The authors assessed the effects of single nucleotide polymorphisms (SNPs) in microRNA-related genes on the survival of 220 patients with T-cell lymphoma (TCL). Results showed that 4 of 13 SNPs were significantly associated with TCL survival, suggesting that microRNA-related polymorphisms may be used individually and jointly to predict survival of patients with TCL.
Keywords: MicroRNAs, Polymorphisms, T-cell lymphoma, Survival
Abstract
Objective
Elaborate evaluation of prognosis of T-cell lymphoma (TCL) is vital for current therapy and future stratified and individualized therapy. MicroRNAs (miRNAs) play important roles in cancer development and prognosis. We aimed to assess the effects of single nucleotide polymorphisms (SNPs) in miRNA-related genes on the survival of patients with TCL.
Patients and Methods
We genotyped 13 SNPs selected from 12 miRNA-related genes in 220 TCL patients and explored the association of SNPs with survival.
Results
Among the 13 SNPs, four (DROSHA rs6877842, DICER rs3742330, mir149 rs2292832, and mir499 rs3746444) were significantly associated with TCL survival after adjusting for subtype and International Prognostic Index score. In stratified analyses, all four SNPs remained significantly associated with survival in patients with mature T type. Of the four SNPs, only mir149 rs2292832 was not significantly associated with survival in patients with an International Prognostic Index score of 0–1. Furthermore, a dose-dependent cumulative effect of the four SNPs on TCL survival was observed by counting the number of unfavorable genotypes. Survival tree analysis also showed higher order interactions between these SNPs.
Conclusion
The results suggested that miRNA-related polymorphisms are associated with survival of TCL patients; thus, they may be used individually and jointly to predict survival of patients with TCL.
Implications for Practice:
Besides the characteristics of a tumor itself, genetic polymorphisms in relevant genes are considered to be important in influencing the clinical outcomes of patients. Previous studies have suggested that microRNAs are associated with survival of patients with T-cell lymphoma (TCL); therefore, we performed association analyses of 13 carefully selected polymorphisms in microRNA-related genes. The results suggested that four polymorphisms in microRNA-related genes are associated with survival of patients with TCL. These polymorphisms may be used to predict the survival of patients with TCL individually or collectively. Although the results are exploratory, they provide clues to warrant further investigation.
Introduction
T-cell non-Hodgkin lymphomas (TCLs) are uncommon malignancies accounting for approximately 12% of all lymphomas [1]. They are composed of a heterogeneous group of diseases and usually have poor prognosis [2, 3]. Currently, the International Prognostic Index (IPI) is widely used to predict prognosis of TCL. The survival probability decreases as the IPI score increases, and patients are divided into low-risk (IPI score 0–1), intermediate-risk (IPI score 2–3), and high-risk (IPI score 4–5) groups according to their survival prospects [2]. In addition, histological subtype is strongly associated with survival of TCL patients [3]. However, these parameters are far from satisfactory for guiding stratified or individualized therapy. It is obvious that complementary prognostic indices for TCL using pretherapeutic characteristics are urgently needed.
MicroRNAs (miRNAs) are a class of endogenous, small, noncoding RNA molecules that participate in diverse biological processes by regulating gene expression. miRNAs are generated by a two-step pathway. They are first transcribed by RNA polymerase II into primary miRNAs (pri-miRNAs). These pri-miRNA transcripts are processed in the nucleus by DROSHA RNase to produce the precursor miRNAs (pre-miRNAs) [4, 5]. The pre-miRNAs are then translocated to the cytoplasm where the pre-miRNAs are cleaved by DICER, leading to the production of mature miRNAs [6]. The mature miRNAs cooperate with the RNA-induced silencing complex comprising GEMIN3, GEMIN4, TRBP, AGO1, and AGO2, resulting in messenger RNA cleavage or translational repression [7]. Several recent studies have demonstrated that aberrations of key genes in the miRNA biogenesis pathway can modify the response of TCL cells to chemotherapy and then affect the survival of patients with TCL [8–10].
Single nucleotide polymorphisms (SNPs) have been shown to be associated with the survival of patients with TCL [11, 12]. However, data are sparse for miRNA biogenesis pathway genes and miRNA genes. Recent studies have suggested that SNPs in these genes can affect gene functions and miRNA expression [13] and, thus, are associated with survival of cancer patients. To provide potential new biomarkers for predicting prognosis of TCL, we therefore evaluated the effects of 13 SNPs in miRNA biogenesis pathway genes and miRNA genes on the survival of patients with TCL individually and jointly.
