Abstract
Objective
The lymphotoxin-α (LTA), as one of the mediators of inflammation, may play an important role in the pathogenesis of myocardial infarction (MI). Genetic association studies (GAS) that have investigated the association between three common polymorphisms (A252G, G10A and C804A) of the LTA gene and susceptibility to MI have produced contradictory and inconclusive results. The aim of this meta-analysis is to provide a relatively comprehensive account of the association of these polymorphisms with susceptibility to MI.
Methods
A literature search for eligible GAS published before October 15, 2013 was conducted in the PubMed, Embase, Web of Science, Cochrane Library, and CNKI (China National Knowledge Infrastructure) databases. We performed a meta-analysis of fifteen case-control studies with a total of 22,549 MI patients and 16,105 healthy controls.
Results
For LTA A252G, a borderline significant overall association was found, indicating that GG genotype may confer an increased susceptibility to MI compared to AA and AG genotypes. Based on an ethnicity stratification analysis, a significant association was observed in Asians, but not in Caucasians. For LTA G10A, no significant overall association was found. However, subgroup analysis based on ethnicity suggested that the 10A allele may confer a significant increased susceptibility to MI only in Asian populations. For LTA C804A, the combined results revealed a significantly increased susceptibility to MI for carriers of the 804A allele in both overall analysis and stratified analyses.
Conclusion
This meta-analysis shows that LTA C804A may be associated with an increased susceptibility to MI, whereas LTA A252G and G10A may confer a significant increased susceptibility to MI only in Asians. Thus, these polymorphisms of the LTA gene can probably be used with other genetic markers together to identify individuals at high susceptibility to MI especially in Asians.
Introduction
Myocardial infarction (MI) remains the principal cause of death in many countries despite improvements in lifestyle and the development of new pharmacologic approaches [1]. According to data from NHANES (National Health and Nutrition Examination Survey) 2003 to 2006 the overall prevalence of MI is 3.6% in US adults over the age of 20, with rates of 4.7% for men and 2.6% for women [2]. The estimated average number of years of life lost due to MI is 15 [3]. MI, which is widely accepted as a chronic inflammatory disease, usually results from the rupture of atherosclerotic plaque with thrombus formation and the occlusion of the coronary vessel, resulting in an acute reduction of blood supply to a portion of the myocardium [4]. Plaque rupture with thrombosis is well established as a critical factor in the pathogenesis of MI [5]. Inflammatory mediators such as cytokines are involved in atheroma formation and rapid evolution of the atheromatous injury, leading to plaque rupture and MI [6]. Furthermore, epidemiological studies have revealed that MI is a complex multifactorial disease and is clearly influenced by environmental factors and genetic predisposition [7]–[9]. Functionally relevant polymorphisms in genes involved in the inflammatory pathways may cause acute thrombus formation over the plaque with abrupt vessel closure and affect an individual's susceptibility to MI [10].
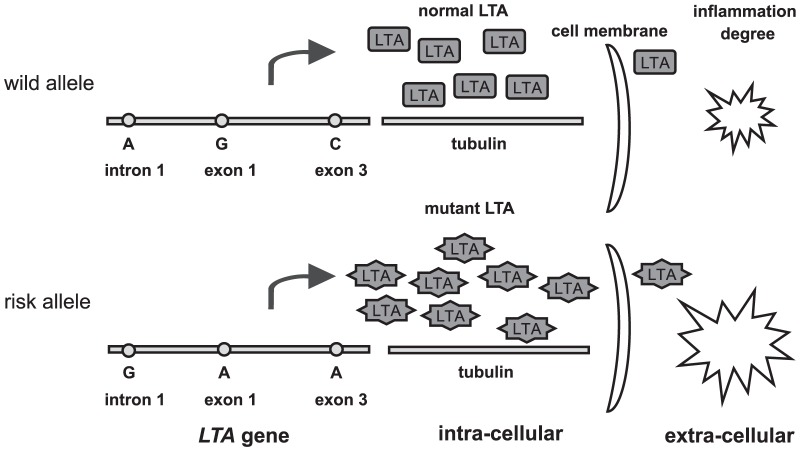
Lymphotoxin-α (LTA) is one of the cytokines produced in the early stages of vascular inflammatory processes [11]. LTA has been implicated in the pathogenesis of atherosclerosis and coronary heart disease (CHD) [12]. Since LTA is an inflammatory mediator, it is likely that functional variations in the gene encoding this protein confer a high susceptibility to MI by affecting the degree of inflammation at the lesion, as shown in Figure 1. Normal LTA protein, trafficking by binding to intra-cellular tubulins, can induce adhesion molecules and cytokines from vascular endothelial cells, vascular smooth-muscle cells, and several kinds of leukocytes, thereby contributing to the inflammatory process [13]. However, these inflammatory biological activities could be influenced by amino-acid substitutions in LTA, such as A-to-C in intron 1, G-to-A in exon 1, and C-to-A in exon 3. Thus, it seems that increased level of functionally mutant LTA protein is associated with increased degree of inflammation, thereby conferring a higher susceptibility to MI.
