Abstract
Rhobtb2 is a candidate tumor suppressor located on human chromosome 8p21, a region commonly deleted in cancer. Rhobtb2 is homozygously deleted in 3.5% of primary breast cancers, and gene expression is ablated in ∼50% of breast and lung cancer cell lines. RhoBTB2 is an 83-kD, atypical Rho GTPase of unknown function, comprising an N-terminal Rho GTPase domain and two tandem BTB domains. In this report, we demonstrate that RhoBTB2 binds to the ubiquitin ligase scaffold, Cul3, via its first BTB domain and show in vitro and in vivo that RhoBTB2 is a substrate for a Cul3-based ubiquitin ligase complex. Moreover, we show that a RhoBTB2 missense mutant identified in a lung cancer cell line is neither able to bind Cul3 nor is it regulated by the ubiquitin/proteasome system, resulting in increased RhoBTB2 protein levels in vivo. We suggest a model in which RhoBTB2 functions as a tumor suppressor by recruiting proteins to a Cul3 ubiquitin ligase complex for degradation.
Keywords: Rho, DBC2, cullin, ubiquitin, cancer
Homozygous deletions or loss of heterozygousity in human chromosome 8p21 are found in many cancers including those of the breast, lung, ovary, and prostate (Brown et al. 1999; Wistuba et al. 1999; Hamaguchi et al. 2002). Rhobtb2 was first identified as a candidate tumor suppressor gene on 8p21 in breast cancer and initially called DBC2 (deleted in breast cancer 2; Hamaguchi et al. 2002). Rhobtb2 is homozygously deleted in 3.5% of breast cancers, gene expression is ablated in ∼50% of breast and lung cancer cell lines, and several somatic missense mutations in Rhobtb2 have been isolated from primary tumors and cancer cell lines. Furthermore, reintroduction of Rhobtb2 into a breast cancer cell line lacking endogenous Rhobtb2 leads to growth arrest (Hamaguchi et al. 2002). Rhobtb2 encodes an 83-kD, atypical Rho GTPase, comprising an N-terminal Rho GTPase domain followed by two BTB domains. Rho family GTPases act as molecular switches, binding effector molecules only when in the GTP-bound state. BTB domains were initially identified in the Drosophila transcriptional repressors broad complex, tramtrack, and bric-a-brac but have since been identified in ∼190 human proteins of various function (Collins et al. 2001). Recently, the BTB domains of several proteins have been shown to interact with the Cul3 ubiquitin ligase scaffold protein (Furukawa et al. 2003; Geyer et al. 2003; Pintard et al. 2003; Xu et al. 2003). Although Rhobtb2 is absent from the yeast and Caenorhabditis elegans genomes, mammals and fish each have three highly homologous Rhobtb family members and Drosophila and Dictyostelium each have a single gene (Ramos et al. 2002). However, the physiological function of RhoBTB homologs from any organism remains undetermined.
The ubiquitin/proteasome system tightly controls the levels of signaling proteins in a variety of biological contexts (Pickart 2001). Proteins are targeted for proteasomal destruction by the covalent attachment of polyubiquitin chains via the activity of substrate-specific ubiquitin ligases. The cullin-based E3 ligases are one major class of ubiquitin ligase (Deshaies 1999; Pickart 2001). Mammals have six distinct cullin proteins (Cul1, Cul2, Cul3, Cul4A, Cul4B, and Cul5), all of which bind the ring finger protein Roc1 (which in turn recruits the E2 ubiquitin-conjugating enzyme) and are modified covalently by the activator Nedd8. This cullin/Roc complex constitutes the core ubiquitin ligase module. Substrate selection is determined by the binding of substrate-specific adaptors to the N-terminal region of the cullin. In the case of Cul1, this consists of the invariant Skp1 and a substrate-specific F-box protein, whereas Cul2 and Cul5 use a combination of elongin C and a BC-box-containing protein (Deshaies 1999). Cullin-based ubiquitin ligases are closely linked to cancer. The pVHL (von Hippel-Lindau) tumor suppressor is a substrate-recognition adaptor for Cul2-based ubiquitin ligases (Pause et al. 1997; Lisztwan et al. 1999; Maxwell et al. 1999; Ohh et al. 2000). Cul4 is part of a protein complex whose aberrant function causes Xeroderma pigmentosa and Cockayne syndrome, both of which can lead to cancer (Groisman et al. 2003). In addition, Cul1-based complexes control the protein levels of many cell cycle regulators, tumor suppressors, and oncogenes (Deshaies 1999).