Materials and Methods
Study Patients
TCL patients (n = 220) were recruited from Chongqing Southwest Hospital and Beijing Cancer Hospital between January 1992 and September 2009. One hundred fifty-eight of these patients were the subject of a previous report [14]. The diagnosis of TCL was based on pathological examination and clinical manifestations. All patients were non-blood-related Han Chinese and were mainly treated with a CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone)-based regimen as the first-line chemotherapy. Here, we classified TCL as natural killer/T-cell lymphoma, peripheral T-cell lymphoma not otherwise specified, T-lymphoblastic lymphoma/leukemia, angioimmunoblastic T-cell lymphoma, subcutaneous panniculitis-like T-cell lymphoma, mycosis fungoides, enteropathy-type T-cell lymphoma, and Lennert lymphoma. The stage of tumor was assessed according to the Ann Arbor system [15]. At recruitment, demographic information and clinical characteristics were collected from each patient via clinical record. Data on whether and when a patient had died were obtained from inpatient and outpatient records, patient or family contact, or local Public Security Census Register Office. All patients provided written informed consent, and the study was approved by the Institutional Review Boards of both the Southwest Hospital of Third Military Medical University and Chinese Academy of Medical Sciences Cancer Institute.
Gene/SNP Selection
We selected 13 SNPs in miRNA biogenesis pathway genes and miRNA genes according to the following criteria: (a) they have a reported minor allele frequency >0.01 in the Chinese population; (b) they reside in functional regions, including exons, untranslated regions, and promoters; (c) for SNPs in miRNA biogenesis pathway genes, they must have been reported to be significantly associated with the survival of cancer; and (d) for SNPs in pre-miRNA and pri-miRNA, their mature counterparts must have been reported to be implicated in cancer etiology or prognosis (supplemental online Table 1).
Genotyping
Genomic DNA was extracted from peripheral blood samples collected at the time of diagnosis with the RelaxGene Blood DNA System according to the manufacturer’s protocol (Tiangen Biotech, Beijing, People’s Republic of China, http://www.tiangen.com) and stored at −80°C until used. All polymorphisms were genotyped by the polymerase chain reaction (PCR)-restriction fragment length polymorphism method. The PCR primers used to amplify DNA fragments and restriction enzymes applied to digest the PCR products for each SNP are summarized in supplemental online Table 2. The digested products were analyzed using 3% agarose gel electrophoresis.
Statistical Analysis
The endpoint of the study was overall survival (OS), defined as the time from the date of TCL diagnosis to the date of death by any cause or last follow-up. Allelic distributions of all SNPs were tested for Hardy-Weinberg equilibrium. The Cox proportional hazard model was used to assess the association of SNPs with OS after the assumption of proportionality was met. Hazard ratios (HRs) with 95% confidence intervals (CIs) on the risk of death were estimated adjusting for covariates that might influence survival, including subtype and IPI score. Three different genetic models, including dominant model (comparing homozygous wild-type genotype with variant allele-carrying genotypes), recessive model (comparing wild-type allele-carrying genotypes with homozygous variant genotype), and additive model (p for trend), were tested and the model that yielded the smallest p value was considered the best-fitting model and was ultimately applied to the analysis [16]. The Kaplan-Meier method was used to plot the OS curves and the log-rank test was used to compare survival difference between the genotypes. Stratified analyses by TCL subtype (precursor T type vs. mature T type) and IPI score (0–1 vs. 2–3 vs. 4–5) were performed to investigate the association of miRNA-related SNPs with OS in the subsets of TCL patients.
The cumulative effects of SNPs significantly associated (p < .05) with TCL survival were assessed by counting the numbers of unfavorable genotypes in each subject and were analyzed using the multivariate Cox proportional hazard model adjusting for subtype and IPI score after the proportionality assumption was satisfied. We performed survival tree analyses with the recursive partitioning method to build a decision tree using the STREE program (http://c2s2.yale.edu/software/stree) [17]. The root node of the survival tree included all 220 patients. The log-rank test statistic was used as node-splitting criteria. The recursive procedure continues to produce offspring nodes until no further statistically significant split is obtained. The resulting tree was binary, and each terminal node represented a group of patients with different survival outcomes depending on different genotype combinations. HRs and their 95% CIs for each terminal node were calculated using the multivariate Cox proportional hazard model adjusting for subtype and IPI score after the proportionality assumption was satisfied.
Statistical analyses were carried out using SPSS version 20 (SPSS, Chicago, IL). All tests were two-sided, with a significance level set at p < .05.
Results
Patient Characteristics
Detailed characteristics of the 220 patients are listed in Table 1. The median age at the time of diagnosis was 37 years (range = 6–80), and 151 (68.6%) patients were men. The most common subtype of TCL was natural killer/T-cell lymphoma (25.0%), followed by peripheral T-cell lymphoma not otherwise specified (22.3%), T-lymphoblastic lymphoma/leukemia (20.5%), anaplastic large cell lymphoma (17.7%), angioimmunoblastic T-cell lymphoma (6.8%), subcutaneous panniculitis-like T-cell lymphoma (4.5%), mycosis fungoides (1.4%), enteropathy-type intestinal T-cell lymphoma (0.9%), and Lennert lymphoma (0.9%). One hundred seventeen (54.4%) patients had IPI scores of 0–1, 87 (40.5%) had IPI scores of 2–3, and 11 (5.1%) had IPI scores of 4–5. By the time of final analysis on July 1, 2013, 89 (40.5%) patients had died, resulting in a 5-year OS rate of 43.0%.