Figure 1. The potential roles of SNPs in the LTA gene in inflammatory process and the pathogenesis of myocardial infarction.
The LTA gene, which encodes LTA on chromosome 6p21, has been linked with the risk of MI [14]. The single nucleotide polymorphisms (SNPs) in the LTA gene seem to be involved in inflammation by both qualitatively and quantitatively modifying the function of the LTA protein [15]. In this respect, many researchers have tested three common polymorphisms in the LTA gene for genetic association with MI risk, including A252G (dbSNP: rs909253) in intron 1, G10A (dbSNP: rs1800683) in exon 1 and C804A (dbSNP: rs1041981) in exon 3. Their results, however, have proven conflicting. In an initial genome-wide case-control screen involving 65,671 single nucleotide polymorphisms (SNPs) from 13,738 genes in 1,133 MI cases and 1,006 controls in Japan, susceptibility to MI appeared to be associated with the LTA A252G polymorphism [16]. Associations between G10A and C804A and susceptibility to MI have subsequently been observed in some studies [17]–[19], but not others [20]–[23]. To further clarify these inconsistent association findings and to identify possible pathogenic polymorphisms in the LTA gene in relation to MI, we performed a meta-analysis using published data from observational studies.
Materials and Methods
Identification of Relevant Studies
A literature search for GAS that investigated the association between the LTA genetic polymorphisms and susceptibility to MI published before October 15, 2013 was conducted in the following electronic databases: PubMed, Embase, Web of Science, Cochrane Library, and CNKI (China National Knowledge Infrastructure) databases. The following combinations of main keywords were used: (‘lymphotoxin-alpha’ or ‘LTA’ or ‘tumor necrosis factor beta’ or ‘TNF-beta’) and (‘myocardial infarction’ or ‘myocardial infarct’ or ‘MI’) and (‘genetic polymorphism’ or ‘single nucleotide polymorphisms’ or ‘SNP’). The search was done without limitations on language but only included those studies that were conducted on human subjects. All references in eligible articles were extensively reviewed to identify additional published articles.
Inclusion and Exclusion Criteria
To be included in the analysis, eligible studies had to meet the following criteria: (1) case-control studies on the association between the LTA genetic polymorphisms and susceptibility to MI; (2) all patients in the candidate studies meet the diagnostic criteria for MI; (3) studies with sufficient available data to calculate ORs with corresponding 95%CIs. The major reasons for excluding studies were: (1) not case-control study; (2) duplicate publications with overlapping subjects from the same study; and (3) no available data reported. For multiple studies using overlapping cases or controls, the most recent study with the largest sample size was included in the meta-analysis. This meta-analysis was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidance with only slight modification, and did not require ethics board approval (Supplement S1) [24].
Data Extraction
According to the PRISMA guidance, two investigators independently checked each full-text report for eligibility and extracted the following data from eligible studies: surname of first author, year of publication, country of origin, ethnicity, definition and number of case and control, age, sex ratio, genotyping method, allele and genotype frequency, etc. Disagreements were solved by discussion between all authors until consensus was reached. For data not provided in table form or in the main text, required information was obtained by contacting corresponding authors when possible.
