
Figure 4.
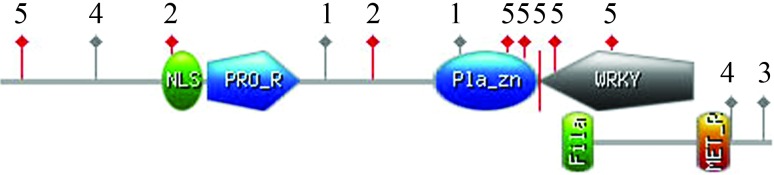
Sequence features of the TaWRKY68 protein. Domains or motifs: WRKY(WRKY domain), Pla_Zn (Plant Zinc Clust domain), PRO_R (Pro_Rich region), MET_R (Met_Rich region), NLS (Nuclear Localization Signal) and Fila (Filamin domain; Active sites: 1, Asn_Glycosylation site; 2, CK2 _Phospho_Site; 3, Microbodies_Cter site; 4, Myristyle site; 5, PKC_Phospho_Site.
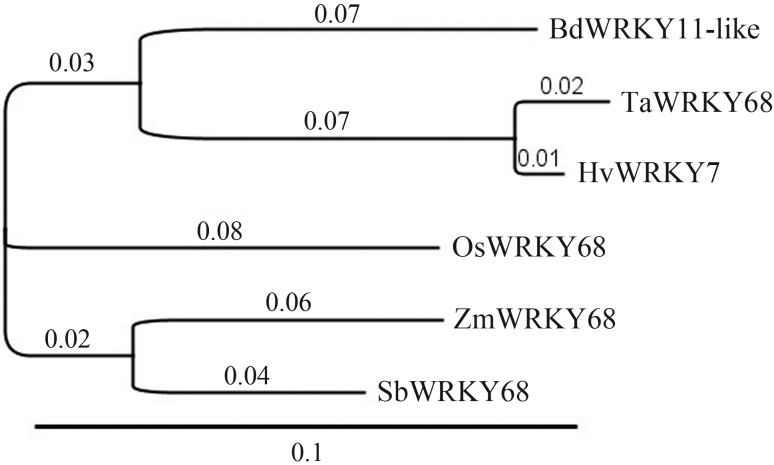
To expand the available resources for the prediction of the TaWRKY68 gene function, we searched for homologous genes in the NCBI databases using the newly developed DELTA-BLAST tool. Genes from sequenced monocot species (rice, maize, sorghum, barley and Brachypodium) were selected for multi-alignment using the COBALT tool and establishing a phylogenetic tree based on the Neighbour-Joining method. The results showed that the six proteins can be classified into three groups: one group with TaWRKY68, HvWRKY7 and BdWRKY11-like, another group including ZmWRKY68 and SbWRKY68, and OsWRKY68 which out-grouped from the other proteins (Figure 5), this clearly showing protein relationships based on sequence similarity.
Figure 5.
Relationship between TaWRKY68 and orthologs revealed by phylogenetic analysis. Numbers stands for substitutions per site, and the scale bar (0.1) is marked.
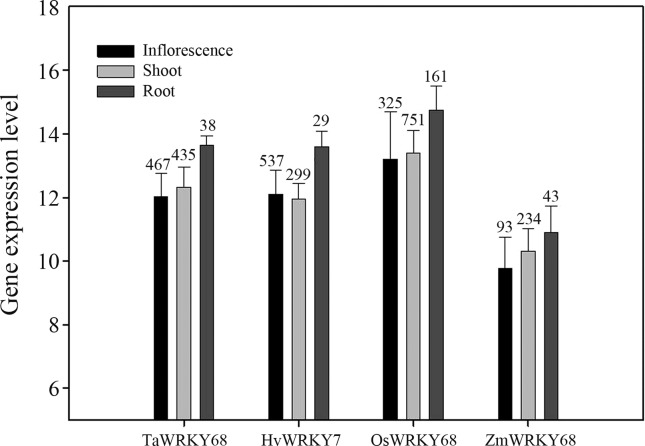
Because homologous genes share high similarity in sequence, they can also be expected to function in a similar way. This means that gene expression data from homologous genes could provide more evidence for functional predictions of TaWRKY68. Genevestigator online tools collect high quality gene expression data including those from monocot plants, such as wheat, barley, maize and rice. Sequences obtained from the phylogenetic analysis were used to query the Affymetrix probeset database and probes were identified as follows: TaAffx.128870.1.S1_at (TaWRKY68, E-value 0.0; Score 795), Contig7798_at (HvWRKY7, E-value 0.0; Score 880), Zm.4272.1.S1_at (ZmWRKY68, E-value 0.0; Score 1301), and Os.49549.1.S1_at (OsWRKY68, E-value 0.0; Score 2179). Genevestigator analysis with these probes revealed similar spatio-temporal expression patterns among TaWRKY68 and homologous genes. The genes were expressed in roots, leaves and flowers, but the expression levels in roots were higher than in other tissues (Figure 6). To verify the results from the Genevestigator analysis, we carried out RT-PCR tests using wheat leaves, roots and inflorescences. The RT-PCR results showed that TaWRKY 68 was expressed in all the samples, with the highest level in wheat roots (Figure 7), which was consistent with the bioinformatic analysis.
