Abstract
Homozygosity for Slc25a21tm1a(KOMP)Wtsi results in mice exhibiting orofacial abnormalities, alterations in carpal and rugae structures, hearing impairment and inflammation in the middle ear. In humans it has been hypothesised that the 2-oxoadipate mitochondrial carrier coded by SLC25A21 may be involved in the disease 2-oxoadipate acidaemia. Unexpectedly, no 2-oxoadipate acidaemia-like symptoms were observed in animals homozygous for Slc25a21tm1a(KOMP)Wtsi despite confirmation that this allele reduces Slc25a21 expression by 71.3%. To study the complete knockout, an allelic series was generated using the loxP and FRT sites typical of a Knockout Mouse Project allele. After removal of the critical exon and neomycin selection cassette, Slc25a21 knockout mice homozygous for the Slc25a21tm1b(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi alleles were phenotypically indistinguishable from wild-type. This led us to explore the genomic environment of Slc25a21 and to discover that expression of Pax9, located 3′ of the target gene, was reduced in homozygous Slc25a21tm1a(KOMP)Wtsi mice. We hypothesize that the presence of the selection cassette is the cause of the down regulation of Pax9 observed. The phenotypes we observed in homozygous Slc25a21tm1a(KOMP)Wtsi mice were broadly consistent with a hypomorphic Pax9 allele with the exception of otitis media and hearing impairment which may be a novel consequence of Pax9 down regulation. We explore the ramifications associated with this particular targeted mutation and emphasise the need to interpret phenotypes taking into consideration all potential underlying genetic mechanisms.
Introduction
Knockout mice are invaluable tools for studying the function of genes both during embryonic development and in the adult. Classically, gene disruption in mice is achieved by replacing a part of the target gene with a selectable marker, e.g. a neo cassette, leading to constitutive ablation of the targeted gene. More contemporary approaches include the creation of conditional alleles and targeted gene traps, as well as small hairpin RNA (shRNA) and the use of lentiviral transgenesis, reviewed in [1]. The Sanger Institute Mouse Genetics Project [2] is contributing to an international effort to conduct systematic, large-scale gene function analysis in a mammalian system, through the generation of mice using targeted ES cells available from the EUCOMM and KOMP resources [3]. The typical EUCOMM and KOMP knockout first conditional ready targeted trap [tm1a(EUCOMM) and tm1a(KOMP)] is a powerful allele configured with the potential to convert to a conditional allele or a lacZ tagged null allele [3]. This tm1a allele was selected for pragmatic reasons, including there being no requirement for additional breeding to modify the allele, as targeted traps typically disrupt expression of the targeted gene [3], [4], and the major advantage that the allele, with full potential for conversion, would be cryopreserved for subsequent community use. However, it has previously been reported that the presence of selectable markers used for gene targeting, such as the neo cassette in the EUCOMM and KOMP tm1a allele, can interfere with the expression of neighbouring genes [5], [6], [7], [8], [9]. It is, therefore, important that phenotypes discovered in mice carrying the knockout first conditional ready allele, are verified using the derived deletion allele.
SLC25A21 has previously been reported to be ubiquitously expressed in all tissues tested [10], and codes for the 2-oxoadipate mitochondrial carrier (ODC) that catalyses the movement of several substrates, primarily 2-oxoadipate and 2-oxoglutarate, across inner mitochondrial membranes [10], [11], [12]. It is suspected that ODC functions by a counter exchange mechanism, catalysing the uptake of 2-oxoadipate into the mitochondrial matrix, in exchange for internal 2-oxoglutarate [10]. The physiological role of the ODC suggests that it may be involved in the human disease 2-oxoadipate acidaemia (OMIM 204750), characterised by an accumulation of 2-oxoadipate and associated metabolites in urine [11]. This inborn error of metabolism shows clinical symptoms of intellectual disability varying from mild to severe, hypotonia, seizures, motor and developmental delay, and cerebellar ataxia [13], [14], [15], [16].
Here we report characterization of the allelic series derived from the KOMP targeted allele of Slc25a21. From this, three distinct observations were made. Firstly, ablation of Slc25a21 was not found to cause symptoms of 2-oxoadipate acidaemia in mice; animals homozygous for Slc25a21tm1b(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi null alleles were normal for all parameters tested. Secondly, we report that Slc25a21tm1a(KOMP)Wtsi homozygous mice present with profound dental, orofacial and hearing/middle ear phenotypes caused by off-target effects on the expression of the neighbouring gene Pax9. Finally, the resulting novel, hypomorphic allele of Pax9 opens the door to further studies into how Pax9 is involved in the patterning of the palatal rugae and may represent a novel model of otitis media in the mouse. With the growing resource of knockout first conditional ready targeted ES cells created by the EUCOMM and KOMP initiatives, these results represent a timely reminder that the phenotype observed in the knockout first conditional ready mice should be verified by creating and analysing the deletion allele.
Materials and Methods
Ethics Statement
The care and use of all mice in this study were in accordance with UK Home Office regulations, UK Animals (Scientific Procedures) Act of 1986, and were approved by the Wellcome Trust Sanger Institute Ethical Review Committee. All efforts were made to minimize suffering including housing mice in a specific pathogen-free unit at a density of 3–5 animals per cage in polysulfone individually ventilated cages [Tecniplast Seal Safe 1284L (overall dimensions of caging: (L×W×H): 365×207×140 mm, floor area = 530 cm2)]. Sterilised Aspen bedding substrate and standard environmental enrichment of nestlet, cardboard tunnel, and three wooden chew blocks were provided. The light cycle was maintained at 12 h light/12 h dark with lights off at 19:30 hours and no twilight period. Room temperature was 21±2°C and humidity was regulated at 55±10%. Mice were given food and water ad libitum unless otherwise stated [2]. Animal welfare is our primary consideration and was assessed routinely for all mice involved in this study. Adult mice were killed by terminal anaesthesia followed by exsanguination and either cervical dislocation or removal of the heart. Embryos were killed by decapitation or cooling followed by immersion in cold tissue fixative. Experimental work was performed in accordance with the Arrive guidelines [17].
Animals
The Slc25a21tm1a(KOMP)Wtsi mice were created by blastocyst injection of targeted ES-cell clone EPD0085_1_D04 from the NIH-funded Knockout Mouse Project (KOMP) [3], [18], [19]. Generation of mice carrying the Slc25a21tm1b(KOMP)Wtsi, Slc25a21tm1c(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi alleles was achieved by breeding to mice expressing Cre (Hprt1Tg(CMV-Cre)Brd) and/or Flp (ROSA26Fki) recombinases. For brevity, the allelic series will be referred to hereafter in figures as tm1a, tm1b, tm1c or tm1d. The Slc25a21tm1a(KOMP)Wtsi, Slc25a21tm1b(KOMP)Wtsi, Slc25a21tm1c(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi colonies were maintained on a C57BL/6N;C57BL/6-Tyrc-Brd genetic background.
Mice were fed Mouse Breeders Diet (Lab Diets 5021-3, Richmond, Indiana, USA) unless otherwise specified, including Slc25a21tm1d(KOMP)Wtsi mice processed through the Mouse Genetics Project screening protocol [2]. Slc25a21tm1a(KOMP)Wtsi, Slc25a21tm1b(KOMP)Wtsi and Slc25a21tm1c(KOMP)Wtsi mice which went through the Mouse Genetics Project screening protocol were transferred to a high-fat (21.4% fat by crude content) dietary challenge (Special Diet Services Western RD-829100, Witham, UK) at 4 weeks of age. Cages were processed randomly and different genotypes could be housed together, hence there was no pattern to the order in which animals were processed. The experimental unit throughout the study was a single mouse.
Viability at postnatal day 14 was assessed from heterozygous (het) intercross matings. Furthermore, viability and embryo morphology for Slc25a21tm1a(KOMP)Wtsi mutant mice was assessed at E14.5 and E18.5.
Validation of targeting event
To confirm the integrity and validity of each allele studied, detailed molecular characterisation was performed using short-range, long-range and quantitative PCR strategies, as described previously [20] and detailed in File S1 and Table S1.
Genotyping for Slc25a21tm1a(KOMP)Wtsi animals was performed using a combination of three short range assays (Table S1) and the neo count qPCR assay (0 = wt, 1 = het, 2 = hom). Conversion to Slc25a21tm1b(KOMP)Wtsi, Slc25a21tm1c(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi was detected using short range PCR assays (Table S1).
Work by Santagati [21] indicated the presence of a regulatory element, named conserved non-coding sequence +6 (CNS+6), for Pax9 within the allele design for Slc25a21tm1a(KOMP)Wtsi. A PCR assay was designed (Table S1) to amplify Slc25a21tm1a(KOMP)Wtsi samples and the resulting products sequenced to determine if the CNS+6 element was disrupted.