The substrate-recognition elements of Cul3 have only just begun to be understood. Several recent reports identified BTB domains as binding to the N-terminal region of Cul3 (Furukawa et al. 2003; Geyer et al. 2003; Pintard et al. 2003; Xu et al. 2003). From these studies, two substrates for Cul3-dependent ubiquitylation were identified: Btb3p from Schizosaccharomyces pombe, which binds directly to Cul3, and MEI-1 from C. elegans, which does not bind Cul3 directly, but instead is recruited by the BTB-domain-containing adaptor MEL26. Although several mammalian BTB proteins have been shown to interact with Cul3, none have been shown to be either substrates or substrate-specific adaptors. In this report, we demonstrate in vitro and in vivo that the tumor suppressor RhoBTB2 binds to Cul3 and is a substrate for a Cul3-based ubiquitin ligase complex. Moreover, we show that a RhoBTB2 mutant, identified in a lung cancer cell line, is both unable to bind Cul3 and is not regulated by the ubiquitin/proteasome system, leading to overexpression of RhoBTB2 in vivo. We speculate that RhoBTB2 functions as a tumor suppressor by recruiting target proteins to a Cul3 ubiquitin ligase complex for degradation.
Results and Discussion
RhoBTB2 interacts with Cul3 but not other cullin family members
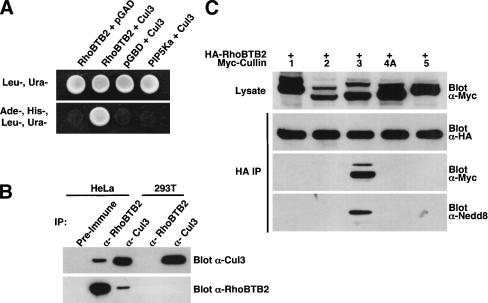
As part of our ongoing investigation of RhoBTB2 function, we screened a mouse embryo yeast two-hybrid library for RhoBTB2-interacting proteins and identified the N-terminal region of murine Cul3 (residues 1–199) as an interacting protein. We then confirmed that the full-length human Cul3 could also mediate a two-hybrid interaction with RhoBTB2 but not the empty bait plasmid or the irrelevant bait, phosphatidylinositol-4-phosphate 5-kinase α (PIP5Kα; Fig. 1A).
Figure 1.
RhoBTB2 interacts with Cul3 but not other cullin family members. (A) Interaction of RhoBTB2 and Cul3 in the yeast two-hybrid system. PJ69-4A yeast were cotransformed with the indicated plasmids and grown in the absence of leucine and uracil (Leu–, Ura–) to maintain both plasmids. Protein/protein interactions were determined by growth in the absence of adenine and histidine (Leu–, Ura–, Ade–, His–). For photography, 10 μL of appropriate clonal yeast suspensions were spotted on the indicated agar plates and grown for 3–5 d before being photographed against a black background. (B) Interaction of endogenous RhoBTB2 and Cul3. HeLa or 293T cell lysates were subject to immunoprecipitation by preimmune, RhoBTB2, or Cul3 antibodies and immune complexes probed for both Cul3 and RhoBTB2 by Western blotting. (C) RhoBTB2 binds specifically to Cul3. 293T cells were transfected with the indicated plasmids, then anti-HA immune complexes isolated and analyzed sequentially for the presence of RhoBTB2, cullin, and Nedd8 by Western blotting.
To verify that endogenous RhoBTB2 and Cul3 physically interact, we subjected HeLa and 293T cell lysates to immunoprecipitation by anti-Cul3, anti-RhoBTB2, or preimmune sera and probed for Cul3 or RhoBTB2 by Western blotting (Fig. 1B). It has previously been shown that HeLa cells express RhoBTB2 (Hamaguchi et al. 2002), whereas 293T lack RhoBTB2 expression (Ramos et al. 2002). We found that Cul3 and RhoBTB2 coprecipitated from HeLa lysates when immunoprecipitation was performed with either anti-RhoBTB2 or anti-Cul3 antibody but not with preimmune serum. As expected, RhoBTB2 antibodies failed to precipitate RhoBTB2 or Cul3 from 293T cells because of the lack of endogenous RhoBTB2, confirming that there was no cross-reaction between the RhoBTB2 antibody and the Cul3 protein. Thus, RhoBTB2 interacts with Cul3 in vivo.
We next sought to determine if RhoBTB2 binds only to Cul3. 293T cells were transfected with HA-RhoBTB2 and various Myc-tagged cullins. RhoBTB2 was immunoprecipitated and immune complexes analyzed by Western blotting for the presence of the cullin. We found that RhoBTB2 bound specifically to Cul3 but not other cullin family members (Fig. 1C). Cullins run as doublets on SDS-PAGE gels, the higher mobility species being covalently linked to the 8.5-kD Nedd8 protein. RhoBTB2 appeared to precipitate both forms of Cul3 (Fig. 1C). The identity of the upper species was confirmed as neddylated Cul3 by reprobing blots with a Nedd8 antibody (Fig. 1C, bottom panel).
