Abstract
Liver tumor cells arise from normal hepatocytes that escape negative control of proliferation. The transcription factor C/EBPα maintains quiescence of hepatocytes through two pathways: inhibition of cdks and repression of E2F. Nevertheless, liver tumors and cultured hepatoma cell lines proliferate in the presence of C/EBPα. In this paper, we present evidence that the activation of the PI3K/Akt pathway in liver tumor cells blocks the growth inhibitory activity of C/EBPα through the PP2A-mediated dephosphorylation of C/EBPα on Ser 193, leading to a failure of C/EBPα to interact with and inhibit cdks and E2F. Mutation of Ser 193 to Ala also abolishes the ability of C/EBPα to cause growth arrest because of a lack of interactions with cdk2 and E2F–Rb complexes. These data provide a molecular basis for the development of liver tumors in which the activation of PI3K/Akt pathway neutralizes C/EBPα growth inhibitory activity.
Keywords: Hepatoma, C/EBPα, cell proliferation, Akt, insulin
Maintenance of the growth-arrested state is a crucial step for normal functions of many tissues. The loss of negative control of proliferation can promote tumorigenesis. The best example of tumor development due to loss of inhibitory functions is the identification of tumors containing mutations within the retinoblastoma protein, Rb (Zheng and Lee 2002; Sage et al. 2003). In the liver and in myeloid cells, CCAAT/enhancer binding protein alpha (C/EBPα) is a key inhibitor of cell proliferation. A number of studies have demonstrated that liver quiescence is mediated by growth inhibitory activity of C/EBPα. C/EBPα knockout livers have an increased rate of proliferation and express high levels of cell cycle proteins (Timchenko et al. 1999). Examination of cultured hepatocytes derived from C/EBPα knockout mice also showed that the lack of C/EBPα leads to increased proliferation (Soriano et al. 1998). A detailed analysis of the mechanisms by which C/EBPα inhibits liver growth revealed that protein–protein interactions are the major pathway of C/EBPα-mediated growth arrest in liver (Timchenko et al. 1999; Wang et al. 2001; Iakova et al. 2003). C/EBPα inhibits the proliferation of myeloid cells through mechanisms that differ from those operating in liver and involve the transcriptional activity of C/EBPα (Porse et al. 2001; Keeshan et al. 2003; Wang et al. 2003). Although mutations within the DNA binding region of C/EBPα cause the development of leukemia, no liver tumors were associated with mutations within the DNA binding region of C/EBPα (Pabst et al. 2001). The lack of proliferation abnormalities in the liver of patients with mutations/deletions within the DNA binding region of C/EBPα is consistent with findings showing that, in liver, C/EBPα inhibits proliferation through protein–protein interactions and that this inhibition does not require the transcriptional activity of C/EBPα (Wang et al. 2001; Iakova et al. 2003).
C/EBPα interacts with a number of cell cycle proteins and potentially might inhibit cell proliferation via different pathways, including inhibition of cdk2 and cdk4, upregulation of p21, and repression of E2F transcription (Timchenko et al. 1996; Johansen et al. 2001; Porse et al. 2001; Wang et al. 2001; Iakova et al. 2003). Given these multiple pathways, one can suggest that the growth inhibitory activity of C/EBPα is tightly regulated in tissues during prenatal development and in adults. Despite the fact that significant progress in the identification of downstream targets of C/EBPα has been made, very little is known about molecular mechanisms that regulate the ability of C/EBPα to cause growth arrest. A growing number of observations suggest that posttranslational modifications of C/EBPα or interactions of C/EBPα with other proteins influence its growth inhibitory activity. Several publications suggested that biological activities of C/EBPα might be controlled on the level of posttranslational modifications (Hemati et al. 1997; Ross et al. 1999). These studies showed that insulin signaling leads to a dephosphorylation C/EBPα via the PI3K/Akt pathway, suggesting that C/EBPα is a downstream target of this pathway. A recent paper with Akt1/Akt2 double knockout animals showed that the ablation of these kinases reduces expression and activities of C/EBPα, which correlates with impaired adipogenesis in these animals (Peng et al. 2003).
Here we present evidence that the growth inhibitory activity of C/EBPα is blocked in liver tumors and in cultured hepatoma cells by activation of the PI3K/Akt pathway. We found that the activation of PI3K/Akt pathway leads to the nuclear accumulation of PP2A, which dephosphorylates mouse C/EBPα on Ser 193, leading to a failure of C/EBPα to cause growth arrest. Phosphorylation of C/EBPα on Ser 193 is required for the interactions of C/EBPα with cdks and with E2F–Rb–Brm complexes and for the inhibition of these targets. Both mutation of Ser 193 and dephosphorylation of Ser 193 block the growth inhibitory activity of C/EBPα. Our data provide a molecular basis for the development of liver tumors: neutralization of C/EBPα growth inhibitory activity by the PI3K/Akt pathway.
Results
Human liver tumors and hepatoma cells are resistant to C/EBPα growth arrest
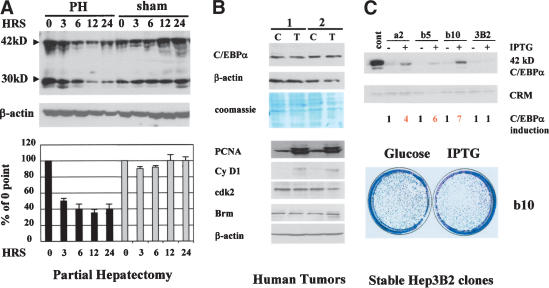
C/EBPα inhibits liver growth through multiple pathways, suggesting that liver should not proliferate in the presence of C/EBPα. However, under several biological situations, liver proliferates and expresses high levels of C/EBPα. As can be seen in Figure 1A, protein levels of C/EBPα remain high in liver proliferating after partial hepatectomy (PH). Surprisingly, we found that liver tumors also proliferate in the presence of C/EBPα. Western blotting of two tumor samples with antibodies to C/EBPα showed no reduction of C/EBPα in tumor sections of the livers (Fig. 1B). A parallel examination of cell cycle proteins in the tumor samples revealed that livers proliferate, because they expressed high levels of cell cycle proteins such as cyclin D1 and S-phase-specific protein PCNA (Fig. 1B). In addition to these observations, our group and others previously showed that hepatoma cell lines derived from liver tumors Hep3B2 and HepG2 express endogenous C/EBPα and may be arrested by C/EBPα only when C/EBPα is expressed to very high levels from strong RSV or CMV promoters (Timchenko et al. 1996; Wang et al. 2001). However, biologically relevant levels of C/EBPα do not cause inhibition of proliferation in these cells (see following). How can liver tumors and hepatoma cell lines proliferate in the presence of C/EBPα? We hypothesized that, during malignant transformation, cells in liver tumors may have developed a mechanism to neutralize the inhibitory activity of C/EBPα. To determine the molecular basis of the resistance of hepatoma cells to C/EBPα growth arrest, we generated stable Hep3B2 clones expressing different levels of C/EBPα. Figure 1C shows that the levels of IPTG-induced expression of C/EBPα in these clones are significantly higher than endogenous levels of C/EBPα. However, the examination of growth in these clones showed that these lines still proliferate in the presence of exogenous C/EBPα (Fig. 1C). A comparison of these Hep3B2 clones and stable clones previously generated in human fibrosarcoma cells HT1080 and in SAOS2 cells showed that similar levels of C/EBPα are sufficient to cause growth arrest in the latter cell lines (Timchenko et al. 1996; Wang et al. 2001). The failure of C/EBPα to inhibit Hep3B2 cells under physiological conditions confirmed the hypothesis that hepatoma cells developed a mechanism to block growth inhibitory activity of C/EBPα.
Figure 1.
Regenerating liver, liver tumors, and hepatoma cells proliferate in the presence of C/EBPα. (A) Expression of C/EBPα in rat livers after PH. Nuclear extracts were isolated from rat livers at different time points after PH or sham surgery and analyzed by Western blotting with C/EBPα antibodies. Levels of C/EBPα were calculated as ratios of two isoforms (42 and 30 kD) to β-actin control. Bar graphs present a summary of three independent experiments. (B) C/EBPα levels are not reduced in tumors. Nuclear extracts from tumor (T) and control (C) sections of livers of two patients were examined by Western blotting with antibodies to C/EBPα. β-Actin and Coomassie stain of the same membrane show protein loading. Expression of cell cycle proteins PCNA, cyclin D1, cdk2, and Brm was examined by Western blotting in control (C) and tumor (T) cells. (C) Hepatoma cells are resistant to C/EBPα growth arrest. Western blotting shows levels of C/EBPα induced by IPTG in stable Hep3B2 clones (shown on the top). Cross-reactive material (CRM) shows protein loading. The bottom image shows a colony growth assay with Hep3B2-b10 clone.