Expression analysis by quantitative PCR and RNA sequencing
E13.5 embryo heads [Slc25a21tm1a(KOMP)Wtsi (wt, n = 4; hom, n = 4), Slc25a21tm1b(KOMP)Wtsi (wt, n = 4; hom, n = 3), Slc25a21tm1c(KOMP)Wtsi (wt, n = 4; hom, n = 3), and Slc25a21tm1d(KOMP)Wtsi (wt, n = 4; hom, n = 3)] were processed for expression analysis by qPCR. For Slc25a21 a custom FAM-labelled TaqMan assay (Applied Biosystems) spanning the junction of exons 8–9, 3′ to the floxed exon (Slc25a21_E8-9_F: CTGCTTCAAAACAATGGAGATGAT, Slc25a21_E8-9_R: GGGACCAGGCCTTTGTATAAGG, Slc25a21_E8-9_M: CGGGAAGAAGGGATTT) was used. For Pax9 a pre-designed TaqMan assay was used (Mm00440629_m1, Applied Biosystems). Details of the assays and methods used are described in File S1.
RNA sequencing was performed using 5 µg of total RNA from a subset of the above E13.5 embryo head samples [3 homozygotes and 2 wild-types for each allele (12 mutant and 8 control samples in total)] as detailed in File S1.
Physical assessment, body composition, radiography and clinical chemistry
Homozygous mice (7M and 7F) from each of the alleles (Slc25a21tm1a(KOMP)Wtsi, Slc25a21tm1b(KOMP)Wtsi, Slc25a21tm1c(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi), as well as age, sex and genetic background matched controls, were analysed using the standard Sanger Institute Mouse Genetics Project phenotyping screen [2]. Assays of particular relevance are described in File S1, whilst those not part of the standard phenotyping screen are described below.
At necropsy, skulls were collected and the teeth (both upper and lower jaw) and palate were reviewed and imaged (MZ16A dissecting microscope, Leica, Wetzlar, Germany; DFC490 digital camera, Canon Powershot G5, Japan). A range of 42 additional tissues and organs from homozygous Slc25a21tm1a(KOMP)Wtsi mice (2M and 2F), and age, sex and genetic background matched controls, were fixed in 10% neutral buffered formalin (Leica Biosystems, Peterborough, UK) for 15–20 hrs and processed to wax. 5 µm sections were stained with haematoxylin and eosin then reviewed by an experienced pathologist.
Micro-CT (computed tomography) images of skulls from Slc25a21tm1a(KOMP)Wtsi mice (wt, n = 2; hom, n = 2) were collected at 18 µm resolution (SkyScan 1176, SkyScan, Kontich, Belgium).
Auditory Brainstem Response (ABR) recordings
ABRs were recorded in Slc25a21tm1a(KOMP)Wtsi mice aged 4 weeks (wt, n = 17; het, n = 27; hom, n = 7), 8 weeks (wt, n = 20; het, n = 33; hom, n = 8), 14 weeks (wt, n = 11; hom, n = 14), and 26 weeks (wt, n = 17; hom, n = 10), using the methods described in detail in Ingham et al. [22]. In addition, mice from the Slc25a21tm1b(KOMP)Wtsi colony (hom, n = 8), Slc25a21tm1c(KOMP)Wtsi colony (hom, n = 5) and Slc25a21tm1d (KOMP)Wtsi colony (hom, n = 8) were tested at 14 weeks of age.
Anaesthetised mice were placed on a heating blanket inside a sound attenuating booth and recording electrodes inserted in the skin to record responses of the left ear. For ABR threshold determination, click (0.01 ms duration) and tone pip (6, 12, 18, 24, and 30 kHz of 5 ms duration, 1 ms rise/fall time) stimuli over a range of intensity levels from 10–95 dB sound pressure level (SPL) in 5 dB steps were presented in free-field. Averaged responses to 256 stimuli, presented at 42.2/s, were analysed and thresholds established as the lowest sound intensity giving a visually-detectable ABR response.
Middle ear analysis
Following ABR testing, Slc25a21tm1a(KOMP)Wtsi mice aged 9 weeks (wt, n = 8; het, n = 11; hom, n = 8) were killed by cervical dislocation and the right external and middle ears were examined in detail using a dissecting microscope for any signs of malformation or inflammation, including: appearance of excess cerumen in the external ear canal; thickening, whitening, sponginess or excess vascularisation of the bulla wall; clarity and vascularisation of the tympanic membrane; presence of fluid or white inflammatory material in the middle ear cavity; and solid masses or bony outgrowths of the middle ear wall or ossicles. In addition, the left side of the head of these mice was fixed in 10% formalin for 48 hours, and decalcified in 10% EDTA diluted in phosphate buffered saline (PBS) for 10 days, dehydrated, embedded in paraffin wax, sectioned at 8 µm and stained with haematoxylin and eosin.
Reporter gene analysis
LacZ reporter gene wholemount expression analysis was performed on Slc25a21tm1a(KOMP)Wtsi adults [aged 15–18 weeks (wt, n = 2; het, n = 3; hom, n = 9)] as described previously [23] and detailed in File S1.
Bone and cartilage staining
Bone and cartilage staining of Slc25a21tm1a(KOMP)Wtsi E18.5 embryos (wt, n = 10; het, n = 27; hom, n = 12) was performed using a protocol based on the Cold Spring Harbour method for Alcian blue/Alizarin Red staining [24], and detailed in File S1.
Data analysis and statistics
A reference range approach was used to assess continuous data, including time course. For categorical data, a Fisher's exact test was used to identify phenotypic variants. More details of both approaches are presented in File S1. Since both of the above approaches to automatically identify significant calls are known to be conservative and assess each parameter in isolation, they were complemented by a manual assessment made by a biological expert who used knowledge of events on the day, or across sexes, or between related variables, to highlight additional potentially abnormal phenotypes (Table S2).
To assess the ABR data, a one-way Kruskall-Wallis ANOVA on Ranks was used where three genotypes were assessed and Mann-Whitney rank sum test where only homozygote and wild-type animals were assessed. The qPCR gene expression results for mutant mice were compared to littermate wild-type mice using a Student's t-Test assuming two-sample unequal variance. P-values for the RNA sequencing were adjusted for multiple testing with the Benjamini-Hochberg procedure. Pre-weaning lethality was assessed using the Test for One Proportion.
Results
Characterisation of Slc25a21tm1a(KOMP)Wtsi homozygous mice
Slc25a21tm1a(KOMP)Wtsi mice were generated and characterised on a C57BL/6N;C57BL/6-Tyrc-Brd background by the Sanger Mouse Genetics Project. Homozygous Slc25a21tm1a(KOMP)Wtsi mice were under-represented at post natal day 14 (P14), as a result of pre-weaning lethality, with 25 homozygous mutants detected among 304 progeny from heterozygous intercrosses (Table 1. Test for one proportion; χ2 = 44.74, df = 1, p = 1.124e-11).
Table 1. Genotype distribution for Slc25a21tm1a(KOMP)Wtsi mice.
| Allele and Stage | Number of Progeny | Number of Wild-types | Number of Heterozygotes | Number of Homozygotes | Percentage Homozygote Viability |
| tm1a E14.5 | 35 | 3 | 22 | 8 | 23% |
| tm1a E18.5 | 44 | 10 | 27 | 7 | 16% |
| tm1a P14 | 304 | 111 | 168 | 25 | 8% |
| tm1b P14 | 130 | 24 | 68 | 38 | 29% |
| tm1c P14 | 145 | 26 | 84 | 35 | 24% |
| tm1d P14 | 193 | 44 | 113 | 36 | 19% |
From heterozygous inter-crossing, homozygous progeny for Slc25a21tm1a(KOMP)Wtsi were recorded at a sub-Mendelian ratio at post natal day 14 (P14), but a normal ratio at two embryonic time points (E14.5 and E18.5). Slc25a21tm1b(KOMP)Wtsi, Slc25a21tm1c(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi homozygotes were detected at the expected ratio.