The first BTB domain of RhoBTB2 binds to the N-terminal region of Cul3
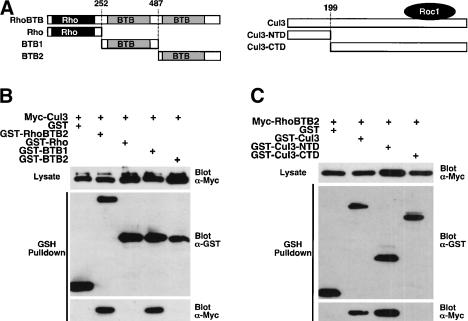
We next used deletion mutants of RhoBTB2 and Cul3 to map their interaction more precisely. Various RhoBTB2 or Cul3 deletion mutants (Fig. 2A) were expressed as GST fusion proteins in 293T cells together with Myc-Cul3 or Myc-RhoBTB2, respectively. The association of transfected RhoBTB2 and Cul3 was determined by Western blotting after precipitation with GSH-agarose. For RhoBTB2, the Rho domain and second BTB domain showed no binding to Cul3, whereas the first BTB domain binds as well as full-length RhoBTB2 to Cul3 (Fig. 2B). The C-terminal domain of Cul3 (Cul3-CTD) failed to bind RhoBTB2, whereas the N-terminal domain (Cul3-NTD) bound at least as well as full-length Cul3 to RhoBTB2 (Fig. 2C). Thus, the first BTB domain of RhoBTB2 binds to the N-terminal region of Cul3. This is in agreement with recent data from four other groups who report the interaction of BTB domains with the N-terminal region of Cul3 (Furukawa et al. 2003; Geyer et al. 2003; Pintard et al. 2003; Xu et al. 2003).
Figure 2.
RhoBTB2 interacts with the N terminus of Cul3 via its first BTB domain. (A) A schematic representation of the RhoBTB2 and Cul3 mutants used. The exact residue numbers where truncations are made are indicated. The approximate binding site of the ring finger protein Roc1 on Cul3 is also shown. (B) Mapping the binding site of Cul3 on RhoBTB2. 293T cells were transfected with the indicated plasmids. GST complexes were isolated and analyzed sequentially for the presence of Cul3 and GST–RhoBTB2 proteins by Western blotting. (C) Mapping the binding site of RhoBTB2 on Cul3. 293T cells were transfected with the indicated plasmids. GST complexes were isolated and analyzed sequentially for the presence of RhoBTB2 and GST–Cul3 proteins by Western blotting.
A BTB domain mutant of RhoBTB2 isolated from a lung cancer cell line fails to bind Cul3
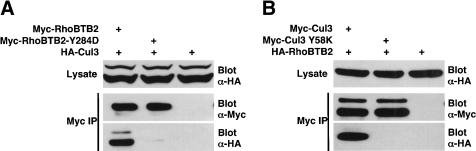
Analysis of primary tumors and cell lines identified four missense mutations in RhoBTB2, three of which (Y284D, D299N, and D368A) were in the first BTB domain (Hamaguchi et al. 2002). Because RhoBTB2 binds Cul3 through this BTB domain, we investigated whether these mutations might affect the binding of RhoBTB2 to Cul3. To this end, we transfected 293T cells with HA-Cul3 and either Myc-RhoBTB2 or one of the Myc-RhoBTB2 mutants (Y284D, D299N, or D366A). RhoBTB2 proteins were immunoprecipitated and their ability to bind Cul3 was assessed by Western blotting. We found that wild-type RhoBTB2 binds robustly to Cul3, as expected, whereas the Y284D RhoBTB2 mutation almost completely abolished binding to Cul3 (Fig. 3A). In contrast, the D299N and D368A mutants of RhoBTB2 bind to Cul3 as well as wild-type RhoBTB2 (data not shown). Interestingly, the Y284D RhoBTB2 mutant was consistently expressed at levels three to fivefold higher than wild type (see Fig. 4A). Consequently, in Figure 3A, four times less RhoBTB2-Y284D precipitate than wild type was loaded to allow easier assessment of the relative amounts of Cul3 binding of the two proteins.
Figure 3.