PI3K/Akt pathway blocks growth inhibitory activity of endogenous C/EBPα in cultured hepatoma cells
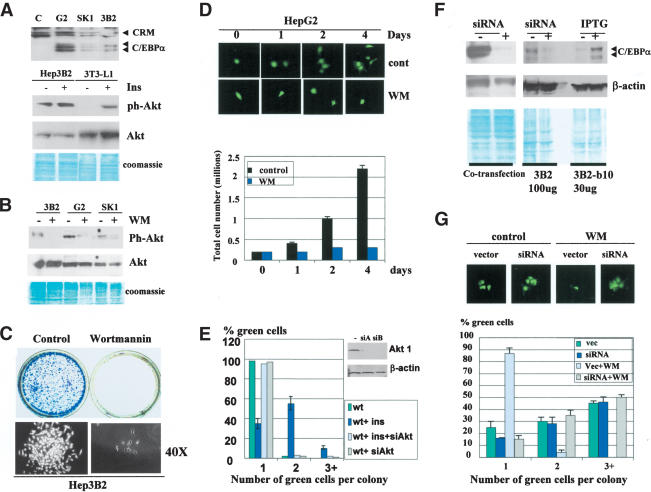
If our hypothesis is correct, one would expect that other hepatoma cell lines also escape negative control of proliferation by the block of C/EBPα growth inhibitory activity. Therefore, we used three hepatoma cell lines, Hep3B2, HepG2, and SK-Hep1, to examine this hypothesis. Figure 2A (upper) shows that all three hepatoma cell lines express C/EBPα. Parallel examinations of growth inhibitory activity of C/EBPα in 3T3-L1 cells indicated that insulin blocks growth inhibitory activity of C/EBPα in these cells (data not shown; see Fig. 6A, below). Because insulin affects many biological processes through activation of the PI3K/Akt pathway (Lawlor and Alessi 2001; Shamji et al. 2003), we examined whether PI3K/Akt is active in hepatoma cells. Western blotting with antibodies to ph-Akt showed that the active form of Akt is abundant in hepatoma cells, whereas in 3T3-L1 cells ph-Akt is not detectable, but can be activated by insulin (Fig. 2A,B). The activation of Akt in hepatoma cells is mediated by PI3K, because the treatment of these cells with the PI3K inhibitor wortmannin (WM) leads to the reduction of the active Akt (Fig. 2B). We next examined whether the inhibition of PI3K/Akt pathway by specific inhibitors might restore growth inhibitory activity of C/EBPα. Colony formation assay (Fig. 2C) and cell counting (Fig. 2D) showed that hepatoma cell lines are arrested by treatment with WM. Because WM is a specific inhibitor of PI3K and because WM restores growth inhibitory activity of C/EBPα (see Fig. 6C, below), this result suggests that hepatoma cells block growth inhibitory activity of C/EBPα via the PI3K/Akt pathway. To confirm the role of Akt in the PI3K-mediated blocking C/EBPα, we applied an additional approach: inhibition of Akt by siRNA technique. It has been recently demonstrated that the inhibition of both Akt1 and Akt2 by siRNA is required for efficient blockage of downstream targets of Akts (Jiang et al. 2003). Therefore, we expressed siAkt1 and siAkt2 RNA oligomers in 3T3-L1 cells, and then transfected these cells with C/EBPα. 3T3-L1 cells were chosen for these experiments because the PI3K/Akt pathway is not active in these cells, but might be activated by insulin (Ross et al. 1999; see Fig. 2A). Growth inhibitory activity of C/EBPα was measured in untreated cells and in cells treated with insulin. As can be seen, the inhibition of Akts by si RNAs abolishes the ability of insulin to block C/EBPα growth arrest (Fig. 2E).
Figure 2.
Hepatoma cell lines block growth inhibitory activity of C/EBPα by activation of PI3K/Akt pathway. (A) PI3K/Akt pathway is active in hepatoma cell lines. The upper image shows expression of C/EBPα in three hepatoma cell lines (shown on the top) detected by Western blotting. The bottom image shows Western blotting of ph-Akt and total Akt with proteins isolated from Hep3B2 cells. 3T3-L1 cells were used as a control in which Akt is activated by insulin. (B) Inhibition of PI3K by WM blocks Akt pathway in hepatoma cells. Treatment of Hep3B2, HepG2, and SK-Hep1 cells with WM blocks the activation of Akt. (C) WM-mediated block of PI3K/Akt leads to inhibition of Hep3B2 cells. Hep3B2 cells were stained at day 7 after plating with or without WM. The bottom image shows the size of colonies under 40× magnification. (D) HepG2 cells are arrested by WM. HepG2 cells were transfected with GFP and treated with WM. The upper images show size of green colonies at 0, 1, 2, and 4 d after transfection. (Bottom bar graphs) HepG2 cells (200,000) were plated and grown in the presence or in the absence of WM. The total number of cells was counted at days 0, 1, 2, and 4 after plating. (E) Inhibition of Akt by siRNA blocks insulin-mediated release of C/EBPα growth arrest. Western blotting shows levels of Akt1 in insulin-treated 3T3-L1 cells after transfections with Akt siRNAs (siA and siB). Bar graphs show C/EBPα growth arrest in cells treated with insulin in the presence of Akt siRNA. (F) Inhibition of C/EBPα by siRNA. (Left) pcDNA–C/EBPα and siRNA were cotransfected into Hep3B2 cells. C/EBPα was examined by Western blotting. β-Actin shows protein loading. (Right) Inhibition of endogenous C/EBPα by siRNA. IPTG-induced C/EBPα in stable Hep3B2-b10 serves as a positive control. (G) WM-mediated growth arrest of hepatoma cells requires C/EBPα. Hep3B2 cells were transfected with C/EBPα-siRNA, treated with wortmannin, and examined for the formation of colonies at day 3 after transfections. Bar graphs show a summary of three independent experiments. The upper image shows a typical picture of colonies.
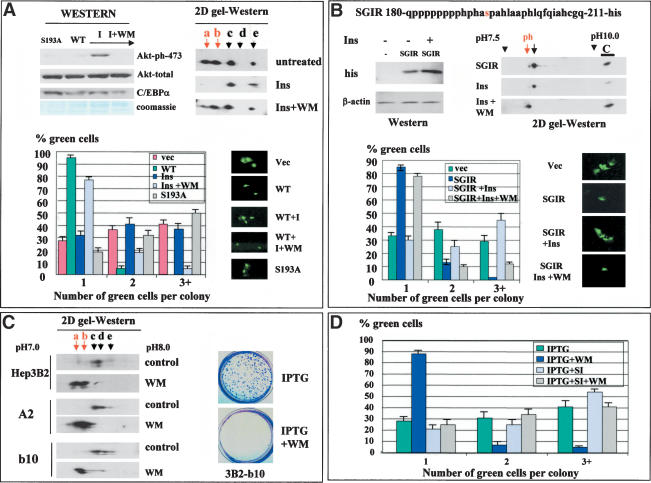
Figure 6.
Inhibition of PI3K/Akt pathway restores phosphorylation of Ser 193 and causes C/EBPα-mediated growth arrest in Hep3B2 cells. (A) Insulin blocks C/EBPα growth arrest by activation of PI3K/Akt pathway. 3T3-L1 cells, transfected with pAdTrack–C/EBPα, were treated with insulin (100 mM) or insulin + wortmannin (WM, 150 mM). Proteins were isolated from the plates at the end of each experiment and analyzed by Western blotting with Abs to ph-S473–AkT and antibodies to total Akt and to C/EBPα (Western). C/EBPα was immunoprecipitated from extracts and examined by 2D gel electrophoresis (2D gel-Western). (Bottom image) Colony growth assay. Bar graphs show a summary of three independent experiments. The image at right shows a typical picture of cells. (B) Insulin blocks the growth inhibitory activity of C/EBPα through dephosphorylation of Ser 193. His-tagged SGIR of C/EBPα (top) was transfected into 3T3-L1 cells. Cells were treated with insulin or insulin + WM and examined for growth arrest (bar graphs). Proteins were isolated from each plate, and expression of SGIR was examined by Western blotting with Abs to his tag and by 2D gel-Western. Two isoforms of SGIR are detectable in untreated cells by 2D gel. To determine a precise position of each isoform, we loaded a parallel lane with the control sample (C, SGIR overexpressed in cultured cells) and determined the position of the SGIR as a distance from the C lane. (C) The inhibition of PI3K/Akt pathway in Hep3B2 cells leads to the accumulation of a growth inhibitory isoform, C/EBPα-S193-ph, and to growth arrest. C/EBPα was immunoprecipitated from Hep3B2 cells and from stable clones Hep3B2-A2 and Hep3B2-b10 induced by IPTG. C/EBPα was examined by 2D gel electrophoresis. The image on the right shows staining of Hep3B2-b10 plates induced by IPTG alone or by IPTG with WM. (D) The inhibition of C/EBPα expression by siRNA abolishes WM-mediated growth arrest in stable clones. Stable clone Hep3B2-b10 was transfected with pAdTrack-siRNA and IPTG was added to the plates. Proliferation of green (transfected) cells was calculated at day 3. A summary of three independent experiments is shown.