Quality Assessment
The strengthening report of genetic association studies (STREGA) quality score system and the Newcastle-Ottawa Scale (NOS) criteria were used to assess the qualities of all included studies [25], [26]. The STREGA system includes twenty-two quality assessment items with scores ranging from 0 to 22 (Supplement S2). Studies are classified into three levels based on their scores: low quality (0–12), moderate-high quality (13–17), and high quality (18–22). The NOS criteria use a “star” rating system to judge methodological quality based on three aspects of a study: selection, comparability, and exposure (Supplement S3). Scores range from 0 stars (worst) to 9 stars (best), with a score of 5 or higher indicating a moderate-high methodological quality. Two authors independently assessed the quality of included studies. Discrepancies over quality scores were resolved by discussion with all authors and subsequent consensus.
Statistical Analysis
Genotype distributions in the controls were tested for conformation to Hardy-Weinberg equilibrium (HWE) using the chi-square test. HWE in the controls was tested by comparing the expected and observed genotype frequencies using the Pearson chi-square test for goodness of fit. The association between the LTA genetic polymorphisms and susceptibility to MI was assessed by the pooled odds ratios (ORs) with their corresponding 95% confidence intervals (95%CIs) under five genetic models, including the allele model, the dominant model, the recessive model, the homozygous model and the heterozygous model. Taking into consideration possible between-study heterogeneity, a statistical test for heterogeneity was first conducted using Cochran's Q statistic and the I2 metric [27], [28]. We considered the presence of significant heterogeneity at the 10% level of significance and values of I2 exceeding 50% as an indicator of significant heterogeneity. When no heterogeneity was found with P>0.10 or I2<50%, a fixed-effects model was used to estimate the pooled ORs and 95%CIs. Otherwise, a random-effects model was applied. In addition to an overall comparison, stratified analyses, based on ethnicity, source of control, HWE status, and genotyping method where applicable, were also performed to explore possible explanations of between-study heterogeneity and to investigate whether overall reported associations were present in subgroups. Univariate and multivariate meta-analyses were conducted to explore potential sources of heterogeneity, considering publication year, ethnicity, genotyping method, source of control, and quality score. Sensitivity analyses were conducted by omitting individual studies in turn to reflect the influence of individual datasets on the pooled results [29]. Begg's funnel plot and Egger's linear regression test were used to assess the potential for publication bias [30], [31]. All two-tailed P<0.05 were considered statistically significant, and all analyses were performed using the STATA 12.0 software (Stata Corp., College Station, TX, USA).
Results
Baseline Characteristics of Included Studies
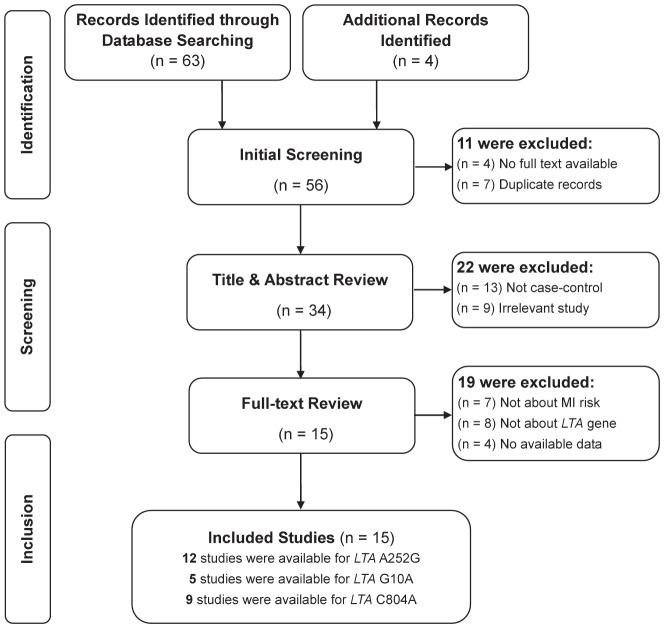
Figure 2 presents a flow chart of retrieved and excluded studies with their reasons for exclusion. A total of 67 relevant papers were identified using the pre-specified search strategy. In accordance with the inclusion criteria, fifteen case-control studies [14], [16]–[23], [32]–[37] were included in this meta-analysis, with a total of 22,549 MI patients and 16,105 healthy controls. Of the included studies, twelve examined associations with A252G (one study reported data separately for Japanese and Korean), five examined associations with G10A, and nine examined associations with C804A. Studies were conducted in two major ethnic populations, with seven on Asians and eight on Caucasians. The publication years of included studies ranged from 2000 to 2010. All studies in this meta-analysis had controls in HWE, except one study conducted by Ozaki et al., which was excluded in further stratified analyses. The quality scores of all included studies were moderate-high, with STREGA scores higher than 13 and NOS stars more than 5. The characteristics and methodological quality of all included studies are summarized in Table 1.