Table 1.
Patient characteristics and their association with overall survival
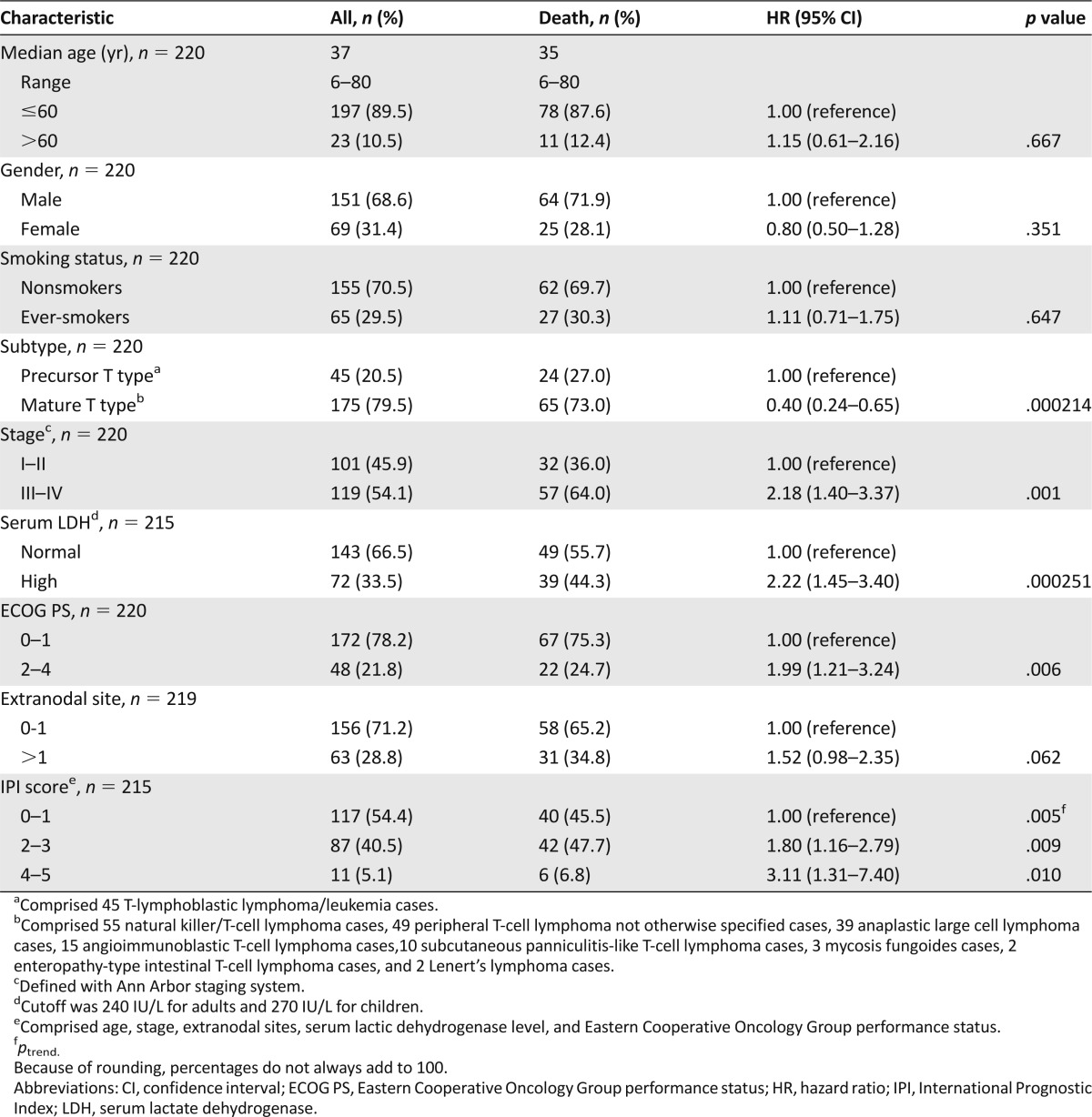
Overall Survival by Clinical Characteristics of TCL Patients
We first investigated whether various clinical characteristics had contributed to OS. We stratified patients by age (≤60 years or >60 years), gender, smoking status (nonsmokers or ever-smokers), subtype (precursor T type or mature T type), stage (I–II or III–IV), lactic dehydrogenase (LDH) level (normal or high), Eastern Cooperative Oncology Group (ECOG) performance status (0–1 or 2–4), extranodal site (0–1 or >1), and IPI score (0–1 or 2–3 or 4–5) and compared OS between groups using the univariate Cox proportional hazard model (Table 1). We found that subtype, individual item of IPI score (including stage, LDH level, and ECOG performance status), and total IPI score significantly affected the survival of patients. The 5-year OS rate was 24.7% for patients with precursor T type and 48.6% for those with mature T type (p = .000214). The 5-year OS rates for patients with IPI scores of 0–1, 2–3, and 4–5 were 48.3%, 36.5%, and 16.7%, respectively (p = .003).