To determine whether WM-mediated growth arrest in hepatoma cells occurs through C/EBPα, we inhibited C/EBPα Hep3B2 cells by siRNA, as shown in Figure 2F. siRNA-mediated inhibition of C/EBPα abolishes WM-dependent growth arrest in Hep3B2 cells (Fig. 2G). Similar results were obtained for HepG2 and SK-Hep1 cells (data not shown). Thus, these data demonstrate that hepatoma cells block the growth inhibitory activity of C/EBPα by activating the PI3K/Akt pathway.
A mutation of Ser 193 to Ala blocks the interaction of C/EBPα with cdks and abolishes C/EBPα-mediated growth arrest
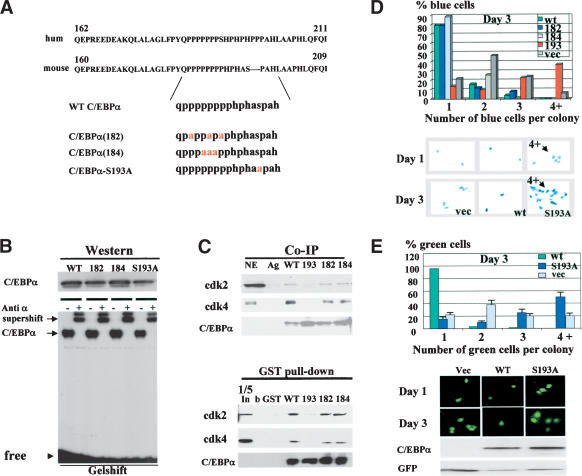
Given that we have identified a potential pathway by which hepatoma cells neutralize C/EBPα, we further determined a precise molecular mechanism by which insulin/PI3K/Akt blocks growth inhibitory activity of C/EBPα. We previously mapped a short region of mouse/rat C/EBPα that interacts with cdk2 and cdk4 and alone is sufficient to cause growth arrest (Wang et al. 2001). Figure 3A shows that amino acid sequences of growth inhibitory regions of human and mouse C/EBPα have a very high level of homology including identical proline-rich sequences and a Ser residue surrounded by the prolines. Given this high level of homology and similar growth inhibitory activities of the mouse and human C/EBPα, we focused our mutational studies on mouse C/EBPα. We generated mouse C/EBPα constructs with mutations that substitute prolines (mut182 and mut184) and a mutation that replaces Ser 193 with Ala (Fig. 3A). The mutant C/EBPα molecules were cloned into the pcDNA vector for expression in mammalian cells and fused to GST to investigate protein–protein interactions. First, we determined whether the C/EBPα mutants interact with C/EBP DNA consensuses by using a gelshift assay. Figure 3B shows that all constructs are expressed in mammalian cells at approximately equal levels and bind to C/EBPα consensus. Then, we examined the interactions of these mutants with cdk2 and cdk4 in cultured cells. 3T3-L1 cells were transfected with C/EBPα constructs, C/EBPα was precipitated with specific antibodies, and cdk2 and cdk4 were examined in C/EBPα IPs. Figure 3C shows a typical picture of these experiments. The mutations of prolines do not affect the interactions of C/EBPα with cdk2 and cdk4; however, the substitution of Ser 193 with Ala abolishes the interactions of C/EBPα with cdks. To verify these observations, we applied a GST pull-down approach by using GST–C/EBPα mutants and nuclear extracts from 3T3-L1 adipocytes. Consistent with Co-IP results, the GST pull-down assay showed that the C/EBPα-S193A mutant fails to interact with cdk2 and cdk4, whereas C/EBPα mut182 and mut184 interact with these kinases. On the basis of these observations, Ser 193 is a key residue that is required for the interactions with cdks.
Figure 3.
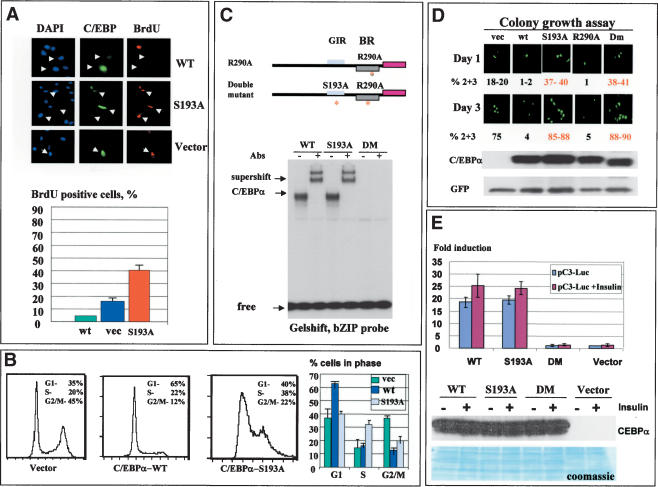
Growth inhibitory activity of C/EBPα is dependent on Ser 193. (A) Amino acid sequences of growth inhibitory regions of human (upper) and mouse (bottom) C/EBPα. C/EBPα constructs with point mutations within the growth inhibitory region of C/EBPα are shown below. (B) Expression and DNA binding activity of the C/EBPα mutant constructs. 3T3-L1 cells were transfected with C/EBPα constructs, and proteins were isolated and examined by Western blotting (upper) or by gelshift assays with C/EBP consensus bZIP. Antibodies to C/EBPα were incorporated into the binding reactions before the probe addition. (C) C/EBPα-S193A mutant does not interact with cdk2 and cdk4. The upper image (Co-IP) shows coimmunoprecipitation (C/EBPα-IP) of cdk2 and cdk4 from cells transfected with C/EBPα mutants shown at the top. (Ag) Mock control. The bottom panel shows C/EBPα in immunoprecipitates. The bottom image (GST pull-down) shows GST pull-down experiments with C/EBPα mutants. (b) Beads only; (In) 1/5 part of the input. (D) The C/EBPα-S193A mutant does not inhibit proliferation of 3T3-L1 cells. C/EBPα mutants were cotransfected with β-gal into 3T3-L1 cells and cells were stained for β-gal activity at day 1 and day 3 after transfections. Proliferation was examined by counting blue cell clusters. A summary of three independent experiments is shown. The bottom image shows a typical picture of blue 3T3-L1 cells at day 3 after transfections. (E) Colony growth assay with pAdTrack–C/EBPα constructs. The bottom image shows a typical picture of cells at days 1 and 3 after transfections. Western blotting indicates levels of C/EBPα at the end of experiment. Bar graphs represent a summary of three independent experiments.
We next examined the growth inhibitory activity of the C/EBPα mutants by using cotransfections with a β-gal plasmid (ratio 10:1) in 3T3-L1 cells. Growth-arrested (single cells) and dividing cells (two, three, and more cells per colony) were counted at day 1 and day 3 after transfections. Results of these studies are shown in Figure 3D. At day 3 after transfection, 85%–90% of cells expressing wild-type C/EBPα, C/EBPα-182, and C/EBPα-184 mutants are arrested and stay as single colonies. However, cells expressing C/EBPα-S193A mutant divide and form colonies containing multiple cells. In the course of these studies, we observed that cells expressing the C/EBPα-S193A mutant proliferate faster than cells transfected with an empty vector. As can be seen in Figure 3D, >50% of cells transfected with C/EBPα-S193A form two and three cell colonies at day 1. At day 3, ∼35% of the colonies contain multiple cells, whereas in control cells only 7%–10% of the colonies contain four-cell clusters. A typical picture of these multicell colonies is shown in Figure 3D (bottom). The increased number of dividing cells in C/EBPα-S193A transfections suggested that this mutant promotes cell proliferation. To better investigate this possibility, we used constructs in which wild-type C/EBPα and C/EBPα mutants were cloned into pAdTrack-CMV plasmid that also expresses a green fluorescent protein (GFP) from a separate mRNA. Using this approach, one can monitor growth arrest without fixation of cells and test the levels of C/EBPα within each experimental plate at the end of experiments. A summary of these studies is shown in Figure 3E (bar graphs). Consistent with previous experiments, this assay confirmed that C/EBPα-S193A mutant promotes cell proliferation. A typical picture of cell images is shown in Figure 3E. Western blotting showed that wild-type C/EBPα and the C/EBPα-S193A are expressed at approximately equal levels. Western blotting for GFP was used as a control for loading. These data and further studies of BrdU uptake and FACS analysis of DNA content (Fig. 4) showed that the C/EBPα-S193A mutant is not capable of inhibiting cell proliferation.
Figure 4.
The C/EBPα-S193A mutant accelerates cell proliferation. (A) BrdU uptake in cells transfected with empty vector, wild-type C/EBPα, or C/EBPα-S193A mutant. The upper image shows a typical picture of incorporation of BrdU in transfected cells. Bar graphs present a summary of three independent experiments. (B) FACS analysis of cells infected with pAdTrack–C/EBPα adenovirus constructs. DNA content was examined in GFP-positive cells. Bar graphs represent summary of three experiments. (C) Generation of a double mutant C/EBPα-S193A, R290A construct that does not bind to DNA. The upper image shows the positions of point mutations within the C/EBPα molecule. The bottom image shows a gelshift assay with bZIP probe. (D) Colony growth assay. pAdTrack–C/EBPα constructs were transfected into 3T3-L1 cells and growth assay was performed as described earlier. Percentage of dividing cells (two or three cells per colony) is shown as a summary of two experiments. (E) Activation of C/EBP reporter construct, pC3-luc, by C/EBPα mutants. pC3-luceferase construct (pC3-luc) contains C3 promoter, which is activated by C/EBPα (Timchenko et al. 1996). pC3-luc was cotransfected with C/EBPα proteins into 3T3-L1 cells treated or untreated with insulin and luciferase activity was examined. Bar graphs represent a summary of three independent experiments. The bottom image shows expression of C/EBPα proteins in transfected cells. The filter was stained with Coomassie to verify protein loading.