Figure 2. Flow diagram of the selection of studies and specific reasons for exclusion from the present meta-analysis.
Table 1. Main characteristics and methodological quality of all eligible studies.
| First author, year | Country | Ethnicity | No. of cases, age(y) | No. of controls, age(y) | Male (%) [Case/Control] | Genotyping method | Variant(s) | STREGA score | NOS star |
| Padovani JC, 2000 | Brazil | Caucasian | 148, mean 43 range (25–55) | 148, mean 42 range (22–55) | 82.5%, 82.5% | PCR-RFLP | A252G | 19/22 | 8/9 |
| Koch W, 2001 | Germany | Caucasian | 793, mean 62.6 (SD 10.2) | 340, mean 63.4 (SD 10.3) | 77.4%, 75.3% | AS-PCR | A252G | 17/22 | 7/9 |
| Iwanaga Y, 2004 | Japan | Asian | 477, mean 56 (SD 8) | 372, mean 59 (SD 9) | 100%, 100% | TaqMan | A252G, G10A, C804A | 18/22 | 7/9 |
| Tobin MD, 2004 | UK | Caucasian | 547, mean 61.9 (SD 9.2) | 505, mean 58.6 (SD 10.7) | 68%, 62% | PCR-RFLP | C804A | 16/22 | 6/9 |
| Yamada A, 2004 | Japan | Asian | 1891, mean 60.6 (SD 10.9) | 1798, mean 58.6 (SD 11.3) | 78.9%, 55.2% | AS-PCR | A252G, C804A | 17/22 | 7/9 |
| Ozaki K, 2005 | Japan | Asian | 1133, mean 62.5 (SD 11.3) | 1006, mean 64.3 (SD 11.3) | NA, NA | PCR-RFLP | A252G, G10A, C804A | 20/22 | 8/9 |
| Clarke R, 2006 | UK | Caucasian | 6928, mean 54.8 (SD 7.3) | 2712, mean 46.2 (SD 9.6) | 82.3%, 44.6% | TaqMan | A252G, G10A, C804A | 19/22 | 8/9 |
| Tanaka T, 2006 | Japan | Asian | 2833, NA | 3399, NA | NA, NA | PCR-RFLP | C804A | 15/22 | 6/9 |
| Kimura A, 2007 | Japan | Asian | 533, mean 59.4 (SD 10.1) | 683, mean 43.3 (SD 10.9) | 83.3%, 67.7% | PCR-RFLP | A252G | 17/22 | 7/9 |
| Kimura A, 2007 | Korea | Asian | 448, mean 61.3 (SD 11.9) | 160, mean 61.3 (SD 8.8) | 82.4%, 83.5% | PCR-RFLP | A252G | 17/22 | 7/9 |
| Koch W, 2007 | Germany | Caucasian | 3657, mean 64.0 (SD 12.0) | 1211, mean 60.3 (SD 11.9) | 75.8%, 50.0% | TaqMan | A252G, G10A, C804A | 19/22 | 8/9 |
| Sedlacek K, 2007 | Germany | Caucasian | 1821, mean 58.7 (SD 8.6) | 2572, mean 57.4 (SD 9.7) | 78.0%, 43.2% | TaqMan | G10A, C804A | 18/22 | 7/9 |
| Panoulas VF, 2008 | UK | Caucasian | 388, mean 61.5 (SD 12.0) | 399, mean 50.2 (SD 15.8) | 26.8%, 39.6% | PCR-MCA | A252G | 16/22 | 6/9 |
| Ryan AW, 2008 | Irish | Caucasian | 835, mean 61.9 (SD 8.5) | 691, mean 52.4 (SD 10.0) | NA, NA | PCR-RFLP | A252G | 15/22 | 6/9 |
| Wang YC, 2010 | Taiwan | Asian | 117, NA | 109, NA | NA, NA | AS-PCR | A252G, C804A | 14/22 | 5/9 |
NA, Not available; SD, Standard deviate; PCR-RFLP, Polymerase chain reaction-restriction fragment length polymorphism; AS-PCR, Allele specific-polymerase chain reaction; PCR-MCA, Polymerase chain reaction-melting curve analysis; STREGA, Strengthening the reporting of genetic association studies; NOS, Newcastle-Ottawa scale.