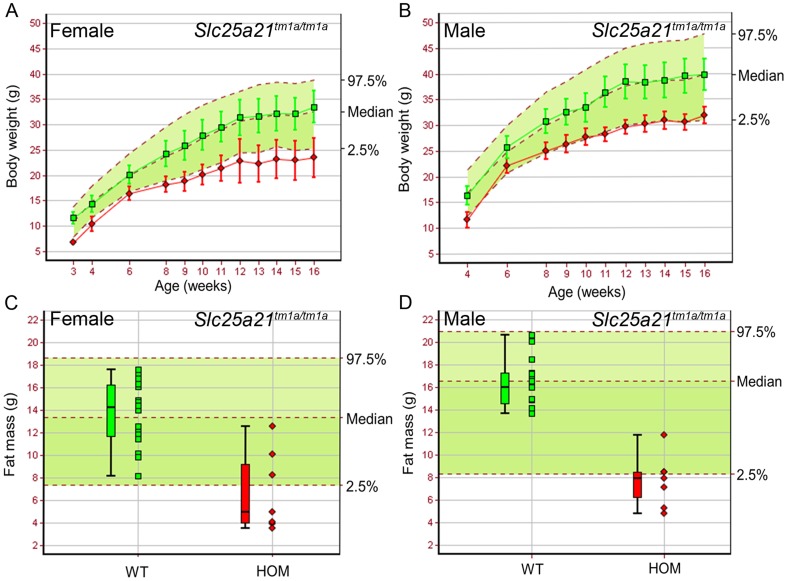
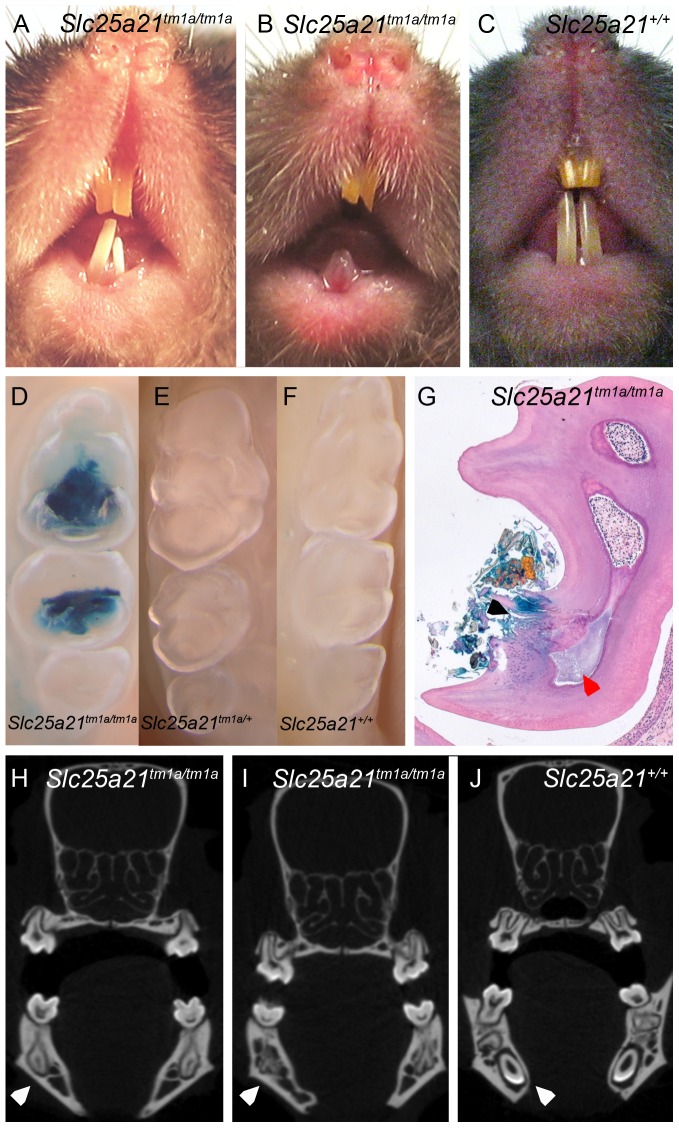
Those homozygous animals surviving to weaning, presented with decreased body weight (Fig. 1A–B), fat mass (Fig. 1C–D) and fat percentage estimate (data not shown), in both sexes, compared to wild-type controls. Furthermore, a range of dental abnormalities were observed in Slc25a21tm1a(KOMP)Wtsi homozygous mice. At 10 weeks of age 13 out of 14 homozygous Slc25a21tm1a(KOMP)Wtsi mice were found to have an abnormally shortened snout (data not shown). At 16 weeks of age all fourteen homozygous Slc25a21tm1a(KOMP)Wtsi mice were found to have macroscopically visible dental abnormalities ranging from white or translucent to severely hypoplastic lower incisors (Fig. 2A–B) compared to wild-type (Fig. 2C). The molars of homozygous Slc25a21tm1a(KOMP)Wtsi adult mice (Fig. 2D), in particular of the lower jaw, were found to be strongly stained with X-gal (requiring β-galactosidase activity) compared to heterozygous and wild-type (Fig. 2E–F). Microscopic examination of sections from representative animals confirmed that in unaffected areas, the molars were fully developed with normal odontoblast morphology. In affected areas of the molars, there was focal fracturing of the surface dentine and necrosis of the pulp with associated inflammation and bacterial colonies (Fig. 2G). Micro-CT scanning revealed that the lower incisor root area in homozygous Slc25a21tm1a(KOMP)Wtsi mice was severely under-developed and that the lower incisors appeared to be lacking the enamel coating (Fig. 2H–I) compared to wild-type (Fig. 2J). Furthermore, two dimensional X-ray images revealed several changes of the manidibular morphology including pronounced pogonion, pronounced anterior tuberosity, under-developed incisor process and alveolus (data not shown), potentially indicating a change in the developmental dynamics of the mandible in homozygous Slc25a21tm1a(KOMP)Wtsi mice. Taken together, the 2D X-ray images, micro-CT and X-gal staining in tooth cavities suggests feeding might be uncomfortable for homozygous Slc25a21tm1a(KOMP)Wtsi mice which may explain their reduced body weight and fat mass.
Figure 1. Body weight and fat mass were reduced in homozygous Slc25a21tm1a(KOMP)Wtsi mice.
Homozygous (red symbols) Slc25a21tm1a(KOMP)Wtsi female (A,C) (n = 7) and male (B,D) (n = 7) mice showed decreased body weight and fat mass compared to local wild-type controls run on the same week (green symbols) (female, n = 22; male, n = 15). Mean (± SD) body weight is plotted against age (A,B). Fat mass is presented as a boxplot, with median, 25th and 75th percentile (box), and the lowest and highest data point within 1.5× inter quartile range (whiskers) shown (C,D). The median and 95% reference range (2.5–97.5th percentiles, dotted lines) for all wild-type mice of the same genetic background and sex (n>750) are displayed on the pale green background.
Figure 2. Dental abnormalities observed in homozygous Slc25a21tm1a(KOMP)Wtsi mice.
Dysmorphology analysis revealed underdeveloped, white incisors (A–B) in Slc25a21tm1a(KOMP)Wtsi homozygous mice as compared to wild-type (C). Wholemount (D–F) and subsequent histological sections (G) demonstrated bacterial lacZ activity in cavities within lower jaw molars of Slc25a21tm1a(KOMP)Wtsi homozygous (D,G) mice compared to heterozygous (E) and wild-type (F) mice. Focal fracturing of the surface dentine is indicated with a black arrow and necrosis of the pulp indicated with a red arrow (G). Micro-CT imaging demonstrated that the root aspect of lower jaw incisors was severely under developed (white arrow) and lacked enamel coating in homozygous Slc25a21tm1a(KOMP)Wtsi mice (H–I) compared to wild-type (J).
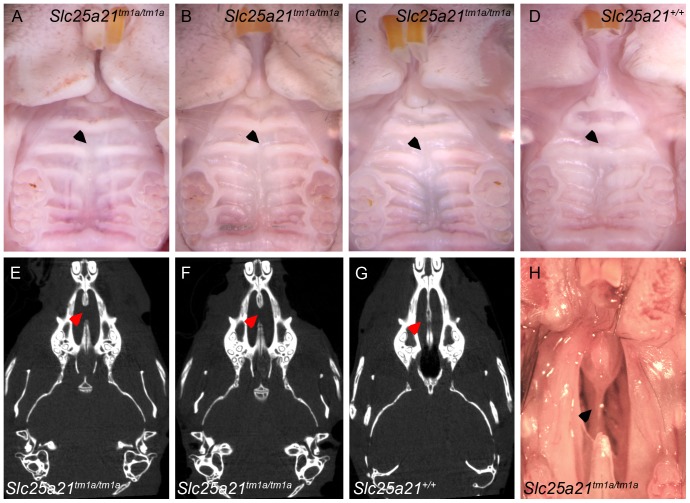
Further characterisation uncovered craniofacial abnormalities in adult homozygous Slc25a21tm1a(KOMP)Wtsi mice. Specifically, they presented with abnormal development of palatal rugae number 3 (Fig. 3A–C) compared to wild-type (Fig. 3D). Micro-CT scanning revealed that the anterior part of the vomer bone was completely absent in homozygous Slc25a21tm1a(KOMP)Wtsi mice (Fig. 3E–F) compared to wild-type (Fig. 3G), this was confirmed by macroscopic observations (Fig. 3H). The vomeronasal organ (VNO) appeared to be intact. Morphological examination of E18.5 embryo heads from homozygous Slc25a21tm1a(KOMP)Wtsi mice did not reveal any further craniofacial abnormalities.