The C/EBPα-S193A mutant accelerates cell proliferation
Because the data for the colony growth assay (Fig. 3D,E) suggested that the C/EBPα-S193A mutant enhances cell proliferation, we performed a detailed analysis of biological activities of this mutant. First, DNA synthesis (BrdU uptake) was measured in cells transfected with empty vector, wild-type C/EBPα, and C/EBPα-S193A mutant. C/EBPα-S193A transfected plates contain ∼40% BrdU-positive cells, whereas cells transfected with an empty vector contain 15%–20% BrdU-positive cells (Fig. 4A). This result confirms data of colony growth arrest, showing that the mutant accelerates proliferation. To obtain more evidence for this conclusion, we examined the cell cycle distribution of transfected cells by using FACS analysis. A typical picture of these studies is shown in Figure 4B. In agreement with colony growth assays and BrdU uptake, the majority of cells transfected with wild-type C/EBPα are arrested in G1. In contrast, the C/EBPα-S193A mutant drives a significant portion of cells (38%–40%) into S phase. The summary of three experiments showed that the number of C/EBPα-S193A transfected cells in the S phase is twofold higher than the number of cells in S phase on the plates transfected with the empty vector. Taken together, these data revealed that the C/EBPα-S193A mutant accelerates cell proliferation by driving a higher percentage of cells into S phase. Thus, the mutation of Ser 193 to Ala not only abolishes growth inhibitory activity of C/EBPα but also accelerates cell proliferation.
Given the acceleration of proliferation by C/EBPα-S193A mutant, we examined mechanisms by which this mutant causes increased proliferation. Because the C/EBPα-S193A mutant binds to DNA, it might accelerate proliferation by transcriptional activity independent of binding and inhibiting cdks. To test this possibility, we incorporated an additional point mutation, R290A, into C/EBPα-S193A. This mutation abolishes the binding of C/EBPα to DNA (Miller et al. 2003). As can be seen in Figure 4C, this additional mutation blocks the binding of the C/EBPα-S193A to DNA. Colony growth assays indicated that the double-mutant (DM) C/EBPα retains the ability to accelerate cell proliferation (Fig. 4D). Examination of the protein levels of C/EBPα showed that wild-type and the mutant C/EBPα are expressed at approximately equal levels (Fig. 4D, bottom image). To further examine the possible role of transcriptional activity of the C/EBPα-S193A mutant in the promotion of proliferation, we examined the activation of C/EBPα-dependent promoters by wild-type, C/EBPα-S193A, and DM. These proteins were cotransfected with a reporter C3-luciferase construct (C3 promoter linked to luciferase; Timchenko et al. 1996) into 3T3-L1 cells. The activation of C3 promoter by C/EBPα mutants was examined in untreated cells and in cells treated with insulin (in which C/EBPα inhibitory activity is blocked, see below). C3 promoter contains a C/EBPα binding site and has been shown to be activated by C/EBPα (Timchenko et al. 1996). As can be seen in Figure 4E, wild-type C/EBPα and C/EBPα-S193A activate C3-luc promoter, whereas the DM construct fails to increase activity of the C3 promoter. We also observed that insulin slightly induces transcriptional activity of C/EBPα; however, this induction is not dependent on Ser 193. Because the DM does not activate transcription, but still accelerates cell growth, these data clearly demonstrate that the transcriptional activity of the C/EBPα-S193A mutant is not required for the acceleration of growth. Our investigations of biological activities of the C/EBPα-S193A mutant suggest that this mutant accelerates proliferation through sequestering of Rb (data not shown).
Dephosphorylation of Ser 193 inhibits the interaction of C/EBPα with cdk2 and Brm
The failure of C/EBPα-S193A mutant to inhibit cell proliferation suggested that Ser 193 plays a key role in the regulation of C/EBPα growth inhibitory activity. Therefore, we examined whether dephosphorylation of C/EBPα might affect its interactions with cdks and Brm and its ability to cause growth arrest. Because C/EBPα inhibits liver proliferation through interactions with cdk2 and cdk4 (Wang et al. 2001, 2002), we tested the phosphorylation status of C/EBPα in young mouse livers, where C/EBPα forms complexes with cdk2 (Wang et al. 2001, 2002). After immunoprecipitation from mouse livers, C/EBPα was separated by 2D gel electrophoresis and examined by Western blotting. Figure 5A shows that C/EBPα in nuclear extracts from mouse livers migrates as five isoforms, three of which are eliminated after treatment with CIP. The shift of these three isoforms after CIP treatment demonstrates that C/EBPα is phosphorylated in livers. Then, we examined the effects of dephosphorylation on the ability of C/EBPα to form C/EBPα–cdk2 complexes. Liver nuclear extracts were treated with CIP, and C/EBPα–cdk complexes were examined by size exclusion chromatography as described in our previous papers (Wang et al. 2001; Iakova et al. 2003). Results of these experiments are presented in Figure 5B. Immunoprecipitation of C/EBPα from gel-filtration fractions of untreated nuclear extracts and Western blotting with Abs to cdk2 showed that C/EBPα forms complexes with cdk2 (Fig. 5B). After the incubation of nuclear extracts with CIP, C/EBPα is dephosphorylated (Fig. 5A) and is shifted to low-molecular-weight fractions of gel filtration. Co-IP studies revealed that the CIP treatment destroys C/EBPα–cdk2 complexes because cdk2 is no longer detectable in C/EBPα IPs (Fig. 5B, bottom).
Figure 5.
Dephosphorylation blocks the interactions of C/EBPα with cdks and Brm in vitro and in vivo. (A) C/EBPα is phosphorylated in livers. 2D gel electrophoresis was performed with C/EBPα precipitated from NE of young mouse livers. (CIP) 2D gel separation of C/EBPα treated with CIP; (Control) C/EBPα precipitated from 3T3-L1 cells transfected with pcDNA–C/EBPα. Five isoforms (a, b, c, d, and e) of C/EBPα are observed in mouse liver. (B) Dephosphorylation of C/EBPα in liver extracts destroys C/EBPα–cdk2 complexes. Untreated (control) and CIP-treated nuclear extracts were fractionated by size exclusion chromatography (HPLC). Positions of C/EBPα and cdk2 within the fractions were determined by Western blotting. To detect C/EBPα–cdk2 complexes, we precipitated C/EBPα from each fraction and examined cdk2 in C/EBPα IPs. (IgG) Heavy chains of immunoglobulins. (C) Insulin signaling leads to a dephosphorylation of C/EBPα on Ser 193. Wild-type C/EBPα or C/EBPα-S193A mutant were transfected in 3T3-L1, cells were treated with insulin, and C/EBPα was isolated and examined by 2D gel electrophoresis. Acidic isoforms (a and b) are not detectable in C/EBPα-S193A mutant and are dramatically reduced in wild-type C/EBPα after treatment with insulin. (D) Insulin-mediated dephosphorylation of C/EBPα blocks the interaction of C/EBPα with cdks and Brm. (Left image) C/EBPα was transfected into 3T3-L1 cells, cells were treated with insulin, and C/EBPα was precipitated with specific antibodies. Cdk2, cdk4, Brm, and C/EBPα were examined in C/EBPα IPs. (Right image) Insulin does not affect the interaction of p21 with cdk2. 3T3-L1 cells were transfected with p21, treated with insulin, and p21 was precipitated with specific Abs. cdk2 and p21 were examined in p21 IPs.
PI3K/Akt pathway triggers dephosphorylation of C/EBPα and blocks the ability of C/EBPα to interact with cdks and Brm
Given the observations that the C/EBPα-S193A mutant does not inhibit cell proliferation (Figs. 3, 4) and that the dephosphorylation destroys cdk2–C/EBPα interactions, we suggested that hepatoma cells might block C/EBPα growth inhibitory activity by dephosphorylation C/EBPα at Ser 193. To test this hypothesis, we used 3T3-L1 cells in which the PI3K/Akt pathway is not detectable, but might be activated by insulin (Ross et al. 1999). We have initially verified that the activation of PI3K/Akt dephosphorylates C/EBPα in these cells. Wild-type C/EBPα and C/EBPα-S193A were immunoprecipitated from insulin-treated and untreated cells and examined by 2D gel electrophoresis. Results of these studies are shown in Figure 5C. Two acidic isoforms of wild-type C/EBPα (a and b) are not detectable in cells treated with insulin. Parallel examination of the C/EBPα-S193A mutant revealed that these acidic isoforms are the result of phosphorylation of wild-type C/EBPα on Ser 193, because they are not detectable within the C/EBPα-S193A mutant. Thus, these data demonstrate that treatment of 3T3-L1 cells with insulin causes dephosphorylation of C/EBPα on Ser 193.