Figure 6.
Spatio-temporal expression patterns of TaWRKY68 and orthologous genes. All data were normalized in the Genevestigator platform and are presented as mean ± SEM, with sample size marked above the error bars.
Figure 7.
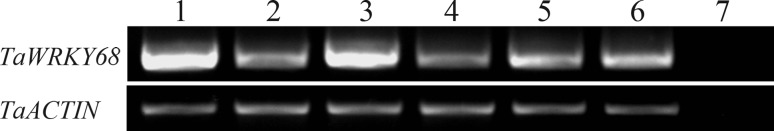
RT-PCR validations of spatio-temporal expression patterns of the TaWRKY68 gene. 1 seedling roots, 2 seedling leaves, 3 roots at the booting stage, 4 leaves at the booting stage, 5 stem at the booting stage, 6 inflorescence at the booting stage, 7 water control.
To clarify the role of TaWRKY68 in plant responses to environmental stimuli, we analyzed gene expression profiles in wheat, barley, rice and maize under various environmental conditions using Genevestigator tools. Genes with a threshold of three-fold expression level change and a 0.01 statistically significant difference were filtered as responding to the respective environmental condition. The results showed that these genes were up-regulated by some biotic stresses (Agrobacterium tumefaciens and Magnaporthe grisea) or elicitor (Deoxynivalenol) (Table 1), indicating that WRKY 68 genes could play important roles in plant responses to biotic stresses. In addition, WRKY68 genes might also be involved in plant responses to abiotic stresses because the expression change of TaWRKY68 (fold change of 2.93, p value = 0.002) nearly approached the threshold in wheat challenged by drought treatment (Ergen et al., 2009).
Table 1.
Processes that TaWRKY68 and its homologous genes are involved in.
| Gene | Processa | References |
|---|---|---|
| HvWRKY7 | Deoxynivalenol treatment (3.31) | Gardiner et al. (2010) |
| OsWRKY68 | Agrobacterium infection (3.75) | Tie et al. (2012) |
| OsWRKY68 | Magnaporthe infection (4.26) | Ribot et al. (2008) |
Fold-change in gene expression is in brackets. Only genes with more than three fold change in gene expression and p values lower than 0.01 are included.
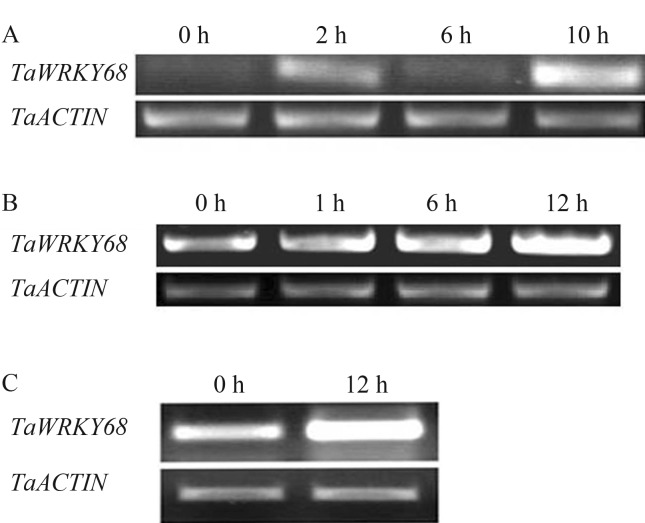
We then conducted RT-PCR tests to validate the results from the Genevestigator analysis. TaWRKY68 expression increased after Deoxynivalenol treatment and reached maximal mRNA levels after 10 h of treatment (Figure 8A). The infection of cells by Agrobacterium tumefaciens stimulated TaWRKY68 expression in wheat cultivar Yangmai 158, and TaWRKY68 mRNA reached its highest level at 12 h (Figure 8B). Similarly, the expression level of TaWRKY68 was increased 12 h after infection with Blumeria graminis (Figure 8C). Because biotic stresses were applied through spores or in water solutions, the mock treatments showed no difference from treatments at 0 h in each experiment.