Point mutations in RhoBTB2 and Cul3 disrupt their interaction. (A) A Y284D mutation in RhoBTB2 disrupts the interaction with Cul3. 293T cells were transfected with the indicated plasmids and anti-HA immune complexes isolated and analyzed sequentially for the presence of RhoBTB2 and Cul3 by Western blotting. The Y284D RhoBTB2 mutant was consistently expressed at levels three to fivefold higher than wild type; consequently, four times less RhoBTB2-Y284D immunoprecipitate than wild type was loaded to allow easier assessment of the relative amounts of Cul3. (B) A Y58K mutation in Cul3 disrupts the interaction with RhoBTB2. 293T cells were transfected with the indicated plasmids and anti-HA immune complexes isolated and analyzed sequentially for the presence of RhoBTB2 and Cul3 by Western blotting.
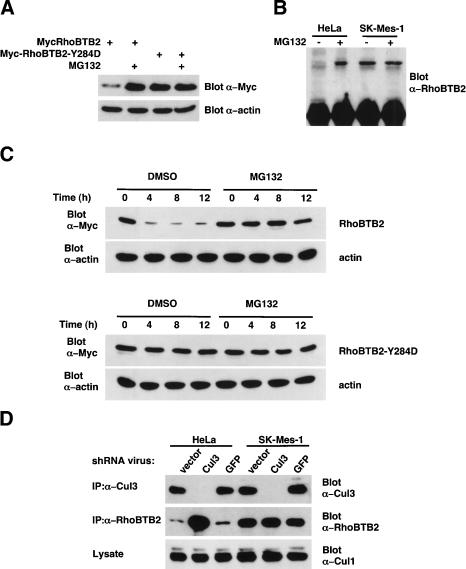
Figure 4.
RhoBTB2 protein levels are regulated by both Cul3 and the ubiquitin/proteasome system in vivo. (A) RhoBTB2 protein levels are negatively regulated by the proteasome in transfected cells. 293T cells were transfected with either wild-type or Y284D RhoBTB2 for 16 h. To control for variability in transfection, we then split cells into two separate dishes and, after cell reattachment, treated them with either 10 μM MG132 or DMSO alone for a further 12 h. Cell lysates were then analyzed for RhoBTB2 expression by Western blotting. As a loading control, blots were also probed for α-actin. (B) Endogenous RhoBTB2 protein levels are negatively regulated by the proteasome. Hela and SK-Mes-1 cells were treated with either 10 μM MG132 or DMSO alone for 12 h. Equal amounts of lysates were then subject to immunoprecipitation by anti-RhoBTB2 antibody and analyzed for the presence of RhoBTB2 by Western blotting. (C) RhoBTB2 has a short half-life that is stabilized by proteasomal inhibition. 293T cells were transfected with either wild-type or Y284D RhoBTB2 for 16 h. To control for variability in transfection, we then split cells into 3-cm dishes and, after cell reattachment, treated them with 100 μM cycloheximide plus either 10 μM MG132 or DMSO alone for the indicated number of hours. Cell lysates were then analyzed for RhoBTB2 expression by Western blotting. As a loading control, blots were also probed for α-actin. (D) shRNA-mediated ablation of Cul3 results in elevated levels of endogenous RhoBTB2 protein. HeLa and SK-Mes-1 cells were infected twice in 24 h with retroviruses expressing shRNA for either Cul3 or GFP. An empty vector virus was used as a negative control. After 48 h of infection, cell lysates were subject to immunoprecipitation and Western blotting by RhoBTB2 or Cul3 antibodies. As a control for the specificity of the Cul3 shRNA, cell lysates were Western blotted for Cul1 levels.
We next investigated the structural elements of Cul3 necessary for RhoBTB2 binding. Extensive crystallo-graphic studies of the interaction of Cul1 and Skp1 revealed that two helices, H2 and H5, from the N-terminal of Cul1 constitute the binding interface for Skp1 (Schulman et al. 2000; Zheng et al. 2002). Inspection of the protein sequence of Cul3 homologs across species revealed two highly conserved regions corresponding to H2 and H5 of Cul1. Moreover, the BTB domain has been postulated to have structural similarity to that of Skp1 (Zheng et al. 2002). To determine if the RhoBTB2/Cul3 interaction was analogous to that of Cul1/Skp1, we made amino acid substitutions in the putative H2 and H5 helices of Cul3 as follows: S53A, E55A/E56A, R59A, Y58K, Y62K, Y125K, R128D, D127A/R128A. To test the ability of these Cul3 mutants to bind RhoBTB2, we transfected 293T cells with HA-RhoBTB2 and either Myc-Cul3 or one of the Myc-Cul3 mutants. Cul3 proteins were immunoprecipitated and their ability to bind RhoBTB2 was assessed by Western blotting. We found that wild-type Cul3 bound robustly to RhoBTB2, as expected, whereas the Y58K Cul3 mutation almost completely abolished binding to RhoBTB2 (Fig. 3B). In contrast, the other mutants of Cul3 did not affect binding to RhoBTB2 (data not shown). This is in agreement with others, who have shown in S. pombe and C. elegans that mutations in the putative H2 and H5 helices of Cul3 disrupt binding to BTB-domain-containing proteins (Geyer et al. 2003; Pintard et al. 2003; Xu et al. 2003).