We next examined whether the insulin-mediated dephosphorylation of Ser 193 affects the interactions of C/EBPα with cdks and Brm. After transfections with wild-type C/EBPα and insulin treatment, C/EBPα was precipitated with specific antibodies. Western blotting of C/EBPα IPs revealed that, in insulin-treated cells, dephosphorylated C/EBPα does not interact with cdks and Brm (Fig. 5D). Because insulin affects several biochemical pathways, the lack of the interactions with cdks might also be due to alterations with cdks. To examine this possibility, we determined whether insulin affects interactions of cdk2 with p21. Co-IP studies showed that p21 interacts equally well with cdk2 in untreated cells and in cells treated with insulin (Fig. 5D, right). These results show that C/EBPα does not interact with cdks and Brm because of dephosphorylation on Ser 193.
Activation of PI3K/Akt pathway blocks C/EBPα growth inhibitory activity in 3T3-L1 cells
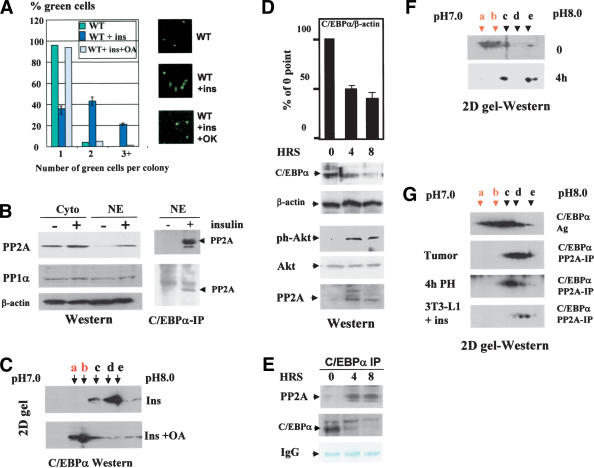
We next determined whether activation of the PI3K/Akt pathway abolishes C/EBPα growth arrest in 3T3-L1 cells in which the PI3K/Akt pathway is not active (see Fig. 2). Western blotting with antibodies specific to Akt–Ser 473-ph showed a dramatic induction of the active, phosphorylated Akt by insulin, whereas there was no change in the total protein levels of Akt (Fig. 6A). Incorporation of a specific inhibitor of PI3K, WM, blocks activation of Akt (Fig. 6A). To examine whether Akt activation leads to a dephosphorylation of C/EBPα and subsequent release of growth arrest, we transfected 3T3-L1 cells with wild-type C/EBPα, treated with insulin (100 nM) or with insulin + WM. Figure 6A (right image) shows that insulin signaling leads to a dephosphorylation of C/EBPα and that WM blocks insulin-mediated C/EBPα dephosphorylation. Examination of the growth inhibitory activity of C/EBPα showed that WM-mediated inhibition of Akt also restores the ability of C/EBPα to cause growth arrest (Fig. 6A, bottom). Thus, these data demonstrate that insulin releases C/EBPα growth arrest in 3T3-L1 cells through activation of PI3K/Akt, which results in dephosphorylation of C/EBPα on Ser 193.
It has been shown that insulin signaling causes a dephosphorylation of C/EBPα on several residues (Ross et al. 1999), suggesting that, in addition to Ser 193, dephosphorylation of other residues might contribute to the lack of growth arrest. To examine whether Ser 193 is the critical target of this pathway, we cloned a short growth inhibitory region (SGIR, previously shown to be sufficient for growth arrest) of C/EBPα into pcDNA6, fusing it to his/myc tag (Fig. 6B), and performed functional analysis of growth inhibitory activity of this region in insulin-treated cells. Figure 6B shows that insulin does not affect the protein levels of the SGIR. Examination of phosphorylation of the SGIR by 2D gel electrophoresis revealed that insulin signaling dephosphorylates the SGIR, and WM causes the accumulation of ph-SGIR. We next examined the growth inhibitory activity of the SGIR in cells treated with insulin or with insulin + WM. Figure 6B (bottom) represents a summary of these studies. Expression of SGIR in 3T3-L1 cells inhibits proliferation, and insulin-mediated de-phosphorylation of the SGIR leads to a block of growth inhibitory activity of the SGIR. WM reverses the growth inhibitory activity of the SGIR of C/EBPα. Because the SGIR contains only one residue (Ser 193) that can be phosphorylated, these data demonstrate that insulin/WM regulates the growth inhibitory activity of C/EBPα through regulation of Ser 193 phosphorylation.
Inhibition of PI3K/Akt in stable Hep3B2 clones leads to the restoration of the growth inhibitory activity of C/EBPα
We next examined whether the block of PI3K/Akt might restore growth inhibitory activity of C/EBPα in stable clones, Hep3B2-A2, and Hep3B2-b10 (see Fig. 1). Examination of C/EBPα in control and in WM-treated Hep3B2 cells by 2D gel-Western shows that, in control samples, C/EBPα is not phosphorylated on Ser 193 and migrates as three isoforms (Fig. 6C). However, WM dramatically increases phosphorylation of C/EBPα and leads to the accumulation of growth inhibitory isoforms of C/EBPα in stable clones and in original Hep3B2 cells. Colony growth assay demonstrated that the accumulation of the growth inhibitory isoform of C/EBPα in WM-treated Hep3B2-b10 and Hep3B2-A2 cells leads to growth arrest (Fig. 6C; data for Hep3B2-A2 clone are not shown). To examine whether WM causes growth arrest in stable clones through activation of C/EBPα we inhibited C/EBPα expression by siRNA as shown in Figure 2. Cells were transfected with pAdTrack-siRNA, and C/EBPα was induced by IPTG. In control samples, cells were transfected with an empty pAdTrack-CMV vector that expresses only GFP. As can be seen in Figure 6D, WM inhibits Hep3B2-b10 cells through the activation of C/EBPα, because WM fails to induce growth arrest in cells that do not express C/EBPα.
PP2A is responsible for insulin/Akt-mediated block of C/EBPα growth inhibitory activity in 3T3-L1 cells
We next determined a phosphatase that dephosphorylates C/EBPα. Because insulin activates PP1α and PP2A (Hemati et al. 1997), we first examined whether these phosphatases might be involved in the insulin-dependent dephosphorylation of C/EBPα. We initially examined whether a specific inhibitor of PP2A and PP1α, okadaic acid, might abolish insulin-mediated effects. Figure 7A shows that insulin is not able to block C/EBPα growth arrest in 3T3-L1 cells treated with okadaic acid. In control cells, the majority of PP2A is located in the cytoplasm (Fig. 7B). Western blotting shows that insulin causes accumulation of PP2A in the nuclei of 3T3-L1 cells, whereas PP1α is not affected in insulin-treated cells (Fig. 7B). Coimmunoprecipitation studies revealed that the accumulation of PP2A in nuclei leads to the interaction of PP2A with C/EBPα and to dephosphorylation of C/EBPα. The inhibition of PP2A by okadaic acid blocks insulin-mediated dephosphorylation of C/EBPα (Fig. 7B,C). Thus, these studies suggested that, in 3T3-L1 cells, the insulin/Akt pathway inhibits C/EBPα activities through PP2A.
Figure 7.
PP2A dephosphorylates C/EBPα on Ser 193. (A) Okadaic acid blocks insulin-mediated release of C/EBPα growth arrest. 3T3-L1 cells were transfected with C/EBPα and treated with insulin or with insulin/okadaic acid. The image on the right shows a typical picture of cells. Bar graphs represent a summary of three independent experiments. (B) Insulin increases concentration of PP2A in nuclear extracts. (Left) Western blotting was performed with antibodies to PP1α and PP2A by using nuclear extracts isolated from untreated and insulin treated 3T3-L1 cells. (Right) C/EBPα was immunoprecipitated from nuclear extracts, and Western blotting was performed with antibodies to PP2A. (C) Inhibition of PP2A by okadaic acid blocks insulin-mediated dephosphorylation of C/EBPα.2D gel-Western of C/EBPα transfected into 3T3-L1 cells treated with insulin or with insulin + okadaic acid was performed. (D) Expression of C/EBPα? ph-Akt and PP2A in mouse livers at 4 and 8 h after partial hepatectomy. Bar graphs show levels of C/EBPα as a summary of three experiments. (Western) Western blotting of cytoplasmic (for Akt) and nuclear (for C/EBPα and PP2A) proteins isolated at 0, 4, and 8 h after PH. (E) Coimmunoprecipitation of C/EBPα and PP2A from NE isolated from liver after partial hepatectomy. The membrane was stained with Coomassie. The section of the membrane with heavy chain IgG is shown. (F) 2D gel-Western. The analysis of C/EBPα isoforms by 2D gel electrophoresis in quiescent livers (0) and in liver 4 h after PH is shown. (G) PP2A dephosphorylates C/EBPα on Ser 193. PP2A was immunoprecipitated from nuclear extracts of liver tumor (patient #2, see Fig. 8) 4 h after PH and from 3T3-L1 cells treated with insulin. C/EBPα was incubated with the PP2A IPs and examined by 2D gel electrophoresis. The control sample (C/EBPα-Ag) was incubated with agarose.