Association Between Individual SNPs and TCL Overall Survival
The genotype distributions of the SNPs were in accordance with Hardy-Weinberg equilibrium (Table 2), except for mir196a 2 rs11614913, which was therefore excluded from further analyses.
Table 2.
Association of individual single nucleotide polymorphisms (SNPs) with overall survival of T-cell lymphoma patients
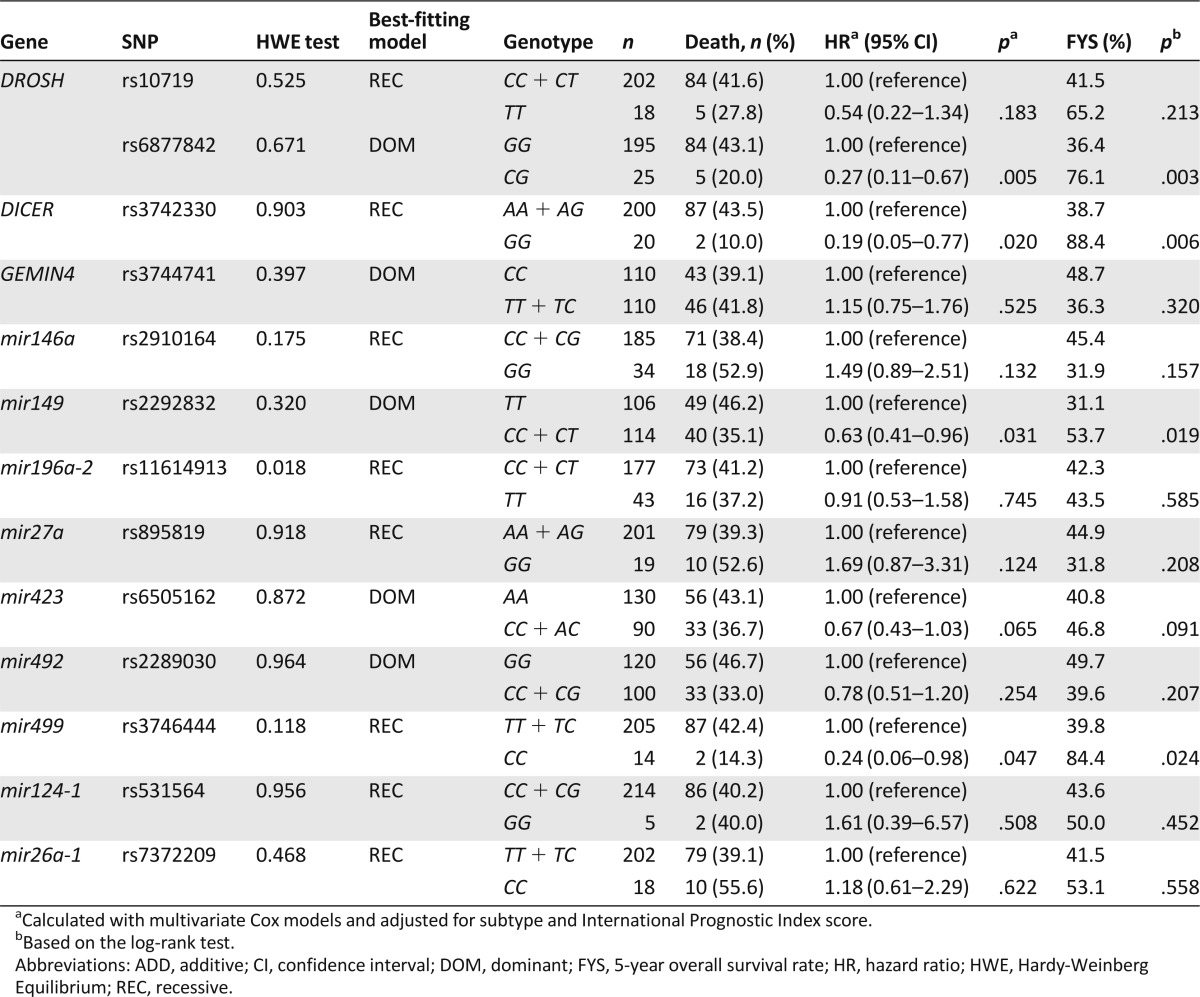
Overall, four SNPs showed a significant association with OS of patients with TCL using the multivariate Cox proportional hazard model after adjustment for subtype and IPI score (Table 2). Among these four SNPs, the dominant model best fit the data for DROSHA rs6877842 and mir149 rs2292832; the recessive model best fit the data for DICER rs3742330 and mir499 rs3746444. Patients with the variant allele-carrying genotypes of DROSHA rs6877842 and mir149 rs2292832 had a significantly increased OS compared with those carrying the homozygous wild-type genotype (HR, 0.27; 95% CI, 0.11–0.67; p = .005; and HR, 0.63; 95% CI, 0.41–0.96; p = .031, respectively). The same was true for the patients with the homozygous variant genotype of DICER rs3742330 and mir499 rs3746444 compared with those carrying the wild-type allele-carrying genotypes (HR, 0.19; 95% CI, 0.05–0.77; p = .020; and HR, 0.24; 95% CI, 0.06–0.98; p = .047, respectively).