Figure 3. Rugae and vomer abnormalities observed in homozygous Slc25a21tm1a(KOMP)Wtsi mice.
Abnormalities in rugae (in particular number 3) were observed in homozygous Slc25a21tm1a(KOMP)Wtsi mice (A–C) compared to wild-type (D), indicated with arrows (wt, n = 11; hom, n = 11). Micro-CT analysis revealed that the anterior part of the vomer bone was absent in Slc25a21tm1a(KOMP)Wtsi homozygous mice (E–F) compared to wild-type (G) (wt, n = 2; hom, n = 12). This was confirmed by dissection of the palate of Slc25a21tm1a(KOMP)Wtsi mice (H).
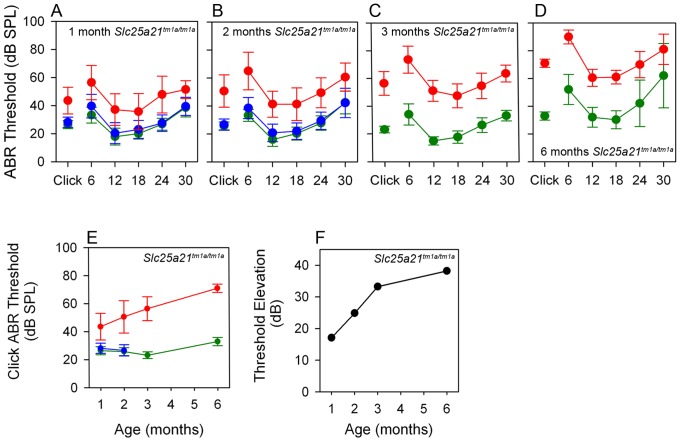
Finally, 14 week old Slc25a21tm1a(KOMP)Wtsi homozygous mice were found to have a hearing impairment as they presented with elevated auditory brainstem response thresholds across all frequencies, with severity of impairment increasing with age (Fig. 4A–D). Slc25a21tm1a(KOMP)Wtsi homozygous mice displayed an approximately parallel pattern of threshold elevation across the range of frequency stimuli used, a profile which is typical of conductive hearing loss. Fig. 4E shows that click threshold in wild-types was stable with increasing age, at least up to 26 weeks of age, whereas a progressive deterioration was seen in Slc25a21tm1a(KOMP)Wtsi homozygous mice over the same age range. At 26 weeks of age, some wild-type mice showed age-related high-frequency hearing loss (indicated by larger standard deviations at 24–30 kHz). The thresholds recorded in the Slc25a21tm1a(KOMP)Wtsi homozygous mice at all ages were significantly elevated above age-matched wild-types (p<0.001) (Fig. 4F).
Figure 4. Auditory brainstem response threshold was elevated in Slc25a21tm1a(KOMP)Wtsi mice.
Auditory brainstem response threshold was elevated across all frequencies in homozygous Slc25a21tm1a(KOMP)Wtsi mice aged 1, 2, 3 and 6 months respectively. Mean ABR thresholds (± SD) are shown for wild-type (green symbols), heterozygote (blue symbols) and homozygote (red symbols) Slc25a21tm1a(KOMP)Wtsi mice (A–D). The click-evoked ABR thresholds (mean ± SD) plotted as a function of age (months) demonstrated a progressive deterioration in Slc25a21tm1a(KOMP)Wtsi mice (E); an observation clearly demonstrated by plotting the mean click-evoked ABR threshold elevation of mutants above wild-types as a function of age (months) (F).
To investigate the morphology of the middle ear, the right temporal bone, containing the entire ear, was dissected from Slc25a21tm1a(KOMP)Wtsi homozygous mice at 9 weeks of age (n = 8) along with wild-type (n = 8) and heterozygous (n = 11) littermates. Compared to wild-types (Fig. 5A), most of the homozygotes (7/8) showed opaque fluid or white debris in the middle ear cavity indicating otitis media (Fig. 5B). Other features of otitis media were more variable, including excessive vascularisation (n = 3) or whitening of the bulla (n = 4) or tympanic membrane (n = 1). The one homozygous Slc25a21tm1a(KOMP)Wtsi mouse in which exudate was not observed in the middle ear also had ABR thresholds only slightly above those seen in wild-type mice. Heterozygotes had normal middle ears.
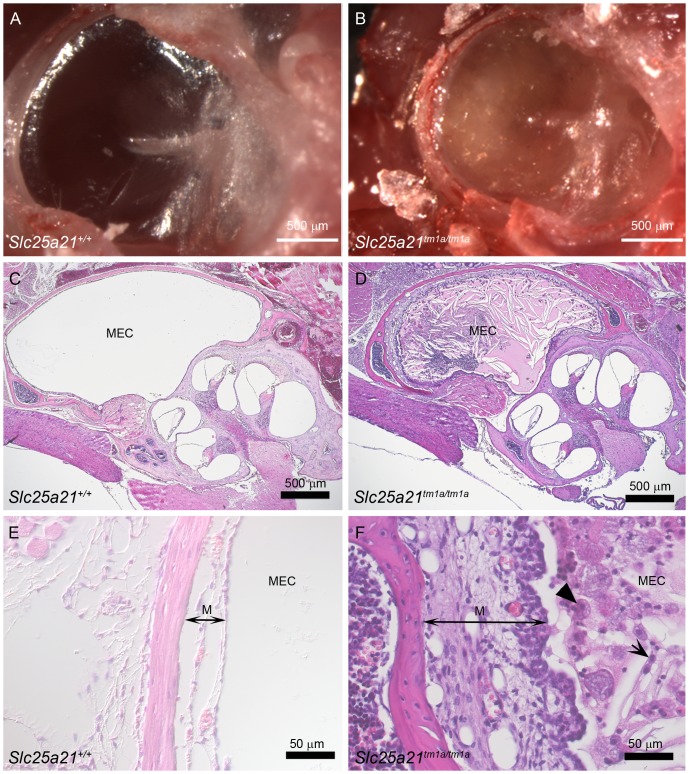
Figure 5. Signs of otitis media in Slc25a21tm1a(KOMP)Wtsi mice.
The tympanic membrane viewed from the external ear canal showed an opaque fluid present in Slc25a21tm1a(KOMP)Wtsi homozygous mice (B) that was not present in wild-type controls (A). The opaque exudate was clearly visible on haematoxylin and eosin stained histology sections through the middle ear of Slc25a21tm1a(KOMP)Wtsi homozygous mice (D) compared to wild-types (C). Examination at higher magnification revealed that the exudate containing inflammatory cells including foamy macrophages (solid arrow head), and neutrophils (short arrow) in the middle ear cavity (MEC), and a thickened mucosa (M, double headed arrow) in Slc25a21tm1a(KOMP)Wtsi homozygous mice due to epithelial hyperplasia, oedema and congestion (F). Wild-type mice appeared normal (E).
The left side temporal bones from the same eight Slc25a21tm1a(KOMP)Wtsi homozygous mice and wild-type controls were sectioned through the middle ear, stained with haemotoxylin and eosin, and examined microscopically. Inflammatory changes and accumulation of exudate consistent with otitis media were observed in six of the eight homozygotes (Fig. 5D, F).
The middle ear cavity of the six affected Slc25a21tm1a(KOMP)Wtsi homozygous mice contained variable amounts of an amorphous eosinophilic exudate containing cholesterol crystals, foamy macrophages and neutrophils, indicating an active inflammatory response. In these mice, the lining epithelium of the middle ear cavity was variably hyperplastic and the underlying lamina propria was oedematous with congestion of blood vessels (Fig. 5D, F). None of the littermate controls showed evidence of inflammation (Fig. 5A, C, E).
Of the two Slc25a21tm1a(KOMP)Wtsi homozygous mice showing little or no effusion one had comparatively normal ABR thresholds, whilst the other had raised ABR thresholds.
The tympanic ring size was normal in Slc25a21tm1a(KOMP)Wtsi homozygous mice; assessment of micro-CT images revealed no change in adults whilst alizarin red/alcian blue stained embryos revealed no changes at E18.5 (data not shown).
Systematic histopathology assessment of a range of tissues from homozygous Slc25a21tm1a(KOMP)Wtsi mice did not reveal any further abnormalities, and in particular, no indication of any inflammation other than in the middle ear was found.