Growth inhibitory activity of C/EBPα is blocked in regenerating livers by Akt–PP2A pathway
We next examined whether the PI3K/Akt/PP2A pathway might be involved in the neutralization of the growth inhibitory activity of C/EBPα in liver when the liver proliferates after surgery. A removal of a portion of the liver by surgery (PH) leads to the initiation of liver proliferation (Iakova et al. 2003). Although expression of C/EBPα is reduced after PH, protein levels of C/EBPα remain relatively high during liver proliferation (Timchenko et al. 1999; Figs. 1A, 7D), suggesting that additional mechanisms are activated to neutralize the growth inhibitory activity of the remaining C/EBPα. We examined whether the Akt-mediated dephosphorylation of C/EBPα might be such a mechanism. Western blotting with ph-Akt-specific antibodies showed that Akt is activated in mouse livers at 4 and 8 h after PH (Fig. 7D). Consistent with data for cultured cells, the activation of Akt also leads to accumulation of PP2A in nuclei after PH. Precipitation of C/EBPα from nuclear extracts and analysis of C/EBPα IPs with antibodies to PP2A revealed that increasing levels of PP2A bind to C/EBPα (Fig. 7E). This suggested that PP2A might dephosphorylate C/EBPα on Ser 193 to allow liver proliferation. Therefore, we next examined C/EBPα status after PH. Examination of C/EBPα isoforms by 2D gel electrophoresis showed that growth inhibitory isoforms of C/EBPα are abundant in quiescent livers; however, these isoforms are not observed at 4 h after PH (Fig. 7F). To examine whether PP2A is responsible for the dephosphorylation of C/EBPα in liver after PH and in insulin-treated 3T3-L1 cells, we immunoprecipitated PP2A from corresponding nuclear extracts and incubated it with C/EBPα overexpressed in untreated 3T3-L1 cells, where it is properly phosphorylated. Examination of C/EBPα isoforms after this incubation shows that PP2A isolated from insulin-treated 3T3-L1 cells and PP2A isolated from livers 4 h after PH dephosphorylates C/EBPα, leading to the disappearance of the growth inhibitory isoforms of C/EBPα (Fig. 7G). Taken together, these data show that the accumulation of PP2A in nuclei causes dephosphorylation of C/EBPα, leading to the neutralization of the growth inhibitory activity of C/EBPα.
Human liver tumors block growth inhibitory activity of C/EBPα through the activation of Akt–PP2A
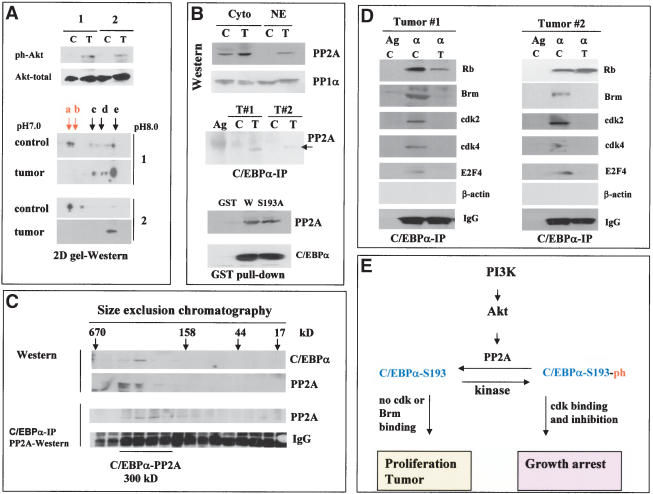
Having established the pathway by which hepatoma cells and regenerating liver neutralize growth inhibitory activity of C/EBPα, we asked whether the Akt/PP2A-mediated block of C/EBPα is relevant for liver tumors in vivo. Therefore, we examined the PI3K/Akt/PP2A pathway and phosphorylation status of C/EBPα in two human liver tumor samples that proliferate and express high levels of C/EBPα (see Fig. 1B). We initially tested whether the tumor samples have mutations/deletions within the growth inhibitory region of C/EBPα by amplifying a fragment of C/EBPα from amino acid 110 to amino acid 200 (which covers the growth inhibitory region of C/EBPα) from each tumor sample. No mutation/deletion was observed in the growth inhibitory region of C/EBPα amplified from the tumor cells (data not shown). Therefore, we next examined the PI3K/Akt/PP2A pathway in control and tumor samples. Western blotting with antibodies to the active, phosphorylated form of Akt showed that Akt is activated in both liver tumor samples (Fig. 8A). As can be seen in Figure 1B, the activation of Akt in the tumors correlates with the induction of the S-phase-specific protein PCNA and cyclin D1, whereas protein levels of cdk2, Brm, and cdk4 are not altered (Fig. 1B; cdk4 data are not shown). Because activation of Akt in liver after PH and in cultured cells neutralizes C/EBPα through dephosphorylation on Ser 193, we next analyzed C/EBPα in liver tumors by 2D gel-Western assay. In control human samples, growth inhibitory isoforms of C/EBPα (a and b) are abundant; however, liver tumors do not contain detectable amounts of these isoforms (Fig. 8A). Because PP2A dephosphorylates C/EBPα in cultured cells and in regenerating livers, we determined protein levels of PP2A in nuclear extracts of human liver tumors and the interactions of PP2A with C/EBPα. Figure 8B shows that, in control samples, the majority of PP2A is observed in the cytoplasm, whereas in liver tumor samples PP2A is accumulated in the nuclei and interacts with C/EBPα (Fig. 8B, C/EBPα IP). GST pull-down assay with nuclear extracts from the livers confirmed that wild-type C/EBPα is interacting with PP2A. Interestingly, the mutant C/EBPα-S193A also interacts with PP2A in GST pull-down assay, suggesting that the presence of a phosphate on Ser 193 is not required for efficient interactions. To further examine C/EBPα–PP2A interactions in liver tumors, we fractionated nuclear extracts from liver tumor by size exclusion chromatography and examined them by Western blotting and Co-IP assays. These studies identified a complex C/EBPα–PP2A that is located in the region of 300 kD (Fig. 8C). Thus, these data demonstrate that PP2A interacts with C/EBPα in tumor cells. We next precipitated PP2A from liver tumors and incubated it with phosphorylated C/EBPα. Examination of C/EBPα by 2D gel electrophoresis after this incubation shows that the interaction of PP2A with C/EBPα leads to dephosphorylation of C/EBPα on Ser 193 (Fig. 7G).
Figure 8.
Human liver tumors block C/EBPα growth inhibitory activity by activating Akt/PP2A. (A) PI3K/Akt pathway is active in tumor cells. The upper image shows Western blotting with phospho-Akt-specific antibodies (ph-Akt) and with antibodies to total Akt. (2D gel-Western) Liver tumors lack the growth inhibitory isoform of C/EBPα. Nuclear proteins from two patients (control and tumor) were separated by 2D gel electrophoresis, and C/EBPα was examined by Western blotting. (B) C/EBPα is associated with PP2A in liver tumors. (Western) Western analysis of PP2A and PP1α in cytoplasmic and nuclear extracts from control and tumor sections of patient #2. (C/EBPα-IP) Examination of PP2A in C/EBPα IPs from nuclear extracts of control (C) and tumor (T) sections. (GST pull-down) GST–C/EBPα proteins were incubated with nuclear extracts from tumor #2, and PP2A was examined in GST pull-down samples. (C) C/EBPα forms a high MW complex with PP2A in liver tumors. Nuclear extracts from liver tumor were fractionated by gel filtration. The location of C/EBPα and PP2A within gel-filtration fractions was determined by Western blotting. (C/EBPα-IP) C/EBPα was precipitated from each fraction, and PP2A was examined in C/EBPα IPs. (D) C/EBPα does not interact with cdks and Brm in liver tumors. Rb, Brm, cdk2, cdk4, and E2F4 were examined in C/EBPα IPs. (IgG) Heavy chains of immunoglobulins. (E) A hypothetical pathway for PI3K/Akt-mediated regulation of growth inhibitory activity of C/EBPα in liver.
We next examined the interactions of C/EBPα with cdks and Brm in control and tumor samples by coimmunoprecipitation assay. Figure 8D shows a typical result of these studies. In control samples, C/EBPα forms complexes with cdks and with Brm; however, no interactions with these proteins were detected in protein extracts isolated from liver tumors. The failure of C/EBPα to interact with cdks and Brm in liver tumor cells correlates with the lack of a growth inhibitory form of C/EBPα, C/EBPα-S193-ph, and with the proliferative status of the livers. Thus, these investigations suggest that liver tumors in humans proliferate because growth inhibitory activity of C/EBPα is neutralized by dephosphorylation on Ser 193.