To investigate the effects of the four SNPs on OS in subsets of patients with TCL, we performed stratified analyses based on TCL subtype (precursor T type vs. mature T type) and IPI score (0–1 vs. 2–3 vs. 4–5). The variant allele-carrying genotypes of DROSHA rs6877842 and mir149 rs2292832 and the homozygous variant genotype of DICER rs3742330 and mir499 rs3746444 were also associated with improved OS in patients with mature T type but not in patients with precursor T type (supplemental online Table 3). In a stratified analysis by IPI score, we found that the association with better OS remained significant for DROSHA rs6877842, DICER rs4742330, and mir499 rs3746444 in patients with an IPI score of 0–1 but not in patients with IPI scores of 2–3 and 4–5 (supplemental online Table 4).
Cumulative Effects of the Unfavorable Genotypes on Overall Survival
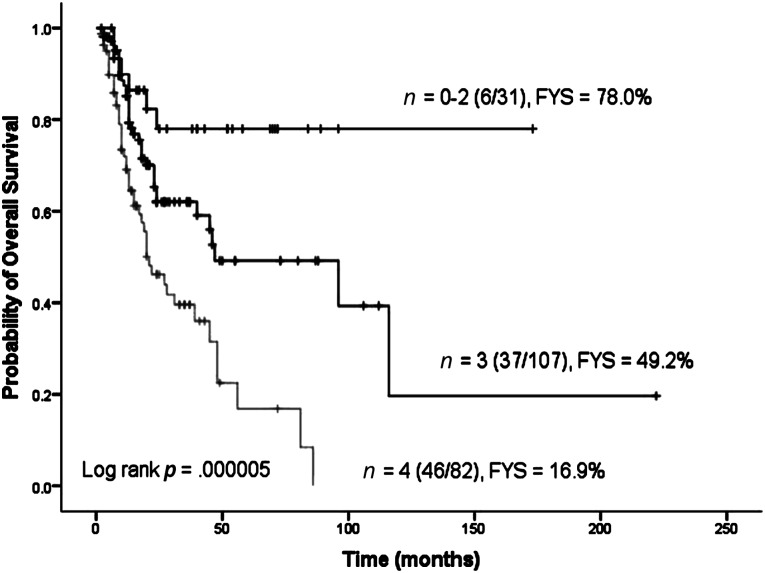
We further evaluated the cumulative effects of SNPs on TCL survival by counting the unfavorable genotypes of the four SNPs identified. We found that compared with patients with zero, one, or two unfavorable genotypes, those with three unfavorable genotypes were at a 2.53-fold (95% CI, 1.06–6.00; p = .036) increased risk of death, and the risk further increased to 5.31-fold (95% CI, 2.24–12.57; p = .00015) for those with all four unfavorable genotypes (ptrend = .00004) (Table 3). The 5-year OS rates were 78.0%, 49.2%, and 16.9% for the above three groups of patients, respectively (p = .000005) (Fig. 1).
Table 3.
Cumulative effects of unfavorable genotypes on T-cell lymphoma survival
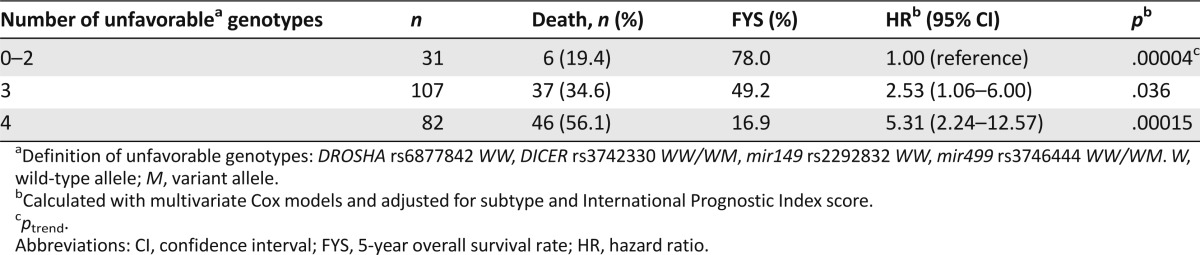
Figure 1.