Regulation of RhoBTB2 expression by the ubiquitin/proteasome system and Cul3
Because RhoBTB2 binds to the N-terminal region of Cul3, we reasoned that it may be a substrate of a Cul3-based ubiquitin ligase complex. If this were true, RhoBTB2 protein levels should be increased by inhibition of the ubiquitin/proteasome system. Moreover, protein levels of the Y284D RhoBTB2 mutant should be unaffected by proteasomal inhibition because it cannot bind to Cul3. We therefore determined the effect of proteasomal inhibition on transfected wild-type and Y284D mutant RhoBTB2. 293T cells were transiently transfected with either wild-type or Y284D RhoBTB2 for 16 h. To control for variability in transfection efficiency, we then split cells into two separate dishes and, after cell reattachment, treated them with either the proteasome inhibitor MG132 or DMSO alone. The level of Myc-RhoBTB2 in cell lysates was determined by Western blotting. As a control for loading, blots were also probed for α-actin. As predicted, MG132 treatment of cells increased the levels of wild-type RhoBTB2 protein at least fourfold (Fig. 4A). Moreover, the Y284D mutant of RhoBTB2, which is defective in Cul3 binding (Fig. 3B), was more highly expressed than wild-type RhoBTB2 (three- to fivefold higher) and this level was not further increased by proteasomal inhibition (Fig. 4A).
We also examined the effect of proteasomal inhibition on the levels of endogenous RhoBTB2 in HeLa cells and SK-MES-1 (the lung cancer cell line that expresses the Y284D RhoBTB2 mutant). Although we were unable to detect RhoBTB2 by Western blotting of total cell lysates, we were able to immunoprecipitate endogenous RhoTB2 and determine relative expression levels. In agreement with the results from transient transfection experiments, proteasomal inhibition dramatically increased protein levels of wild-type RhoBTB2 in HeLa cells, whereas the level of mutant RhoBTB2-Y284D protein in SK-Mes-1 cells was higher basally than in HeLa cells and was unresponsive to proteasome inhibitor (Fig. 4B).
We next determined the relative half-lives of RhoBTB2 and the Y284D RhoBTB2 mutant. If RhoBTB2 was regulated by the ubiquitin/proteasome system, as a direct consequence of binding to Cul3, then the half-life of RhoBTB2 should be significantly shorter than that of the Y284D RhoBTB2 mutant. To test this, we transiently transfected 293T cells with either wild-type or Y284D RhoBTB2 for 16 h. To control for variability in transfection efficiency, we then split cells into four separate dishes and, after cell reattachment, treated them with the protein synthesis inhibitor cycloheximide and either the proteasome inhibitor MG132 or DMSO alone. The level of Myc-RhoBTB2 in cell lysates was determined by Western blotting. As a control for loading, blots were also probed for α-actin. Following cyclohexamide treatment, the levels of wild-type RhoBTB2 protein decline rapidly (half-life <4 h) and this is prevented by proteasomal inhibition (Fig. 4C). In contrast, the protein levels of the Y284D RhoBTB2 mutant are stable for >12 h and unaffected by proteasomal inhibition.
If RhoBTB2 is degraded as a result of binding to Cul3, then ablation or down-regulation of Cul3 should lead to a concomitant increase in wild-type RhoBTB2 protein levels but have little or no effect on the levels of the Y284D RhoBTB2 mutant. To address this, we reduced the levels of endogenous Cul3 protein by using retrovirally expressed shRNA specific to Cul3. HeLa and SK-Mes-1 cells were infected with retrovirus expressing shRNA for Cul3. A retrovirus expressing shRNA specific for GFP (Jin et al. 2003) and an “empty vector” retrovirus were used as controls for specificity. After 48 h of infection, cells were lysed and the levels of Cul3 and RhoBTB2 determined by immunoprecipitation and Western blot. As a further control for specificity of the Cul3-shRNA, the levels of Cul1 in cell lysates was also determined by Western blotting. For both HeLa and SK-Mes-1 cells, Cul3 shRNA almost completely ablated endogenous Cul3 protein relative to “empty vector” and GFP shRNA controls, but Cul1 protein levels were unchanged. As predicted, in HeLa cells down-regulation of Cul3 correlated with a dramatic increase in endogenous RhoBTB2 levels (Fig. 4D). In contrast, in SK-Mes-1 cells there was no change in RhoBTB2 levels in the absence of Cul3.