Discussion
In this paper, we show that the PI3K/Akt pathway is active in human liver tumors and in cultured hepatoma cells and that this activation causes dephosphorylation of C/EBPα on Ser 193, leading to proliferation of cells. Akt was originally identified as an oncogene that can transform rodent cells (Bellacosa et al. 1991). Examination of mouse transgenic models overexpressing Akt in heart showed that Akt causes an increased proliferation and increased heart size (Shioi et al. 2002). Interestingly, Akt promotes proliferation of cells under conditions in which cells should normally be growth arrested (Lawlor and Alessi 2001; Shamji et al. 2003). Our findings suggest a pathway by which Akt displays its growth promotion function in liver: through inactivation of C/EBPα. In agreement with this hypothesis, a number of previous investigations indicated that the inhibition of Akt in many hepatoma cell lines leads to growth arrest (Lin and Chou 1998; Zou et al. 2002; Shi et al. 2003). Examination of liver tumors in humans revealed that C/EBPα growth inhibitory activity is also blocked in tumors by activation of Akt. Our hypothetical model for the mechanism by which tumor cells block C/EBPα activity is shown in Figure 8E. We suggest that the activation of the PI3K/Akt pathway in liver tumors causes a translocation of PP2A into nuclei, where PP2A binds to and dephosphorylates C/EBPα. As a result, liver loses a negative control of proliferation and may proliferate or develop tumors. Although our experimental data favor the hypothesis that the PI3K/Akt pathway activates PP2A to dephosphorylate C/EBPα, it is also possible that inhibition of an additional kinase might contribute to the dephosphorylation of C/EBPα.
C/EBP family proteins regulate transcription of genes in specific tissues. Recent studies demonstrated that two members of the C/EBP family, C/EBPα and C/EBPβ, control cell proliferation and cell survival through protein–protein interactions (Buck et al. 2001; McKnight 2001; Wang et al. 2001; Timchenko 2003). These new activities of C/EBP proteins are mainly regulated at the level of posttranslational modifications. In this paper, we determined mechanisms that block the growth inhibitory activity of C/EBPα in hepatoma cells. We have previously shown that a short region of C/EBPα interacts with cdks and alone is sufficient to inhibit cell proliferation (Wang et al. 2001). Within this region, we found a residue (Ser 193 for mouse/rat protein and Ser 190 for human protein) that is crucial for the regulation of growth inhibitory activity of C/EBPα and for the interactions with cdk2 and with Rb–E2F complexes. A mutation of Ser 193 to Ala makes C/EBPα unable to cause growth arrest. Surprisingly, this mutant molecule displays an opposite activity, which is the acceleration of proliferation. Our unpublished observations suggest that the C/EBPα-S193A accelerates cell proliferation via sequestering of Rb. Further work is required to understand whether this acceleration is relevant to in vivo conditions.
C/EBPα interacts with several cell cycle proteins and might cause growth arrest through multiple pathways (Timchenko et al. 1996; Johansen et al. 2001; Porse et al. 2001; Wang et al. 2001; Iakova et al. 2003). In addition to growth arrest, C/EBPα is also a critical regulator of expression of adipose- and liver-specific genes. How might all of these activities (pathways) be regulated in vivo? Our previous papers showed that one possible mechanism is a switch of protein partners that interact with C/EBPα (Iakova et al. 2003; Timchenko 2003). In this paper, we show that substitution of Ser 193 with Ala abolishes C/EBPα-mediated growth arrest. On the other hand, the S193A mutation does not affect the ability of C/EBPα to interact with DNA and to activate transcription of target genes (Fig. 4). This finding suggests a pathway by which cells might distinctly regulate growth inhibitory and transcriptional activities of C/EBPα. Insulin-mediated dephosphorylation of Ser 193 specifically blocks the growth inhibitory function of C/EBPα, but does not affect the binding to DNA and presumably would not affect the activation of corresponding promoters.
Materials and methods
Materials and plasmids
Antibodies to C/EBPα (14AA and N19), cdk4 (C-22), cdk2 (M2), Brm, and Rb (C-15) were purchased from Santa Cruz Biotechnology. Antibodies to PP2A and PP1α are from Signal Transduction Laboratories. Expression vectors for wild-type mouse CEBPα and mutations were generated by PCR-based amplification of the coding region of C/EBPα from genomic clone pBS-Bam#6 (Wang et al. 1995). The full-length C/EBPα cDNA was subcloned into the pcDNA3.1(+) expression vector. Based on the pcDNA3.1(+)-mCEBPα construct, mutations (shown in Fig. 3A) were created using point mutational PCR. For growth arrest assays, wild-type CEBPα and mutants were cloned into a pAdTrack-CMV vector that expresses GFP from a separate CMV promoter. Generation of pcDNA6/myc-His(B)-SGIR: An SGIR of C/EBPα (see Fig. 6) was generated by fusing a synthetic DNA oligomer (amino acids 180–211) to the AUG codon on the 5′ end, and cloned it into KpnI and Not I sites of pcDNA6/myc-His expression vector. Generation of mCEBPα recombinant adenoviruses: pAdTrack–C/EBPα constructs were cotransformed into BJ 5183 cells. Recombinant virus DNAs were selected from the kanamycin plates. The recombinant adenoviruses were packaged and produced in HEK 293 cells. Purified high-titer viral supplies were used in culture cell infections as described (He et al. 1998). Constructs with small interfering RNAs against C/EBPα were generated as follows: Two siRNA primers (specific for mouse and human CEBPα) were synthesized. Each primer contained 19 nt of CEBPα sequences (828–846 in mouse and 825–843 in human). The annealed fragment was cloned into BglII/HindIII sites of pSUPER vector. For growth arrest assay, the siRNA fragment was subcloned into pAdTrack-CMV expression vector. All constructs were verified by sequencing.
Human liver tumor samples and liver regeneration
Human liver samples were obtained as part of the IRB approved protocol, where tumor and normal sections were collected from resected samples. Liver regeneration and examination of C/EBPα were performed as described in our previous paper (Iakova et al. 2003).
Transient transfection assay
Analysis of C/EBPα growth arrest was performed in Hep3B2, HepG2, SK-Hep1, HT1080, and 3T3-L1 cells by using C/EBPα mutants described earlier. Transient transfection assay was performed with two approaches: cotransfections of pcDNA–C/EBPα with β-gal and single transfection of pAdTrack–C/EBPα, which expresses both GFP and C/EBPα from distinct mRNAs. C/EBPα vectors were cotransfected into cells with CMV-β-gal at a ratio of 10:1. Cells were stained for β-gal activity at days 1 and 3 following transfection. Cell growth was calculated by counting the number of blue-stained cells in each colony. In experiments with pAdTrack–C/EBPα, the inhibition was calculated by counting the number of green cells in each colony.
BrdU uptake
Cells were transfected with pAdTrack–C/EBPα constructs. Control cells were transfected with an empty pAdTrack vector. Twenty-four hours later, BrdU was added for 1 h, and cells were fixed and stained with monoclonal Abs to BrdU. DAPI staining was performed to visualize untransfected cells.
Gel-filtration analysis of C/EBPα–cdk2 complexes in mouse liver
The detailed procedure for the analysis of C/EBPα complexes is described in our previous papers (Wang et al. 2001; Iakova et al. 2003). Nuclear extracts were isolated from livers as described earlier (Timchenko et al. 1999) and fractionated by size-exclusion column SEC-400 (HPLC, BioLogic HR, Bio-Rad). To examine the effects of dephosphorylation on C/EBPα–cdk2 complexes, we treated nuclear extracts with CIP and fractionated them as described earlier. Gel-filtration fractions were loaded on denaturing gradient (4%–20%) PAAG, blotted onto membrane, and probed with antibodies to C/EBPα and cdk2. To detect C/EBPα–cdk2 complexes, we immunoprecipitated C/EBPα from each fraction and probed IPs with antibodies to cdk2.
Protein isolation and Western blotting
Nuclear extracts were isolated as described in previous papers (Timchenko et al. 1996; Wang et al. 2001). Stable C/EBPα clones in Hep3B2 cells were generated as described (Timchenko et al. 1996). C/EBPα was induced by IPTG, and proteins were isolated 18 h after C/EBPα induction. Proteins (50 μg) were loaded on gradient (4%–20%) PAAG, transferred on the membrane, and probed with antibodies to C/EBPα, cdk2, cdk4, Rb, E2F4, or Brm. To verify protein loading, we reprobed each filter with β-actin and then stained it with Coomassie.
2D gel-Western
C/EBPα was immunoprecipitated from transfected cells or from livers with specific antibodies (N19, Santa Cruz Biotechnology). IPs were separated by 2D gel electrophoresis (Protean IEF, Bio-Rad) and C/EBPα was transferred on the membrane and probed with rabbit antibodies (14AA, Santa Cruz Biotechnology).
Gelshift
Conditions for gelshift assay with bZIP probe are described in our earlier papers (Timchenko et al. 1996, 1999).