Overall survival of patients with T-cell lymphoma according to different numbers of unfavorable genotypes. Abbreviations: FYS, 5-year overall survival rate; n, number of unfavorable genotypes.
Survival Tree Analysis
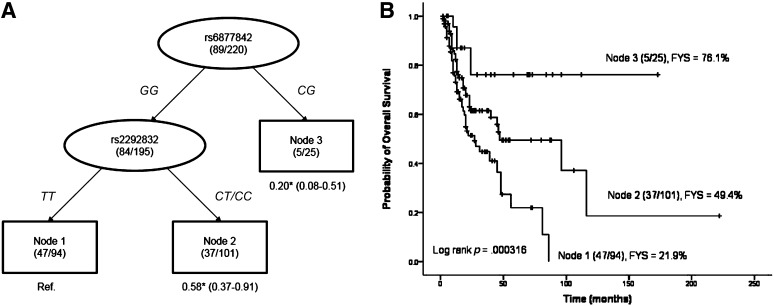
To explore potential high-order interactions between SNPs and to define subgroups that have distinct survival prospects, we performed a survival tree analysis using the four SNPs identified in the individual SNP analysis. The tree structure resulted in three terminal nodes. The top splitting factor was DROSHA rs6877842, followed by mir149 rs2292832 (Fig. 2A). Taking terminal node 1 (patients with the homozygous wild-type genotypes of both rs6877842 and rs2292832) as the reference group, the HR for terminal node 2 (patients with the homozygous wild-type genotype of rs6877842 and at least one variant allele of rs2292832) was 0.58 (95% CI, 0.37–0.91; p = .017), and the HR for terminal node 3 (patients with the heterozygous genotype of rs6877842) was 0.20 (95% CI, 0.08–0.51; p = .001) (Fig. 2A). The 5-year OS rates were 21.9% for patients in terminal node 1, 49.4% for patients in terminal node 2, and 76.1% for patients in terminal node 3 (p = .000316) (Fig. 2B).
Figure 2.
Survivability defined by high-order interactions between SNPs. (A): Survival tree analysis of survival of patients with T-cell lymphoma. (B): Overall survival according to different nodes identified by survival tree analysis. The number under each node represents the hazard ratio with 95% confidence interval in parenthesis. *, p < .05. Abbreviation: C, cytosine; FYS, 5-year overall survival rate; G, guanine; T, thymine.
Discussion
To apply individualized therapy for TCL, we need to predict the prognosis of each patient according to his/her pretherapeutic features [18]. It is increasingly suggested that SNPs, the most common genetic variants in humans, as a whole are key enablers [19]. MiRNA-related genes have been reported to be associated with biological features of T-cells and clinical outcomes of TCL [10, 20], but whether SNPs in these genes are associated with survival of TCL is still unclear.
Therefore, we carefully selected 13 SNPs in miRNA-related genes based on prespecified criteria and evaluated their roles in the prognosis of patients with TCL. We found that DROSHA rs6877842, DICER rs3742330, mir149 rs2292832, and mir499 rs3746444 were associated with the survival of patients. Furthermore, we observed a dose-dependent cumulative effect of these SNPs on the survival of patients and a higher order interaction between these SNPs.
For individual SNP analysis, DROSHA rs6877842 showed the most statistically significant association with TCL survival. DROSHA initiates the miRNA biogenesis in the nucleus [21]. Previous studies have demonstrated that altered expression of DROSHA may alter the miRNA expression profile associated with survival of cancer patients [22, 23]. SNP rs6877842 is located in the promoter of DROSHA and may affect the expression of DROSHA through affecting the binding of transcription factor to this region; hence, it may be associated with the survival of patients with TCL.
Besides the newly identified significant SNP rs6877842 in DROSHA, we also confirmed the previously reported association of DICER rs3742330 with survival of patients with TCL [14] after more patients were tested and the follow-up period was extended. DICER is essential for the production of mature miRNAs through cleaving the double-strand pre-miRNA, and silencing of DICER in cells can reduce pre-miRNA and mature miRNA sequences [24]. SNP rs3742330 is located in the 3′-UTR, which may affect the expression of DICER through changing the binding capacity of regulatory miRNAs [25, 26], thus affecting the survival rate of TCL.
SNPs in the pri- or pre-miRNA regions were also evaluated, among which mir149 rs2292832 and mir499 rs3746444 were demonstrated to be significantly associated with the survival of patients with TCL. Mir149 rs2292832 and mir499 rs3746444 have been previously reported to be significantly associated with the survival of patients with other cancers [27–30]. The predicted target protein TP63 of mir149 has been reported to be associated with the survival of patients with TCL [31]. The potential targeted protein NOTCH1 of mir499 was a biomarker for predicting the survival of TCL patients [32]. These analyses suggested that it is highly likely that the miRNAs identified in this study are actively involved in affecting the survival of patients with TCL.