Taken together, these data provide compelling evidence for a model in which RhoBTB2 is a direct substrate of a Cul3 ubiquitin ligase complex and is degraded by the ubiquitin/proteasome system in vivo.
RhoBTB2 is a direct substrate of Cul3-based ubiquitin ligase complex in vitro
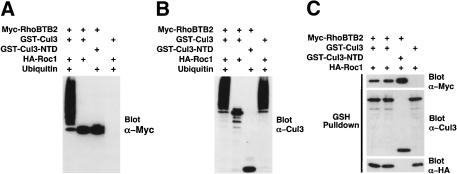
We next directly tested in vitro whether RhoBTB2 was a substrate for a Cul3-based ubiquitin ligase complex. Accordingly, 293T cells were transfected with constructs expressing GST–Cul3 (or Cul3-NTD), Myc-RhoBTB2, and HA-Roc1. Cul3/RhoBTB2/Roc1 complexes were precipitated with GSH-agarose and subjected to an in vitro ubiquitylation assay (Fig. 5). After the reaction, proteins were Western blotted sequentially for RhoBTB2 (Fig. 5A) and Cul3 (Fig. 5B). The constituents of the Cul3 complexes used for each assay were confirmed by Western blotting for Cul3, RhoBTB2, and Roc1 (Fig. 5C). We found that a high-molecular-weight ladder of RhoBTB2, characteristic of polyubiquitin chains, was generated robustly by complexes of RhoBTB2, Cul3, and Roc1 in the presence of ubiquitin (Fig. 5A). However, omission of ubiquitin or RhoBTB2 from the reaction prevented formation of this laddering, confirming the identity of the high-molecular-weight species as polyubiquitylated RhoBTB2. Also, as predicted, complexes of RhoBTB2 and Cul3-NTD showed no polyubiquitylation because of an inability to bind Roc1 or the E2 enzyme. As a further control, the catalytic activity of Cul3 complexes was also assessed via Cul3 auto-polyubiquitylation (Fig. 5B). As expected, complexes of full-length Cul3 auto-polyubiquitylated to produce high-molecular-weight polyubiquitin chains only in the presence of ubiquitin, whereas the Cul3-NTD complexes were catalytically dead. These data confirm that RhoBTB2 is indeed a direct substrate of a Cul3-based E3 ubiquitin ligase complex.
Figure 5.
RhoBTB2 is a target for a Cul3 ubiquitin ligase complex in vitro. 293T cells were transfected with GST–Cul3, Myc-RhoBTB, and Roc1 or with appropriate controls. GST–Cul3 complexes were subsequently isolated with GSH-agarose and subject to in vitro ubquitylation assay (see Materials and Methods). (A) After assay, the reactions were resolved by SDS-PAGE and probed for RhoBTB2. (B) The blot was then stripped and reprobed for Cul3. (C) The constituents of each Cul3 complex were confirmed by Western blotting for Cul3, Roc1, and RhoBTB2. In A and B, the only components of assay that were varied are indicated. In C, the transfected constructs are indicated.
Our results suggest several models for RhoBTB2 function. One possibility is that RhoBTB2 is simply a substrate of a Cul3 ubiquitin ligase complex with the first BTB domain serving as the targeting signal for degradation. However, disruption of binding to Cul3 prevents RhoBTB2 degradation and leads to increased RhoBTB2 expression (Figs. 3A, 4A). Consequently, this model is difficult to reconcile with the role for RhoBTB2 as a tumor suppressor in which deletion of RhoBTB2 is associated with cancer. A more attractive model that encompasses all the available data is that RhoBTB2 functions as a tumor suppressor by binding and recruiting substrates to a Cul3 ubiquitin ligase complex for degradation. In this model, loss of RhoBTB2 would lead to up-regulation of proteins involved in oncogenesis. The differential binding of RhoBTB2 mutants to Cul3 can also be explained in this is model. The Y284D RhoBTB2 mutant would be loss of function in this context because it fails to bind Cul3 and would therefore not recruit target proteins for ubiquitylation. The other somatic RhoBTB2 mutants, although they bind Cul3, could still be loss of function with respect to substrate binding or ubiquitylation. This model would be entirely consistent with the role of RhoBTB2 as a tumor suppressor and has several precedents supporting it. First, the von Hippel Lindau tumor suppressor is a substrate-recognition element for Cul2 (Pause et al. 1997; Lisztwan et al. 1999; Maxwell et al. 1999; Ohh et al. 2000). Second, F-box proteins are both substrate-recognition modules and themselves substrates of Cul1-based ubiquitin ligase complexes (Zhou and Howley 1998). Finally, the C. elegans BTB domain protein Mel26 acts as a substrate-specific adaptor for Cul3, recruiting the MEI-1 protein for ubiquitylation (Furukawa et al. 2003; Pintard et al. 2003; Xu et al. 2003). We are currently attempting to identify more RhoBTB2-interacting proteins to test whether RhoBTB2 is indeed a substrate-specific adaptor for Cul3.