Ablation of Slc25a21 does not cause signs of 2-oxoadipate acidaemia in mice
It has been proposed that loss of function of ODC, the protein encoded by SLC25A21, causes the human disease 2-oxoadipate acidaemia which is clinically characterised by hypotonia, seizures, motor and developmental delay, cerebellar ataxia and varying severities of intellectual disability [13], [14], [15], [16]. Unexpectedly, homozygous Slc25a21tm1a(KOMP)Wtsi mice did not phenocopy this human disease. In particular, lean mass, grip strength and gait, correlates of the human disease manifestation, were all normal for these mice (data not shown), and no seizures were detected.
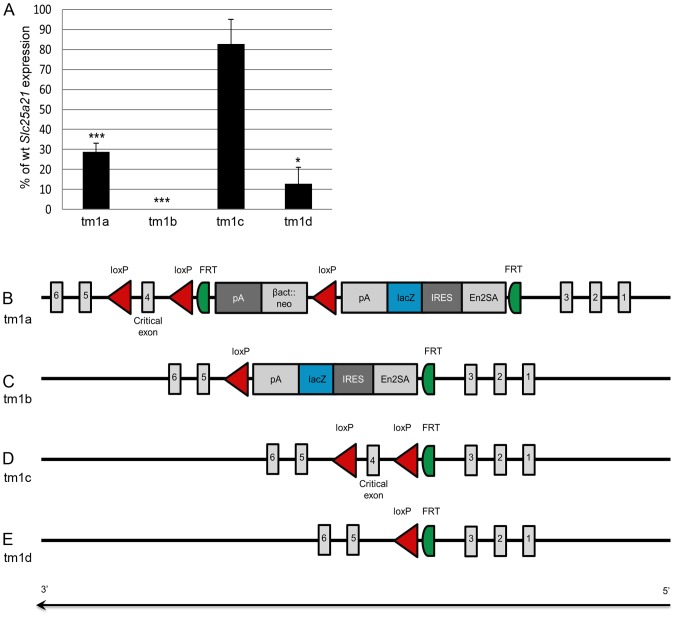
To ascertain the molecular consequence of the targeting event, expression analysis of Slc25a21tm1a(KOMP)Wtsi was performed on RNA extracted from embryo heads (E13.5). When using the Slc25a21 TaqMan assay which spanned exons 8–9, 3′ of the cassette insertion, expression was found to be reduced in homozygous Slc25a21tm1a(KOMP)Wtsi samples (28.7% of wild-type expression, p = 2.0e-05) but not completely ablated (Fig. 6A). The hypomorphic nature of this allele may have explained the absence of the predicted phenotypes, therefore an allelic series was created (Fig. 6B–E) where tm1b was a lacZ reporter tagged deletion with the critical exon (exon 4) removed, tm1c a conditional allele with wild-type function restored, and tm1d the deletion allele where both exon 4 and the lacZ reporter were removed. Expression analysis of each of these Slc25a21 alleles was performed on RNA extracted from embryo heads (E13.5). Slc25a21 RNA expression was found to be ablated in homozygous Slc25a21tm1b(KOMP)Wtsi (0% of wild-type expression, expression was below the limit of detection) and greatly reduced in Slc25a21tm1d(KOMP)Wtsi homozygotes (12.9% of wild-type expression, p = 0.043) (Fig. 6A). As expected, expression levels of Slc25a21 in embryos homozygous for Slc25a21tm1c(KOMP)Wtsi were not significantly different from wild-type expression (Fig. 6A).
Figure 6. Schematic of the Slc25a21tm1(KOMP)Wtsi allelic series and the associated RNA expression levels.
Quantitative PCR on RNA from E13.5 embryo heads demonstrated that Slc25a21 RNA expression was significantly reduced in Slc25a21tm1a(KOMP)Wtsi, Slc25a21tm1b(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi homozygotes (* p<0.05; *** p<0.001), but equivalent to wild-type levels in Slc25a21tm1c(KOMP)Wtsi homozygotes (A). A schematic of the Slc25a21tm1(KOMP)Wtsi allelic series is presented, consisting of: (B) Slc25a21tm1a(KOMP)Wtsi, the knockout first reporter (lacZ) tagged insertion allele; (C) Slc25a21tm1b(KOMP)Wts i, the lacZ tagged deletion allele generated by breeding with mice ubiquitously expressing Cre recombinase; (D) Slc25a21tm1c(KOMP)Wtsi, the conditional allele generated by breeding mice carrying the Slc25a21tm1a(KOMP)Wtsi allele with mice ubiquitously expressing Flp recombinase; and (E) Slc25a21tm1d(KOMP)Wtsi, the deletion allele arising after breeding mice carrying the Slc25a21tm1c(KOMP)Wtsi allele with mice ubiquitously expressing Cre recombinase (E).
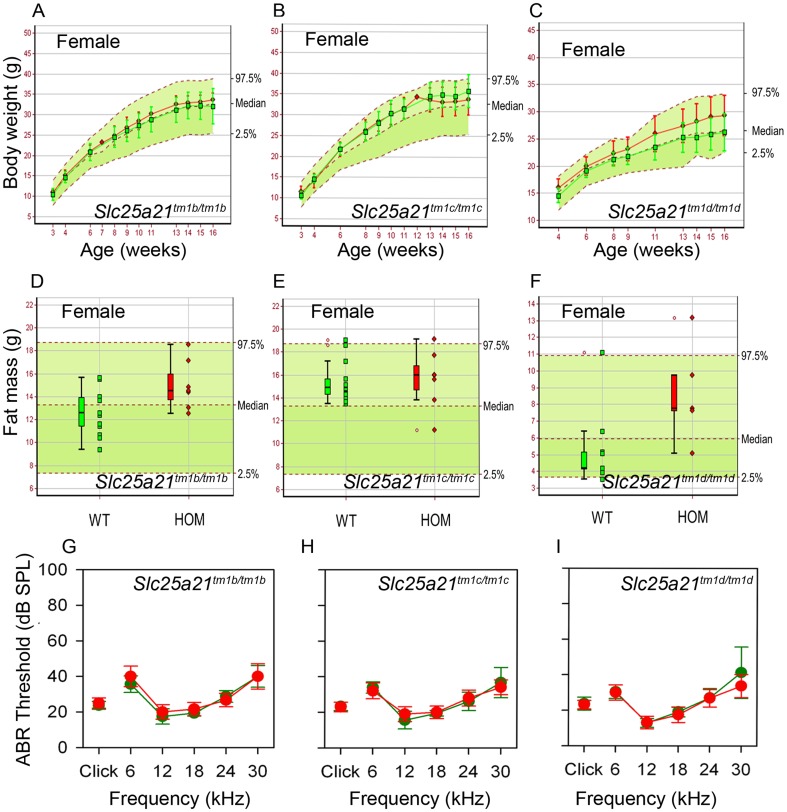
We proceeded by phenotyping animals homozygous for the Slc25a21tm1b(KOMP)Wtsi, Slc25a21tm1c(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi alleles. Following heterozygous intercrossing, homozygous animals for all three alleles were detected at the expected Mendelian ratio. Furthermore, body weight, fat mass and auditory brainstem response thresholds (Mann-Whitney Rank Sum Tests; tm1b, p = 0.12; tm1c, p = 0.48; tm1d, p = 0.59) (Fig. 7A–I) were all found to be normal compared with wild-types, as were the teeth and palatal rugae (data not shown). Full data from the phenotyping pipeline can be viewed online (www.sanger.ac.uk/mouseportal).
Figure 7. Normal phenotype observed in Slc25a21tm1b(KOMP)Wtsi, Slc25a21tm1c(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi homozygous mice.
Body weight and fat mass were normal in females homozygous (red symbols) for Slc25a21tm1b(KOMP)Wtsi (A,D) (n = 7), Slc25a21tm1c(KOMP)Wtsi (B,E) (n = 7), and Slc25a21tm1d(KOMP)Wtsi mice (C,F) (body weight n = 7, fat mass n = 5) compared to local wild-type controls run on the same week (green symbols) (n≥9). Male homozygotes for each of these three alleles and two parameters were also phenotypically normal (data not shown). Mean (± SD) body weight is plotted against age (A–C). Fat mass is presented as a boxplot, with median, 25th and 75th percentile (box), and the lowest and highest data point within 1.5× inter quartile range (whiskers) shown (D–F). The median and 95% reference range (2.5–97.5th percentiles, dotted lines) for all wild-type mice of the same genetic background and sex (n>80) are displayed on the pale green background. ABR thresholds were normal in homozygous Slc25a21tm1b(KOMP)Wtsi (G), Slc25a21tm1c(KOMP)Wtsi (H) and Slc25a21tm1d(KOMP)Wtsi mice (I) (red symbols) compared to wild-type controls (green symbols).