Histological subtype and IPI score are major clinical characteristics that are associated with the survival of patients with TCL as demonstrated in this and previous studies [2, 33]. We performed stratified analyses according to subtype or IPI score to investigate the role of identified SNPs in different subsets of patients. When the patients were divided by subtype, the four SNPs remained significantly associated with survival in patients with mature T type, but not in patients with precursor T type. When patients were divided by IPI score, three SNPs remained significantly associated with survival in patients with IPI scores of 0–1, but not in patients with IPI scores of 2–3 or 4–5. This discordance between different subsets of patients may be due to the limited number of patients in the study. Another possible explanation is that the poor prognostic role of unfavorable genotypes is inferior to that of subtype or IPI score; thus, the effect of these SNPs can be prominent only in patients with better potential prognosis.
Prognosis of patients is usually predicted by combinations of multiple factors. In order to explore the combined effects of these miRNA-related SNPs on the survival of TCL, we performed cumulative effect analysis by counting the number of unfavorable genotypes. Using the Cox proportional hazard model, we identified a trend of poor survival with an increasing number of unfavorable genotypes in a dose-dependent manner.
Cox proportional hazard models are used to investigate the impact of certain factors on prognosis, and the survival tree method is to define subgroups that have distinct survival prospects. The graphical output of the latter facilitates the visualization of prognostic groups, reflecting multimarker interactions. This method is usually used in a complementary fashion with the Cox proportional model [34]. In our survival tree analysis, we found a higher order interaction between these SNPs and identified that the subgroup of patients with the homozygous wild-type genotypes of both rs6877842 and rs2292832 had the lowest survival probability, whereas those with the heterozygous genotype of rs6877842 had the highest probability of survival. These results suggested that the clinical progression or remission of TCL may be a polygenic process and the pathway-based multiple gene approach has high predictive power.
Conclusion
miRNA-related SNPs DROSHA rs6877842, DICER rs3742330, mir149 rs2292832, and mir499 rs3746444 are associated with the survival of TCL patients and can be used to predict the prognosis of TCL individually and collectively. Nonetheless, these results are to be validated in larger cohorts of patients, and the biological functions of these SNPs remain to be explored.
See http://www.TheOncologist.com for supplemental material available online.
Supplementary Material
Acknowledgments
We thank Dr. Weihuang Zhang for polishing this manuscript and Dr. Yanqi Zhang for statistical assistance.
This work was supported by the National Natural Science Fund of China (30971066, 81270605), Chongqing Natural Science Fund Project in China (2008BA5001), Third Military Medical University Clinic and Science Great Fund Project of China (2012XLC03), and the Military Emphasis Medical Scientific Research Project Fund of China.
Author Contributions
Conception/Design: Jieping Chen, Xi Li, Xiaobo Tian
Provision of study material or patients: Jieping Chen, Xiaobo Tian, Bo Zhang
Collection and/or assembly of data: Jieping Chen, Xi Li, Xiaobo Tian
Data analysis and interpretation: Jieping Chen, Xi Li
Manuscript writing: Jieping Chen, Xi Li
Final approval of manuscript: Jieping Chen, Xi Li, Xiaobo Tian, Bo Zhang
Disclosures
The authors indicated no financial relationships.
References
- 1.Rizvi MA, Evens AM, Tallman MS, et al. T-cell non-Hodgkin lymphoma. Blood. 2006;107:1255–1264. doi: 10.1182/blood-2005-03-1306. [DOI] [PubMed] [Google Scholar]
- 2.The International Non-Hodgkin’s Lymphoma Prognostic Factors Project. A predictive model for aggressive non-Hodgkin’s lymphoma. N Engl J Med. 1993;329:987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 3.Jaffe ES, Harris NL, Stein H, et al. World Health Organization classification of tumours. Pathology and genetics of tumours of hematopoietic and lymphoid tissues. Lyon, France: IARC Press; 2001. [Google Scholar]
- 4.Han J, Lee Y, Yeom KH, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 5.Denli AM, Tops BB, Plasterk RH, et al. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 6.Lee YS, Nakahara K, Pham JW, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 7.Redfern AD, Colley SM, Beveridge DJ, et al. RNA-induced silencing complex (RISC) proteins PACT, TRBP, and Dicer are SRA binding nuclear receptor coregulators. Proc Natl Acad Sci USA. 2013;110:6536–6541. doi: 10.1073/pnas.1301620110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paik JH, Jang JY, Jeon YK, et al. MicroRNA-146a downregulates NFkB activity via targeting TRAF6 and functions as a tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin Cancer Res. 2011;17:4761–4771. doi: 10.1158/1078-0432.CCR-11-0494. [DOI] [PubMed] [Google Scholar]