Materials and methods
Yeast two-hybrid screening
The PJ694A yeast strain and accompanying vector system were used throughout (James et al. 1996). Full-length human RhoBTB2, Cul3A, and PIP5Kα were cloned into the pGBDU-C1 vector. The mouse whole-embryo cDNA library (Vojtek et al. 1993) was a gift from C. Richardson (Harvard Medical School, Boston, MA) and J. Blenis (Harvard Medical School, Boston, MA). Two-hybrid interactions were selected on synthetic defined medium lacking leucine, uracil, and adenine for 3–5 d at 30°C as described in James et al. (1996).
Plasmids
Human RhoBTB2 was amplified by RT–PCR from fetal human mRNA and cloned into the pEBB vector (a gift from B Mayer, University of Connecticut, Storrs, CT) as a Myc, HA, or GST fusion. The Myc-Cullin and HA-Roc vectors were provided by X. Xiong (University of North Carolina at Chapel Hill, NC). Cul3 and RhoBTB2 mutants were generated using the Quikchange mutagenesis kit (Stratagene). To generate retroviral vectors expressing Cul3-specific shRNA, we cloned double-stranded DNA oligonucleotides into pMSCV-U6 exactly as described (Devroe and Silver 2002). The sequence of the sense strand is as follows: GGTGCTCACGACAGGATATttctgaagaATATCCTGTCGTGAGCACCctt (gene-specific sequences are in capitals and hairpin sequences are underlined). A retroviral vector expressing shRNA specific to humanized Renilla GFP was a gift from J. Wade Harper (Harvard Medical School, Boston, MA; Jin et al. 2003). Exact details of all constructs used are available on request.
Antibodies
Anti-Cul3 (Santa Cruz), anti-Nedd8 (Alexis), anti-Myc 9E10 (Clontech), and anti-HA HA11 (Roche) antibodies were used as suggested by the manufacturers. Rabbit polyclonal RhoBTB2 antibodies were raised against a RhoBTB2 GST fusion protein.
Cell culture, transient transfection, and protein interactions
293T, HeLa, and SK-Mes-1 cells were purchased from ATCC and maintained according to the product literature. 293T cells were transfected with Polyfect (Qiagen) for 24 h unless otherwise stated in the text. Cells were lysed with RIPA buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate). Immunoprecipitation was performed by using protein-A sepharose (Pharmacia), GST precipitation by using GSH-agarose (Pharmacia), and Western blotting by using standard procedures.
Retroviral shRNA
Retroviruses expressing shRNA for Cul3 and GFP were packaged in 293T cells by cotransfection with pCL-Ampho (IMGENEX). HeLa and SK-Mes-1 cells were infected twice in 24 h with the appropriate virus, selected for a further 48 h with 2 μg/mL puromycin (except the shRNA GFP virus, which does not confer puromycin resistance), then lysed and subjected to immunoprecipitation by Cul3 and RhoBTB2 antibodies.
Ubiquitylation assay
293T cells were transfected with GST–Cul3, Myc-RhoBTB, and Roc1 or appropriate controls. Cul3 complexes were isolated with GSH-agarose, washed three times in lysis buffer, once in 50 mM HEPES (pH 7.6), then added to a reaction mixture containing 2 μM ubiquitin aldehyde, 10 μM MG132, 5 mM MgCl2, 1 mM ATP, 0.5 mM DTT, 50 μg ubiquitin, 500 ng yeast E1 enzyme, 500 ng E2 enzyme (Ubc5), and 1 mM phenylmethylsuphonylfluoride in a total volume of 50 μL. The reaction was incubated at 37°C for 1 h, then resolved by SDS-PAGE and Western blotted sequentially for RhoBTB2 and Cul3. A small fraction of the Cul3 complexes were saved and Western blotted simultaneously for Cul3, RhoBTB2, and Roc1. All ubiquitin-related reagents were purchased from Boston Biochemicals.
Acknowledgments
This work was supported by NIH grant RO154389 to C.L.C. and a Wellcome Trust International Prize Travelling Fellowship to A.W. We thank Wade Harper and Nikola Pavletich for helpful discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1177904.