These results led us to conclude that the phenotypes observed in mice homozygous for the Slc25a21tm1a(KOMP)Wtsi targeted allele were not due to ablation of Slc25a21 function, and speculate that off-target effects due to the selection cassette present in the tm1a allele affected expression of neighbouring genes.
Hypomorphic expression of Pax9 may account for observed phenotypes
The 1 Mb genomic interval containing Slc25a21 (500 kb flanking the target gene) is predicted to contain seven known protein coding genes, one non coding RNA, one novel antisense and one putative processed transcript (Fig. S1). Of particular interest is Pax9, located at the 3′ end of Slc25a21 and orientated on the opposite strand. The 3′UTR regions of Slc25a21 and Pax9 overlap by 188 bp. Consistent with the phenotypes observed in Slc25a21tm1a(KOMP)Wtsi homozygous animals, mice carrying a hypomorphic Pax9 allele had previously been reported as causing reduced viability at weaning [25], and presented with a range of dental and craniofacial abnormalities [25], [26] similar to those described herein.
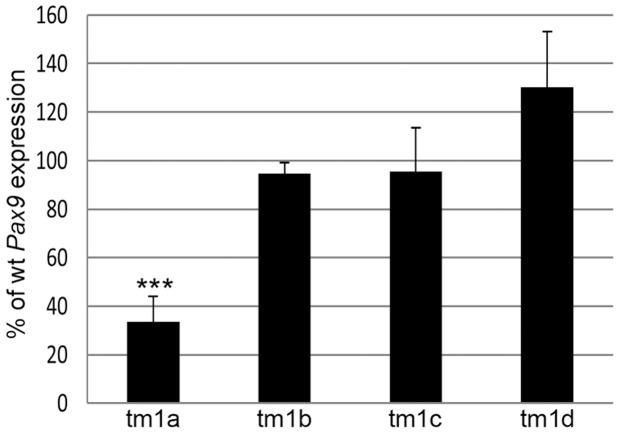
RNA expression analysis of Pax9 was performed on embryo heads (E13.5) for all four alleles in the Slc25a21 series. Pax9 expression was found to be down regulated [approximately 34% of wild-type Pax9 levels (p = 1.1E-06)] in homozygous Slc25a21tm1a(KOMP)Wtsi samples (Fig. 8) confirming our hypothesis that we had inadvertently created a Pax9 hypomorphic allele. Pax9 levels were found to be unaffected in homozygous Slc25a21tm1b(KOMP)Wtsi, Slc25a21tm1c(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi samples (Fig. 8). A known Pax9 regulatory element (CNS+6) was located within the targeting construct used to create the Slc25a21tm1a(KOMP)Wtsi allele [21]. PCR and sequence analysis was used to demonstrate that this sequence was not disrupted during the vector construction or at the homologous recombination stage (Table S1; data not shown). Consequently, we hypothesise that the presence of the promoter driven neomycin cassette in the homozygous Slc25a21tm1a(KOMP)Wtsi mice caused the reduction of Pax9 expression.
Figure 8. RNA expression of Pax9 was reduced in Slc25a21tm1a(KOMP)Wtsi mice.
Quantitative PCR on RNA from E13.5 embryo heads demonstrated that Pax9 RNA expression was significantly reduced in Slc25a21tm1a(KOMP)Wtsi (*** p = 1.1E-06) homozygotes, but equivalent to wild-type levels in Slc25a21tm1b(KOMP)Wtsi, Slc25a21tm1c(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi homozygotes.
RNA sequencing was performed to further explore the impact of the targeting event on the genomic environment around Slc25a21. Comparing RNA from wild-type and homozygous Slc25a21tm1a(KOMP)Wtsi E13.5 embryo heads, the only gene in the 1 Mb interval surrounding Slc25a21 that had significantly altered expression levels was Pax9 [q (adjusted p value) = 7.10E-06 (data not shown)]. Pax9 was not found to be differentially expressed in homozygous Slc25a21tm1b(KOMP)Wtsi, Slc25a21tm1c(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi embryo heads. The low endogenous expression level of Slc25a21 in wild-type mice was below the threshold where significant changes could be detected.
The phenotypes we observed in homozygous Slc25a21tm1a(KOMP)Wtsi mice were broadly consistent with those reported previously in mice carrying hypomorphic Pax9 alleles [26] with the exception that the otitis media and hearing impairment reported in this study have not been assessed previously in Pax9 mutant mice and may represent a novel consequence of Pax9 suppression.
Discussion
Characteristics of 2-oxoadipate acidaemia are absent in mice ablated for Slc25a21 expression
We report the generation and phenotypic characterisation of the Slc25a21 allelic series derived from a typical KOMP-CSD allele [3]. Subject to the sensitivity and scope of the phenotyping performed, ablation of Slc25a21 expression was not found to phenocopy the human disease 2-oxoadipate acidaemia (OMIM 204750) for which it is a candidate gene [11]. There are several possible explanations for this. Whilst no hypotonia, ataxia or seizures were detected in Slc25a21 null mice [homozygous for the tm1b (KOMP)Wtsi or tm1d (KOMP)Wtsi alleles], a subset of other clinical symptoms associated with 2-oxoadipate acidaemia, including intellectual disability and urine metabolites, were not assessed by the standard Mouse Genetics Project primary phenotyping pipeline with which these animals were screened [2]. Therefore it cannot be ruled out that additional tests, or analysis performed with increased sensitivity, may reveal subtle disease similarities. Alternatively, mitochondrial transport of metabolites is a complex process involving many carrier proteins. Unknown compensatory mechanisms may be at play in Slc25a21 null mice that prevent the cytoplasmic build-up of oxoadipate and the associated disruption in the catabolism of key amino acids such as lysine [11]. For example, it is known that the substrate specificity of Slc25a21 itself varies between species [11]. Finally, definitive proof of the candidacy of Slc25a21 for 2-oxoadipate acidaemia is not published and it has been speculated that the disease may be due to defective 2-oxoadipate dehydrogenase, an alternative gene in the same metabolic pathway [11].
Slc25a21tm1a(KOMP)Wtsi allele affects expression of a neighbouring gene
The Slc25a21tm1a(KOMP)Wtsi homozygous mice investigated in this study were found to be sub-viable. Those homozygous animals surviving to weaning presented with growth retardation, orofacial abnormalities, brittle, white incisors and hearing impairment. Given the absence of any detectable phenotype in mice homozygous for the Slc25a21 tm1b (KOMP)Wtsi, tm1c (KOMP)Wtsi or tm1d (KOMP)Wtsi derived alleles, we conclude that the phenotypes observed in mice homozygous for Slc25a21tm1a(KOMP)Wtsi were not due to ablation of Slc25a21 function, and propose that off-target effects of the tm1a allele affected expression of a neighbouring gene. The spatial, temporal and quantitatively correct activity of a gene requires the presence of not only intact coding sequence but also properly functioning regulatory control. Interference of the expression of neighbouring genes by the presence of selectable marker cassettes or short DNA sequences [5], [6], [7], [8], [9] has been reported for genes in close vicinity to the target gene and as far as >100 kb from the targeted gene [6]. Transcriptional silencing of neighbouring genes due to CpG rich sequences within the standard, non-codon optimised, lacZ reporter gene used in this KOMP-CSD allele has also been reported [27]. Furthermore, disruption of cis-regulatory elements has been linked with abnormal phenotypes in both mice [5] and humans [28], [29]. In addition, a wide variety of mechanisms which cause disruption of gene expression by a change in its chromosomal environment, while leaving the transcription unit intact, have been identified [29].
At E13.5, RNA sequencing revealed that the only gene within the 1 Mb genomic interval surrounding Slc25a21 that was differentially expressed, compared to wild-type, in homozygous Slc25a21tm1a(KOMP)Wtsi embryos was Pax9, an observation confirmed by quantitative PCR. Pax9 is located in a 21.06 kb genomic interval directly 3′ of Slc25a21 and orientated on the opposite strand. Evolutionarily conserved non-coding sequences are present both up-and down-stream of Pax9, some of which have been demonstrated to induce Pax9 expression [21]. One such conserved region, CNS+6, [21] was located within the homology arm of the Slc25a21tm1a(KOMP)Wtsi targeting construct, and therefore a candidate for causing altered Pax9 expression. However, sequencing confirmed the integrity of CNS+6 in the Slc25a21tm1a(KOMP)Wtsi allele. Since homozygous Slc25a21tm1b(KOMP)Wtsi mice, which still have part of the targeting cassette present (Fig. 6C), were found to be phenotypically normal, interference by the polyA-β-actin::neo cassette is the most likely cause of the reduction of Pax9 expression seen in homozygous Slc25a21tm1a(KOMP)Wtsi mice. The findings in this study highlight the need to confirm phenotypes found in animals carrying the knockout first conditional ready [tm1a (KOMP)Wtsi or tm1a (EUCOMM)Wtsi] allele by creating and analysing the derived deletion allele. This is a timely reminder, with direct implications on the 3Rs, given the growing resource of knockout first conditional ready targeted ES cells created by the EUCOMM and KOMP initiatives.