Figure 8.
TaWRKY68 gene expressions under biotic stresses. A: Deoxynivalenol treatment; B: infection by Agrobacterium tumefaciens; C: infection by Blumeria graminis.
Discussion
WRKY transcription factors have a conserved WRKY domain and play important roles in plant growth and development and in responses to environmental stimuli (Eulgem et al., 2000; Rushton et al., 2010). WRKY genes have been cloned from various species including wheat. Wu et al. (2008) cloned 15 wheat WRKY genes and, using RT-PCR validation, found that wheat WRKY genes are involved in abiotic stresses and in leaf senescence. Talanova et al. (2009) identified three wheat WRKY genes, Wcor15, Wrab17, and Wrab19, whose expression patterns were significantly altered after cold treatment. Another study on wheat WRKY genes demonstrated that overexpression of two other wheat genes, TaWRKY2 and TaWRKY19, in Arabidopsis plants enhanced plant tolerance to salt, drought and freezing stresses (Niu et al., 2012). These results revealed the importance of WRKY genes in wheat responses to abiotic stresses. Nonetheless, WRKY genes function in wheat responses to biotic stresses are still largely elusive. In this study, we cloned the TaWRKY68 gene and analyzed sequence features and expression patterns of TaWRKY68 and orthologous genes under different conditions and developmental stages using bioinformatic tools, and found that the TaWRKY68 gene might play important roles in wheat responses to biotic stresses.
Amino acid sequences contain important information for protein function. Therefore, the analysis of sequence features is a key step for the prediction and clarification of protein function. We found that TaWRKY68 has 19 sequence features including 6 domains/motifs and 13 active sites: NLS (nuclear Localization Signal) peptides, which lead proteins to the cell nucleus; WRKY domains, which bind WRKY proteins to the W-box motif in the promoter of target genes for transcription regulation; Plant Zinc Cluster domains which function in association with WRKY domains; filamin repeats, which are related to a kind of actin-binding protein (Noegel et al., 1989). Based on the results actin might be a potential player in WRKY-regulated gene transcription, which is consistent with the evidence that nuclear actin interacts with the RNA polymerase for gene transcription (Ye et al., 2008). Proline-rich motifs are reported to bind with actin and are crucial for protein-protein interactions, Methionine-rich motifs are believed to bind with virus ribonucleoprotein (RNP) and to mediate export of the viral RNA complex from the infected cell nucleus to the cytoplasm, Microbodies_Cter is a microbody targeting signal mostly residing in proteins of peroxisomes and other microbodies (Gould et al., 1988; Yanai et al., 2006). The function of glycan sites varies from structural roles to participation in molecular trafficking, self-recognition and clearance (Marino et al., 2010). Protein kinases play important roles in signal transduction pathways, so TaWRKY68 could be a component (substrate) in a kinase mediated signal pathway. There are two Myristyl sites in TaWRKY68 which could function during TaWRKY68-mediated gene responses to stresses, as myristoylation sites play a vital role in membrane targeting and signal transduction in plant responses to environmental stresses (Podell and Gribskov, 2004).