References
- Brown, M.R., Chuaqui, R., Vocke, C.D., Berchuck, A., Middleton, L.P., Emmert-Buck, M.R., and Kohn, E.C. 1999. Allelic loss on chromosome arm 8p: Analysis of sporadic epithelial ovarian tumors. Gynecol. Oncol. 74: 98–102. [DOI] [PubMed] [Google Scholar]
- Collins, T., Stone, J.R., and Williams, A.J. 2001. All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 21: 3609–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies, R.J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15: 435–467. [DOI] [PubMed] [Google Scholar]
- Devroe, E. and Silver, P.A. 2002. Retrovirus-delivered siRNA. BMC Biotechnol. 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, M., He, Y.J., Borchers, C., and Xiong, Y. 2003. Targeting of protein ubiquitination by BTB–Cullin 3–Roc1 ubiquitin ligases. Nat. Cell Biol. 5: 1001–1007. [DOI] [PubMed] [Google Scholar]
- Geyer, R., Wee, S., Anderson, S., Yates, J., and Wolf, D.A. 2003. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12: 783–790. [DOI] [PubMed] [Google Scholar]
- Groisman, R., Polanowska, J., Kuraoka, I., Sawada, J., Saijo, M., Drapkin, R., Kisselev, A.F., Tanaka, K., and Nakatani, Y. 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113: 357–367. [DOI] [PubMed] [Google Scholar]
- Hamaguchi, M., Meth, J.L., von Klitzing, C., Wei, W., Esposito, D., Rodgers, L., Walsh, T., Welcsh, P., King, M.C., and Wigler, M.H. 2002. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proc. Natl. Acad. Sci. 99: 13647–13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J., Shirogane, T., Xu, L., Nalepa, G., Qin, J., Elledge, S.J., and Harper, J.W. 2003. SCFβ–TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes & Dev. 17: 3062–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan, J., Imbert, G., Wirbelauer, C., Gstaiger, M., and Krek, W. 1999. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin–protein ligase activity. Genes & Dev. 13: 1822–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, P.H., Wiesener, M.S., Chang, G.W., Clifford, S.C., Vaux, E.C., Cockman, M.E., Wykoff, C.C., Pugh, C.W., Maher, E.R., and Ratcliffe, P.J. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399: 271–275. [DOI] [PubMed] [Google Scholar]
- Ohh, M., Park, C.W., Ivan, M., Hoffman, M.A., Kim, T.Y., Huang, L.E., Pavletich, N., Chau, V., and Kaelin, W.G. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the β-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2: 423–427. [DOI] [PubMed] [Google Scholar]
- Pause, A., Lee, S., Worrell, R.A., Chen, D.Y., Burgess, W.H., Linehan, W.M., and Klausner, R.D. 1997. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl. Acad. Sci. 94: 2156–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart, C.M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70: 503–533. [DOI] [PubMed] [Google Scholar]
- Pintard, L., Willis, J.H., Willems, A., Johnson, J.L., Srayko, M., Kurz, T., Glaser, S., Mains, P.E., Tyers, M., Bowerman, B., et al. 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425: 311–316. [DOI] [PubMed] [Google Scholar]
- Ramos, S., Khademi, F., Somesh, B.P., and Rivero, F. 2002. Genomic organization and expression profile of the small GTPases of the RhoBTB family in human and mouse. Gene 298: 147–157. [DOI] [PubMed] [Google Scholar]
- Schulman, B.A., Carrano, A.C., Jeffrey, P.D., Bowen, Z., Kinnucan, E.R., Finnin, M.S., Elledge, S.J., Harper, J.W., Pagano, M., and Pavletich, N.P. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1–Skp2 complex. Nature 408: 381–386. [DOI] [PubMed] [Google Scholar]
- Vojtek, A.B., Hollenberg, S.M., and Cooper, J.A. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74: 205–214. [DOI] [PubMed] [Google Scholar]
- Wistuba, I.I., Behrens, C., Virmani, A.K., Milchgrub, S., Syed, S., Lam, S., Mackay, B., Minna, J.D., and Gazdar, A.F. 1999. Allelic losses at chromosome 8p21–23 are early and frequent events in the pathogenesis of lung cancer. Cancer Res. 59: 1973–1979. [PubMed] [Google Scholar]
- Xu, L., Wei, Y., Reboul, J., Vaglio, P., Shin, T.H., Vidal, M., Elledge, S.J., and Harper, J.W. 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425: 316–321. [DOI] [PubMed] [Google Scholar]
- Zheng, N., Schulman, B.A., Song, L., Miller, J.J., Jeffrey, P.D., Wang, P., Chu, C., Koepp, D.M., Elledge, S.J., Pagano, M., et al. 2002. Structure of the Cul1–Rbx1–Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416: 703–709. [DOI] [PubMed] [Google Scholar]
- Zhou, P. and Howley, P.M. 1998. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin–protein ligases. Mol. Cell 2: 571–580. [DOI] [PubMed] [Google Scholar]