Hypomorphic expression of Pax9 may account for orofacial abnormalities
The Pax family of transcription factors plays a pivotal role in embryonic patterning and disease. The expression of each Pax gene is highly regulated in a temporal and spatial manner [30]. During mouse embryogenesis, Pax9 is expressed mainly in the sclerotome of somites, the pharyngeal pouch endoderm and its derivatives, developing limb buds, and in facial mesenchyme of neural-crest cell origin, including nasal and jaw processes and tooth buds [26], [31]. There is strong expression of Pax9 in the medial edge epithelium during the critical time points of palate fusion [32]. In adult mice, Pax9 is expressed in oesophagus and tongue [33]. Looking at published models of Pax9 deficiency, homozygous Pax9 knockout mice die shortly after birth and exhibit a wide range of developmental defects [26] including the absence of teeth. Death has been attributed to cleft palate, affecting the maxillary and palatine shelves, resulting in an inability to suckle and respiratory issues [26]. Two hypomorphic alleles of Pax9 have been published previously [25]. The first, which expressed wild-type Pax9 mRNA at 7% of control levels, presented with hypoplastic upper incisors lacking enamel, missing lower incisors, unilateral or bilateral absence of the third molar in the upper jaw and the second and third molar in the lower jaw, and the lower molars that were present showed severe attrition. In comparison, the hypomorphic allele with 20% of wild-type Pax9 mRNA expression relative to controls presented with less severely affected dentition including relatively normal upper incisors, hypoplastic lower incisors lacking enamel and unilateral or bilateral absence of the upper and lower third molar. We now report homozygous Slc25a21tm1a(KOMP)Wtsi mice which express wild-type Pax9 mRNA at 34% of control levels and present with an even milder dental phenotype including upper incisors that were commonly maloccluded, which may be due to abnormal snout morphology or the fact that the incisors do not wear against each other. We found fragile, damaged or missing lower incisors which were white or translucent; normally incisors in mice are brownish/yellow due to incorporation of iron-containing pigment in the enamel [34]. The lower incisor root area was severely under-developed. Lower jaw molars showed focal fracturing of the surface dentine and necrosis of the pulp with associated inflammation and bacterial accumulation. The presence of lacZ activity within the cavities of the lower molars of homozygous Slc25a21tm1a(KOMP)Wtsi mice but not heterozygous mice, confirms that bacterial β-galactosidase is the cause of the staining, not expression of the lacZ reporter gene within the targeted allele. Molars in the upper jaw were less affected. The more severe findings in the lower jaw are consistent with findings in the other Pax9 hypomorphic alleles [25].
Only limited assessment of the palate has been published for the previous Pax9 hypomorphic lines and no abnormalities were reported [25]. In this study, more extensive analysis was performed and whilst the phenotype was significantly less severe than that reported in Pax9 null mice [26], abnormalities were identified, characterised largely by discordance of rugae 3 and an incomplete fusion of the vomer bone to the maxilla.
This gradated allelic series of Pax9 expression levels, extended by the data reported herein, clearly demonstrates that dosage of this gene affects orofacial development and that lower jaw dentition in the mouse is more susceptible to reduced Pax9 expression [25]. This is consistent with humans where dentition is not uniformly affected by a reduced Pax9 expression [35], [36], [37] and the observation that the minimal Pax9 gene dosage required for the formation of individual teeth varies, the posterior tooth in each tooth family being the most sensitive to a reduction in Pax9 levels.
Otitis media and hearing impairment in mice may be due to disrupted Pax9 expression
A reduction in the size of the tympanic ring has previously been described in Pax9 mutant mice [26], however the effect on hearing has not previously been assessed. The homozygous Slc25a21tm1a(KOMP)Wtsi mice have a normal sized tympanic ring but display a clear hearing impairment with increased thresholds at all frequencies tested. The moderate hearing impairment seen in homozygous Slc25a21tm1a(KOMP)Wtsi mice is robust and reproducible across different cohorts of mice, tested at ages from 4 to 26 weeks of age. The degree of impairment in mutant mice is progressive from 4–14 weeks old, with slower progression to 26 weeks of age. The parallel shifts in audiometric profiles across frequencies at the different ages (Fig. 4) are consistent with an underlying conductive hearing loss. Middle ears of the majority of homozygous Slc25a21tm1a(KOMP)Wtsi mice were filled with fluid of varying viscosity at all ages examined and sections revealed inflammation of the lining of the middle ear. The presence of eosinophilic exudate and the thickening of the mucosa are evident in the homozygous Slc25a21tm1a(KOMP)Wtsi mice, indicating an on-going inflammatory process. Whilst the aetiology of otitis media is complex [38] and was beyond the scope of this study, the correlation between signs of otitis media and elevated ABR thresholds supports our suggestion that the otitis media is a major contributor to the hearing impairment observed in the young adult homozygous Slc25a21tm1a(KOMP)Wtsi mice. However, cochlear dysfunction cannot be ruled out especially in the 26 week-old group of both mutants and controls. ABR data from Slc25a21tm1b(KOMP)Wtsi, Slc25a21tm1c(KOMP)Wtsi and Slc25a21tm1d(KOMP)Wtsi homozygotes indicate that hearing thresholds are normal.
Conclusion
In conclusion, ablation of Slc25a21 expression was not found to phenocopy the human disease 2-oxoadipate acidaemia (OMIM 204750) for which it is a candidate gene. The phenotypes observed in mice homozygous for Slc25a21tm1a(KOMP)Wtsi were not due to ablation of Slc25a21 function, but instead the presence of the selection cassette affected expression of the neighbouring gene, Pax9. The resulting mutant line confirmed and extended the existing knowledge of Pax9 gene dosage effect on orofacial development and represents a novel model of otitis media that may be due to reduced Pax9 expression.
Supporting Information
Ensembl view of 1 Mb genomic interval encompassing Pax9 and Slc25a21 . Ensembl view of the 500 kb of genomic DNA flanking the 5′ and 3′ end of Slc25a21. The Slc25a21tm1a(KOMP)Wtsi targeting construct (shown in blue) designates exon 4 as the critical exon.
(TIF)
Supplementary materials.
(DOC)
The sequence of primers used for molecular characterisation. Molecular characterisation of the targeting event was performed using a combination of PCR assays. The 5′ to 3′ primer sequence along with the expected product size (bp) is presented.
(DOC)
Comparison of the outcomes from automatic data evaluation and manual assessment. Comparison of the outcomes from automatic data evaluation and manual assessment for each parameter included in the dataset is presented. Discrepancies between these two methods of assessment are highlighted, and the rationale behind the manual assessment is provided in each instance there was a discrepancy.
(DOC)
Acknowledgments
We thank the staff from the Sanger Institute's Research Support Facility and Mouse Informatics Group for their excellent support and Darren Logan for useful discussions. We thank Skyscan for use of the SkyScan1176 micro-CT machine. We thank the Illumina Bespoke Team for the RNA-seq data.
The Sanger MGP Slc25a21 Project Team was led by: Hannah Wardle Jones and included Caroline Barnes, Ryan Beveridge, Emma Cambridge, Diane Gleeson, Ozama Ismail, Lee Mulderrig, Hayley J. Protheroe, Laura-Anne Roberson, Grace Salsbury, Mark Sanderson, Daniel Sanger, Agnes Swiatkowska, David Tino Lafont and Valerie E. Vancollie. All members of the Sanger MGP Slc25a21 Project Team were affiliated with the Wellcome Trust Sanger Institute at the time the work was performed.