- 9.Jardin F, Figeac M. MicroRNAs in lymphoma, from diagnosis to targeted therapy. Curr Opin Oncol. 2013;25:480–486. doi: 10.1097/CCO.0b013e328363def2. [DOI] [PubMed] [Google Scholar]
- 10.Valencak J, Schmid K, Trautinger F, et al. High expression of Dicer reveals a negative prognostic influence in certain subtypes of primary cutaneous T cell lymphomas. J Dermatol Sci. 2011;64:185–190. doi: 10.1016/j.jdermsci.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Farre L, Bittencourt AL, Silva-Santos G, et al. Fas-670 promoter polymorphism is associated to susceptibility, clinical presentation, and survival in adult T cell leukemia. J Leukoc Biol. 2008;83:220–222. doi: 10.1189/jlb.0407198. [DOI] [PubMed] [Google Scholar]
- 12.Lee JJ, Kim DH, Lee NY, et al. Interleukin-10 gene polymorphism influences the prognosis of T-cell non-Hodgkin lymphomas. Br J Haematol. 2007;137:329–336. doi: 10.1111/j.1365-2141.2007.06570.x. [DOI] [PubMed] [Google Scholar]
- 13.Iwai N, Naraba H. Polymorphisms in human pre-miRNAs. Biochem Biophys Res Commun. 2005;331:1439–1444. doi: 10.1016/j.bbrc.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Tian X, Zhang B, et al. Variation in Dicer gene is associated with increased survival in T-cell lymphoma. PLoS One. 2012;7:e51640. doi: 10.1371/journal.pone.0051640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carbone PP, Kaplan HS, Musshoff K, et al. Report of the Committee on Hodgkin’s Disease Staging Classification. Cancer Res. 1971;31:1860–1861. [PubMed] [Google Scholar]
- 16.Ye Y, Wang KK, Gu J, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila) 2008;1:460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang HP, Singer BH. Recursive Partitioning and Applications. New York, NY: Springer; 2010. [Google Scholar]
- 18.Ramsdale E, van Besien K, Smith SM. Personalized treatment of lymphoma: Promise and reality. Semin Oncol. 2011;38:225–235. doi: 10.1053/j.seminoncol.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Maciejewski JP, Mufti GJ. Whole genome scanning as a cytogenetic tool in hematologic malignancies. Blood. 2008;112:965–974. doi: 10.1182/blood-2008-02-130435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muljo SA, Ansel KM, Kanellopoulou C, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 22.Lin RJ, Lin YC, Chen J, et al. MicroRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010;70:7841–7850. doi: 10.1158/0008-5472.CAN-10-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 24.Macrae IJ, Zhou K, Li F, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 25.Persson H, Kvist A, Rego N, et al. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res. 2011;71:78–86. doi: 10.1158/0008-5472.CAN-10-1869. [DOI] [PubMed] [Google Scholar]
- 26.Friedländer MR, Mackowiak SD, Li N, et al. MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan X, Sturgis EM, Song X, et al. Pre-microRNA variants predict HPV16-positive tumors and survival in patients with squamous cell carcinoma of the oropharynx. Cancer Lett. 2013;330:233–240. doi: 10.1016/j.canlet.2012.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong MJ, Choi YY, Jang JA, et al. Association between genetic variants in pre-microRNAs and survival of early-stage NSCLC. J Thorac Oncol. 2013;8:703–710. doi: 10.1097/JTO.0b013e318288dc0a. [DOI] [PubMed] [Google Scholar]
- 29.Tu HF, Liu CJ, Chang CL, et al. The association between genetic polymorphism and the processing efficiency of miR-149 affects the prognosis of patients with head and neck squamous cell carcinoma. PLoS One. 2012;7:e51606. doi: 10.1371/journal.pone.0051606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn DH, Rah H, Choi YK, et al. Association of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the Korean population. Mol Carcinog. 2013;52(suppl 1):E39–51. doi: 10.1002/mc.21962. [DOI] [PubMed] [Google Scholar]
- 31.Vasmatzis G, Johnson SH, Knudson RA, et al. Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas. Blood. 2012;120:2280–2289. doi: 10.1182/blood-2012-03-419937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Callens C, Baleydier F, Lengline E, et al. Clinical impact of NOTCH1 and/or FBXW7 mutations, FLASH deletion, and TCR status in pediatric T-cell lymphoblastic lymphoma. J Clin Oncol. 2012;30:1966–1973. doi: 10.1200/JCO.2011.39.7661. [DOI] [PubMed] [Google Scholar]
- 33.Cronin-Fenton DP, Sharp L, Deady S, et al. Treatment and survival for non-Hodgkin’s lymphoma: Influence of histological subtype, age, and other factors in a population-based study (1999–2001) Eur J Cancer. 2006;42:2786–2793. doi: 10.1016/j.ejca.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 34.Segal MR, Bloch DA. A comparison of estimated proportional hazards models and regression trees. Stat Med. 1989;8:539–550. doi: 10.1002/sim.4780080503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.