Sequence is the prime determining factor of function (Eisen, 1998). Homologous genes with similar sequence are likely to have equivalent functions and to play the same functional role in equivalent biological processes. So it is very important to identify homologous genes, especially those which are supported by experimental data. Therefore, we carried out a homologous gene search using the newly developed DELTA blast program, which establishes alignment constraints based on conserved domain RPS blast. A phylogenetic tree from the COBALT alignment of these genes was then constructed. The results showed that TaWRKY68 has a close relationship with orthologous genes from barley and Brachypodium. Genes from sorghum and maize shared the closest similarity, while OsWRKY68 from rice was out-grouped from other monocot plants. Our study fits well with related studies (Bossolini et al., 2007; Bauer et al., 2011) and provides the evidence for the utility of DELTA blast and COBALT for phylogenetic analysis.
There is very high correlation between sequence similarity and functional similarity (Louie et al., 2009). So homologous genes identified by the phylogenetic analysis mentioned above are expected to share similar functions in different species. Therefore, we analyzed expression data of these genes using Genevestigator to explore the extensive information for the prediction of TaWRKY68 function. The results revealed that TaWRKY68 showed similar spatial and temporal expression patterns to those of orthologous genes in barley, rice and maize, and our RT-PCR results were perfectly consistent with the Genevestigator analysis. TaWRKY68 expression in roots was higher than in leaves and inflorescences (Figure 7), a finding that enriches our knowledge on expression patterns of the TaWRKY68 gene, because Wu et al. (2008) only measured mRNA levels of TaWRKY68-a and TaWRKY68-b in leaves.
As for gene expression under environmental stresses, Wu et al. (2008) found that TaWRKY68-a responded to PEG application that stimulates plant drought responses. By querying the wheat microarray data from Ergen et al. (2009) using the TaWRKY68 probe, we found that the TaWRKY68 gene was upregulated by 2.93 times under drought stress. Moreover, Genevestigator analysis also found that HvWRKY7 and OsWRKY68 genes, which are orthologs of TaWRKY68, were up-regulated more than three fold under the influence of pathogens and an elicitor. As expected, the RT-PCR results revealed that the mRNA level of TaWRKY68 increased following the application of Deoxynivalenol, a mycotoxin that accumulates after plants are infected by Fusarium graminearum (Gardiner et al., 2010), and the infections by Agrobacterium tumefaciens and Blumeria graminis. These results further supported our hypothesis that TaWRKY68 gene is involved in wheat responses to both abiotic and biotic stresses.
In conclusion, TaWRKY68 is involved in wheat growth and development and might function as a sharing signal component in wheat responses to various biotic stresses. Sequence features harbored in the protein could provide essential information for clarifying the working model of TaWRKY68 gene. Although homology may not mean conservation in function (Rhee et al., 2006) and bioinformatics results from orthologous genes need to be cautiously validated through laboratory experiments, bioinformatics analyses with massive data resources of orthologous cereal genes did provide significant clues for characterization of gene functions and open a bright prospect for studies on wheat genes.
Acknowledgments
We thank Dr Deirdre Mclachlan and Dr Ioanna Kostaki for critical reading of the manuscript. This project was financially supported by the National High-tech R&D Program of China (2012AA10A309 and 2011AA100104) and the Natural Science Fund of China (31240024) and of Tianjin (11JCYBJC09100).