Funding Statement
This work was supported by the Wellcome Trust (grant no 098051 to WTSI) (http://www.wellcome.ac.uk/) and the European Union's Seventh Framework program under grant agreement number 282510 - BLUEPRINT (NW) (www.blueprint-epigenome.eu/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. van der Weyden L, White JK, Adams DJ, Logan DW (2011) The mouse genetics toolkit: revealing function and mechanism. Genome Biol 12: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, et al. (2013) Genome-wide Generation and Systematic Phenotyping of Knockout Mice Reveals New Roles for Many Genes. Cell 154: 452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, et al. (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474: 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mitchell KJ, Pinson KI, Kelly OG, Brennan J, Zupicich J, et al. (2001) Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat Genet 28: 241–249. [DOI] [PubMed] [Google Scholar]
- 5. Meier ID, Bernreuther C, Tilling T, Neidhardt J, Wong YW, et al. (2010) Short DNA sequences inserted for gene targeting can accidentally interfere with off-target gene expression. FASEB J 24: 1714–1724. [DOI] [PubMed] [Google Scholar]
- 6. Pham CT, MacIvor DM, Hug BA, Heusel JW, Ley TJ (1996) Long-range disruption of gene expression by a selectable marker cassette. Proc Natl Acad Sci U S A 93: 13090–13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hug BA, Wesselschmidt RL, Fiering S, Bender MA, Epner E, et al. (1996) Analysis of mice containing a targeted deletion of beta-globin locus control region 5′ hypersensitive site 3. Mol Cell Biol 16: 2906–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fiering S, Kim CG, Epner EM, Groudine M (1993) An “in-out” strategy using gene targeting and FLP recombinase for the functional dissection of complex DNA regulatory elements: analysis of the beta-globin locus control region. Proc Natl Acad Sci U S A 90: 8469–8473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Olson EN, Arnold HH, Rigby PW, Wold BJ (1996) Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell 85: 1–4. [DOI] [PubMed] [Google Scholar]
- 10. Fiermonte G, Dolce V, Palmieri L, Ventura M, Runswick MJ, et al. (2001) Identification of the human mitochondrial oxodicarboxylate carrier. Bacterial expression, reconstitution, functional characterization, tissue distribution, and chromosomal location. J Biol Chem 276: 8225–8230. [DOI] [PubMed] [Google Scholar]
- 11. Palmieri F (2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch 447: 689–709. [DOI] [PubMed] [Google Scholar]
- 12. Palmieri F (2013) The mitochondrial transporter family SLC25: Identification, properties and physiopathology. Molecular Aspects of Medicine 34: 465–484. [DOI] [PubMed] [Google Scholar]
- 13. Duran M, Beemer FA, Wadman SK, Wendel U, Janssen B (1984) A patient with α-ketoadipic and α-aminoadipic aciduria. Journal of Inherited Metabolic Disease 7: 61–61. [DOI] [PubMed] [Google Scholar]
- 14. Fischer MH, Brown RR (1980) Tryptophan and lysine metabolism in alpha-aminoadipic aciduria. Am J Med Genet 5: 35–41. [DOI] [PubMed] [Google Scholar]
- 15. Przyrembel H, Wendel U, Becker K, Bremer HJ, Bruinvis L, et al. (1976) Glutaric aciduria type II: Report on a previously undescribed metabolic disorder. Clinica Chimica Acta 66: 227–239. [DOI] [PubMed] [Google Scholar]
- 16. Wilson RW, Wilson CM, Gates SC, Higgins JV (1975) Alpha-ketoadipic aciduria: a description of a new metabolic error in lysine-tryptophan degradation. Pediatr Res 9: 522–526. [DOI] [PubMed] [Google Scholar]
- 17. Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG (2010) Improving Bioscience Research Reporting: The ARRIVE Guidelines for Reporting Animal Research. PLoS Biol 8: e1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collins FS, Rossant J, Wurst W (2007) A mouse for all reasons. Cell 128: 9–13. [DOI] [PubMed] [Google Scholar]
- 19. Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, et al. (2009) Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods 6: 493–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ryder E, Gleeson D, Sethi D, Vyas S, Miklejewska E, et al. (2013) Molecular Characterization of Mutant Mouse Strains Generated from the EUCOMM/KOMP-CSD ES Cell Resource. Mamm Genome 24: 286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Santagati F, Abe K, Schmidt V, Schmitt-John T, Suzuki M, et al. (2003) Identification of Cis-regulatory elements in the mouse Pax9/Nkx2-9 genomic region: implication for evolutionary conserved synteny. Genetics 165: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ingham NJ, Pearson S, Steel KP (2011) Using the Auditory Brainstem Response (ABR) to Determine Sensitivity of Hearing in Mutant Mice. Curr Protoc Mouse Biol 1: 279–287. [DOI] [PubMed] [Google Scholar]
- 23. Adams DJ, van der Weyden L (2008) Contemporary approaches for modifying the mouse genome. Physiol Genomics 34: 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ovchinnikov D (2009) Alcian blue/alizarin red staining of cartilage and bone in mouse. Cold Spring Harb Protoc 2009: pdb prot5170. [DOI] [PubMed] [Google Scholar]
- 25. Kist R (2005) Reduction of Pax9 gene dosage in an allelic series of mouse mutants causes hypodontia and oligodontia. Human Molecular Genetics 14: 3605–3617. [DOI] [PubMed] [Google Scholar]
- 26. Peters H, Neubuser A, Kratochwil K, Balling R (1998) Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes & Development 12: 2735–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chevalier-Mariette C, Henry I, Montfort L, Capgras S, Forlani S, et al. (2003) CpG content affects gene silencing in mice: evidence from novel transgenes. Genome Biology 4: R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kleinjan DJ, Coutinho P (2009) Cis-ruption mechanisms: disruption of cis-regulatory control as a cause of human genetic disease. Brief Funct Genomic Proteomic 8: 317–332. [DOI] [PubMed] [Google Scholar]
- 29. Kleinjan DA, Lettice LA (2008) Long-range gene control and genetic disease. Adv Genet 61: 339–388. [DOI] [PubMed] [Google Scholar]
- 30. Lang D, Powell S, Plummer R, Young K, Ruggeri B (2007) PAX genes: Roles in development, pathophysiology, and cancer. Biochemical Pharmacology 73: 1–14. [DOI] [PubMed] [Google Scholar]
- 31. Neubuser A, Koseki H, Balling R (1995) Characterization and developmental expression of Pax9, a paired-box-containing gene related to Pax1. Dev Biol 170: 701–716. [DOI] [PubMed] [Google Scholar]
- 32. Sasaki Y, Okane S, Dixon J, Dixon M, Ferguson M (2007) Temporal and spatial expression of Pax9 and Sonic hedgehog during development of normal mouse palates and cleft palates in TGF-β3 null embryos. Archives of Oral Biology 52: 260–267. [DOI] [PubMed] [Google Scholar]
- 33. Peters H, Schuster G, Neubuser A, Richter T, Hofler H, et al. (1997) Isolation of the Pax9 cDNA from adult human esophagus. Mamm Genome 8: 62–64. [DOI] [PubMed] [Google Scholar]
- 34.Mataki S (1991) Studies on the transport mechanism of iron in rat incisor enamel. In: Ohya K, Ino M, Ogura H, editors. Tokyo: Springer-Verlag. pp. 298–301 [Google Scholar]
- 35. Das P, Hai M, Elcock C, Leal SM, Brown DT, et al. (2003) Novel missense mutations and a 288-bp exonic insertion in Pax9 in families with autosomal dominant hypodontia. American Journal of Medical Genetics 118A: 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Frazier-Bowers SA, Guo DC, Cavender A, Xue L, Evans B, et al. (2002) A Novel Mutation in Human Pax9 Causes Molar Oligodontia. Journal of Dental Research 81: 129–133. [PubMed] [Google Scholar]
- 37. Nieminen P, Arte S, Tanner D, Paulin L, Alaluusua S, et al. (2001) Identification of a nonsense mutation in the Pax9 gene in molar oligodontia. Eur J Hum Genet 9: 743–746. [DOI] [PubMed] [Google Scholar]
- 38. Tyrer HE, Crompton M, Bhutta MF (2013) What have we learned from murine models of otitis media? Curr Allergy Asthma Rep 13: 501–511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ensembl view of 1 Mb genomic interval encompassing Pax9 and Slc25a21 . Ensembl view of the 500 kb of genomic DNA flanking the 5′ and 3′ end of Slc25a21. The Slc25a21tm1a(KOMP)Wtsi targeting construct (shown in blue) designates exon 4 as the critical exon.
(TIF)
Supplementary materials.
(DOC)
The sequence of primers used for molecular characterisation. Molecular characterisation of the targeting event was performed using a combination of PCR assays. The 5′ to 3′ primer sequence along with the expected product size (bp) is presented.
(DOC)
Comparison of the outcomes from automatic data evaluation and manual assessment. Comparison of the outcomes from automatic data evaluation and manual assessment for each parameter included in the dataset is presented. Discrepancies between these two methods of assessment are highlighted, and the rationale behind the manual assessment is provided in each instance there was a discrepancy.
(DOC)