Coimmunoprecipitation and GST pull-down
C/EBPα was immunoprecipitated from nuclear extracts with polyclonal antibodies (14AA, Santa Cruz), and the presence of Rb, Brm, E2F4, cdk4, or cdk2 in C/EBPα IPs was examined by Western blotting with monoclonal antibodies to mentioned proteins. GST–C/EBPα constructs were generated and GST pull-down assay was performed as described in our papers (Wang et al. 2001, 2002).
Acknowledgments
This work was supported by National Institutes of Health Grants AG20752, CA100070, and GM55188 (NAT). We thank Alana Welm and Estela Medrano for critical review of the paper and for useful suggestions. We thank Xiurong She for excellent technical assistance with Western blotting experiments.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1183304.
References
- Bellacosa, A., Testa, J.R., Staal, S.P., and Tsichlis, P.N. 1991. A retroviral oncogene, akt, encoding a serine–threonine kinase containing an SH2-like region. Science 254: 274–277. [DOI] [PubMed] [Google Scholar]
- Buck, M., Poli, V., Hunter, T., and Chojkier, M. 2001. C/EBPβ phosphorylation by RSK creates a functional XEXD caspase inhibitory box critical for cell survival. Mol. Cell 8: 807–816. [DOI] [PubMed] [Google Scholar]
- He, T.-C., Zhou, S., Costa, L.T., Yu, J., Kinzler, K.W., and Vogelstein, B. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. 95: 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemati, N., Ross, S.E., Erickson, R.L., Groblewski, G.E., and MacDougald, O.A. 1997. Signaling pathway through which insulin regulates CCAAT/enhancer binding protein α (C/EBPα) phosphorylation and gene expression in 3T3-L1 adipocytes. J. Biol. Chem. 272: 25913–25919. [DOI] [PubMed] [Google Scholar]
- Iakova, P., Awad, S.S., and Timchenko, N.A. 2003. Aging reduces proliferative capacities of liver by switching pathways of C/EBPα growth arrest. Cell 113: 495–506. [DOI] [PubMed] [Google Scholar]
- Jiang, Z.Y., Zhou, Q.L., Coleman, K.A., Choinard, M., Does, Q., and Czech, M.P. 2003. Insulin signaling through Akt/protein kinase B analyzed by small interfering RNA-mediated gene silencing. Proc. Natl. Acad. Sci. 100: 7569–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, L.M., Iwama, A., Lodie, T.A., Sasaki, K., Felsher, D.W., Golub, T.R., and Tenen, D.G. 2001. C-myc is a critical target for C/EBPα in granulopoiesis. Mol. Cell. Biol. 21: 3789–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeshan, K., Santili, G., Corradini, F., Perroti, D., and Calabretta, B. 2003. Transcription activation function of C/EBPα is required for induction of granulocytic differentiation. Blood 102: 1267–1275. [DOI] [PubMed] [Google Scholar]
- Lawlor, M.A. and Alessi, D.R. 2001. PBK/Akt: A key mediator of cell proliferation, survival and insulin response. J. Cell Sci. 114: 2903–2910. [DOI] [PubMed] [Google Scholar]
- Lin, Y.-L. and Chou, C.-K. 1998. Phosphatidylinositol 3-kinase is required for the regulation of hepatitis B surface antigen production and mitogen-activated protein kinase activation by insulin but not by TPA. Biochem. Biophys. Res. Commun. 246: 172–175. [DOI] [PubMed] [Google Scholar]
- McKnight, S.L. 2001. McBindall—A better name for CCAAT/enhancer binding proteins? Cell 107: 259–262. [DOI] [PubMed] [Google Scholar]
- Miller, M., Shuman, J.D., Sebastian, T., Dauter, Z., and Johnson P.F. 2003. Structural basis for DNA recognition by the basic region leucine zipper transcription factor CCAAT/enhancer binding protein alpha. J. Biol. Chem. 278: 15178–15184. [DOI] [PubMed] [Google Scholar]
- Pabst, T., Mueller, B.U., Zhang, P., Radomska, H.S., Narravula, S., Schnittger, S., Behre, G., Hiddemann, W., and Tenen, D.G. 2001. Dominant-negative mutations of C/EBPA, encoding CCAAT/enhancer binding protein-α (C/EBPα), in acute myeloid leukemia. Nat. Genet. 27: 263–270. [DOI] [PubMed] [Google Scholar]
- Peng, X., Xu, P.-Z., Chen, M.-L., Hahn-Windgassen, A., Skeen, J., Jacobs, J., Sundararajon, D., Chen, W.S., Crawford, S.E., Coleman, K.G., et al. 2003. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impaired adipogenesis in mice lacking Akt1 and Akt2. Genes & Dev. 17: 1352–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porse, B.T., Pederson, T.A., Xu, X., Lindbergh, B., Wewer, U.M, Fris-Hansen, L., and Nerlov, C. 2001. E2F repression by C/EBPα is required for adipogenesis and granulopoiesis in vivo. Cell 107: 247–258. [DOI] [PubMed] [Google Scholar]
- Ross, S.E., Erickson, R.L., Hemati, N., and MacDougald, O.A. 1999. Glycogen synthase kinase 3 is an insulin-regulated C/EBPα kinase. Mol. Cell. Biol. 19: 8433–8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage, J., Miller, A.L., Perez-Mancera, P.A., Wysocki, J.M., and Jacks, T. 2003. Acute mutations of retinoblastoma function is sufficient to cell cycle re-entry. Nature 10: 223–228. [DOI] [PubMed] [Google Scholar]
- Shamji, A.F., Nghiem, P., and Schreiber, S.L. 2003. Integration of growth factor and nutrient signaling: Implications for cancer biology. Mol. Cell 12: 271–280. [DOI] [PubMed] [Google Scholar]
- Shi, D.-Y., Deng, Y.-R., Liu, S.-L., Zhang, Y.-D., and Wei, L. 2003. Redox stress regulates ell proliferation and apoptosis of human hepatoma through Akt protein phosphorylation. FEBS Lett. 542: 60–64. [DOI] [PubMed] [Google Scholar]
- Shioi, T., McMullen, J.R., Kang, P.M., Douglas, P.S., Obata, T., Franke, T.F., Cantley, L.C., and Izumo, S. 2002. Akt/protein kinase B promotes organ growth in transgenic mice. Mol. Cell. Biol. 22: 2799–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano, H.E., Kang, D.C., Finegold, M.J., Hicks, M.J., Wang, N.-D., Harrison, W., and Darlington, G.J. 1998. Lack of C/EBPα gene expression results in increased DNA synthesis and in an increased frequency of immortalization of freshly isolated rat hepatocytes. Hepatology 27: 392–401. [DOI] [PubMed] [Google Scholar]
- Timchenko, N.A. 2003. Old livers: C/EBPα meets new partners. Cell Cycle 2: 445–446. [DOI] [PubMed] [Google Scholar]
- Timchenko, N.A, Wilde, M., Nakanishi, M., Smith, J.R., and Darlington G.J. 1996. CCAAT/enhancer-binding protein α (C/EBPα) inhibits cell proliferation through the p21 (WAF-/CIP-1/SDI-1) protein. Genes & Dev. 10: 804–815. [DOI] [PubMed] [Google Scholar]
- Timchenko, N.A., Wilde, M., and Darlington, G.J. 1999. C/EBP alpha regulates formation of S-phase-specific E2F–p107 complexes in livers of newborn mice. Mol. Cell. Biol. 19: 2936–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N.D., Finegold, M.J., Bradly, A., Ou, C.N., Abdelsayed, S.V., Wilde, M.D., Taylor, R.L., Wilson, D.R., and Darlington, G.J. 1995. Impaired energy homeostasis in C/EBP alpha knockout mice. Science 269: 1108–1112. [DOI] [PubMed] [Google Scholar]
- Wang, H., Iakova, P., Wilde, M., Welm, A., Goode, T., Roesler, W.J., and Timchenko, N.A. 2001. C/EBPα arrests cell proliferation through direct inhibition of cdk2 and cdk4. Mol. Cell 8: 817–828. [DOI] [PubMed] [Google Scholar]
- Wang, H., Goode, T., Iakova, P., Albrecht, J., and Timchenko, N.A. 2002. C/EBPα triggers proteasome-dependent degradation of cdk4 during growth arrest. EMBO J. 21: 930–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q.-F., Cleaves, R., Kummalue, T., Nerlov, C., and Friedman, A.D. 2003. Cell cycle inhibition mediated by the outer surface of the C/EBPα basic region is required but not sufficient for granulopoiesis. Oncogene 22: 2548–2557. [DOI] [PubMed] [Google Scholar]
- Zheng, L. and Lee, W.H. 2002. Retinoblastoma tumor suppressor and genomic stability. Adv. Cancer Res. 85: 13–50. [DOI] [PubMed] [Google Scholar]
- Zou, W., Li, Z.-Y., Li, Y.-L., Ma, K.-L., and Tsui, Z.-C. 2002. Overexpression of PEMT2 downregulates the PI3K/Akt signaling pathway in rat hepatoma cells. Biochem. Biophys. Acta 1581: 49–56. [DOI] [PubMed] [Google Scholar]