Footnotes
Associate Editor: Marcia Pinheiro Margis
References
- Bauer P, Elbaum R, Weiss IM. Calcium and silicon mineralization in land plants: transport, structure and function. Plant Sci. 2011;180:746–756. doi: 10.1016/j.plantsci.2011.01.019. [DOI] [PubMed] [Google Scholar]
- Bossolini E, Wicker T, Knobel PA, Keller B. Comparison of orthologous loci from small grass genomes Brachypodium and rice: Implications for wheat genomics and grass genome annotation. Plant J. 2007;49:704–717. doi: 10.1111/j.1365-313X.2006.02991.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Eisen JA. Phylogenomics: Improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- Ergen NZ, Thimmapuram J, Bohnert HJ, Budak H. Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct Integr Genomics. 2009;9:377–396. doi: 10.1007/s10142-009-0123-1. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Gardiner SA, Boddu J, Berthiller F, Hametner C, Stupar RM, Adam G, Muehlbauer GJ. Transcriptome analysis of the barley-deoxynivalenol interaction: Evidence for a role of glutathione in deoxynivalenol detoxification. Mol Plant Microbe Interact. 2010;23:962–976. doi: 10.1094/MPMI-23-7-0962. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Keller GA, Subramani S. Identification of peroxisomal targeting signals located at the carboxy terminus of four peroxisomal proteins. J Cell Biol. 1988;107:897–905. doi: 10.1083/jcb.107.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator v. 3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinformat. 20082008:420–747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol Gen Genet. 1994;244:563–571. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- Louie B, Higdon R, Kolker E. A statistical model of protein sequence similarity and function similarity reveals overly-specific function predictions. PLoS One. 2009;4:e7546. doi: 10.1371/journal.pone.0007546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: A path through the maze. Nat Chem Biol. 2010;6:713–723. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
- Niu CF, Wei W, Zhou QY, Tian AG, Hao YJ, Zhang WK, Ma BA, Lin Q, Zhang ZB, Zhang JS, et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012;35:1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x. [DOI] [PubMed] [Google Scholar]
- Noegel AA, Rapp S, Lottspeich F, Schleicher M, Stewart M. The Dictyostelium gelation factor shares a putative actin binding site with alpha-actinins and dystrophin and also has a rod domain containing six 100-residue motifs that appear to have a cross-beta conformation. J Cell Biol. 1989;109:607–618. doi: 10.1083/jcb.109.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei HR, Milne J, Page GP, Somerville CR. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA. 2005;102:8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S, Gribskov M. Predicting N-terminal myristoylation sites in plant proteins. BMC Genomics. 2004;5:e37. doi: 10.1186/1471-2164-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proietti S, Bertini L, Van der Ent S, Leon-Reyes A, Pieterse CMJ, Tucci M, Caporale C, Caruso C. Cross activity of orthologous WRKY transcription factors in wheat and Arabidopsis. J Exp Bot. 2011;62:1975–1990. doi: 10.1093/jxb/erq396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Dickerson J, Xu D. Bioinformatics and its applications in plant biology. Annu Rev Plant Biol. 2006;57:335–360. doi: 10.1146/annurev.arplant.56.032604.144103. [DOI] [PubMed] [Google Scholar]
- Ribot C, Hirsch J, Balzergue S, Tharreau D, Notteghem JL, Lebrun MH, Morel JB. Susceptibility of rice to the blast fungus, Magnaporthe grisea. J Plant Physiol. 2008;165:114–124. doi: 10.1016/j.jplph.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Ross CA, Liu Y, Shen QXJ. The WRKY gene family in rice (Oryza sativa) J Integr Plant Biol. 2007;49:827–842. [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996;15:5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Talanova VV, Titov AF, Topchieva LV, Malysheva IE, Venzhik YV, Frolova SA. Expression of WRKY transcription factor and stress protein genes in wheat plants during cold hardening and ABA treatment. Russ J Plant Physiol. 2009;56:702–708. doi: 10.1134/s0012496608060215. [DOI] [PubMed] [Google Scholar]
- Tie W, Zhou F, Wang L, Xie W, Chen H, Li X, Lin Y. Reasons for lower transformation efficiency in indica rice using Agrobacterium tumefaciens-mediated transformation: Lessons from transformation assays and genome-wide expression profiling. Plant Mol Biol. 2012;78:1–18. doi: 10.1007/s11103-011-9842-5. [DOI] [PubMed] [Google Scholar]
- Wu HL, Ni ZF, Yao YY, Guo GG, Sun QX. Cloning and expression profiles of 15 genes encoding WRKY transcription factor in wheat (Triticum aestivum L.) Prog Nat Sci. 2008;18:697–705. [Google Scholar]
- Yanai H, Kobayashi T, Hayashi Y, Watanabe Y, Ohtaki N, Zhang G, de la Torre JC, Ikuta K, Tomonaga K. A methionine-rich domain mediates CRM1-dependent nuclear export activity of Borna disease virus phosphoprotein. J Virol. 2006;80:1121–1129. doi: 10.1128/JVI.80.3.1121-1129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Zhao J, Hoffmann-Rohrer U, Grummt I. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev. 2008;22:322–330. doi: 10.1101/gad.455908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- Myhits, http://myhits.isb-sib.ch/ (July 10, 2012).