Association between the LTA A252G Polymorphism and Susceptibility to MI
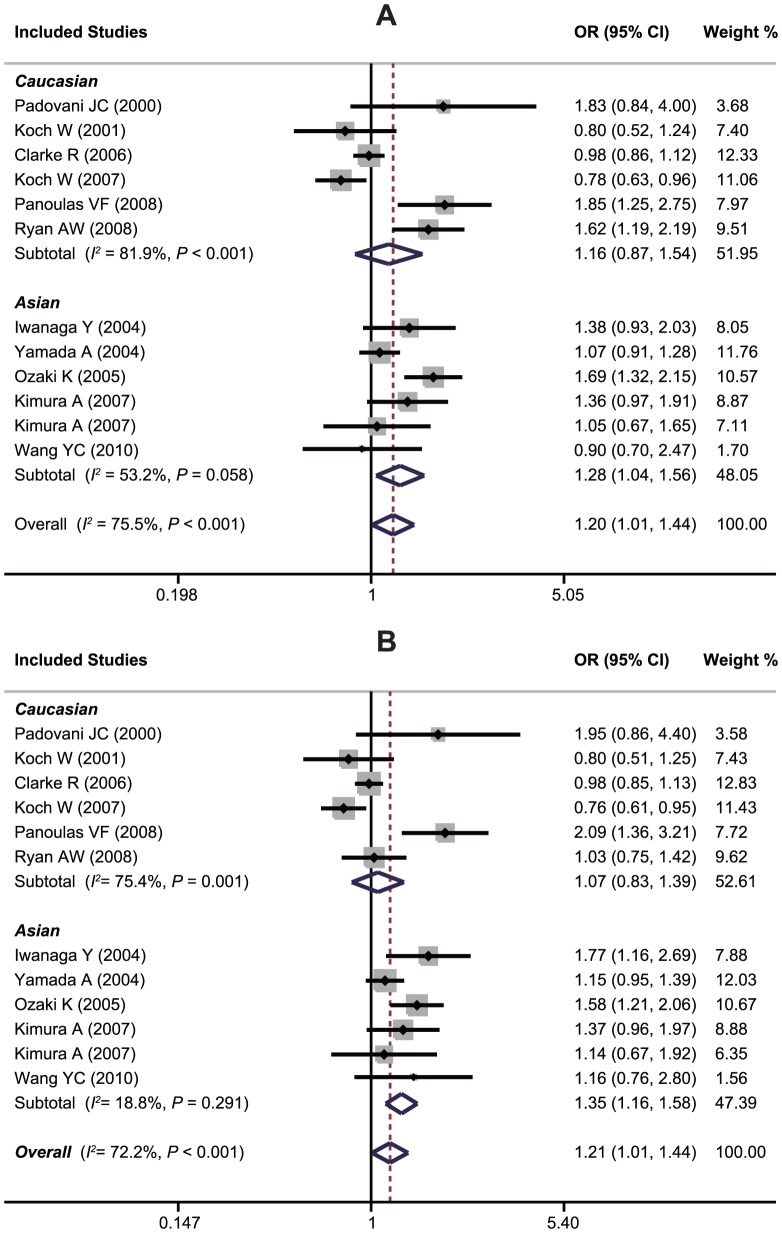
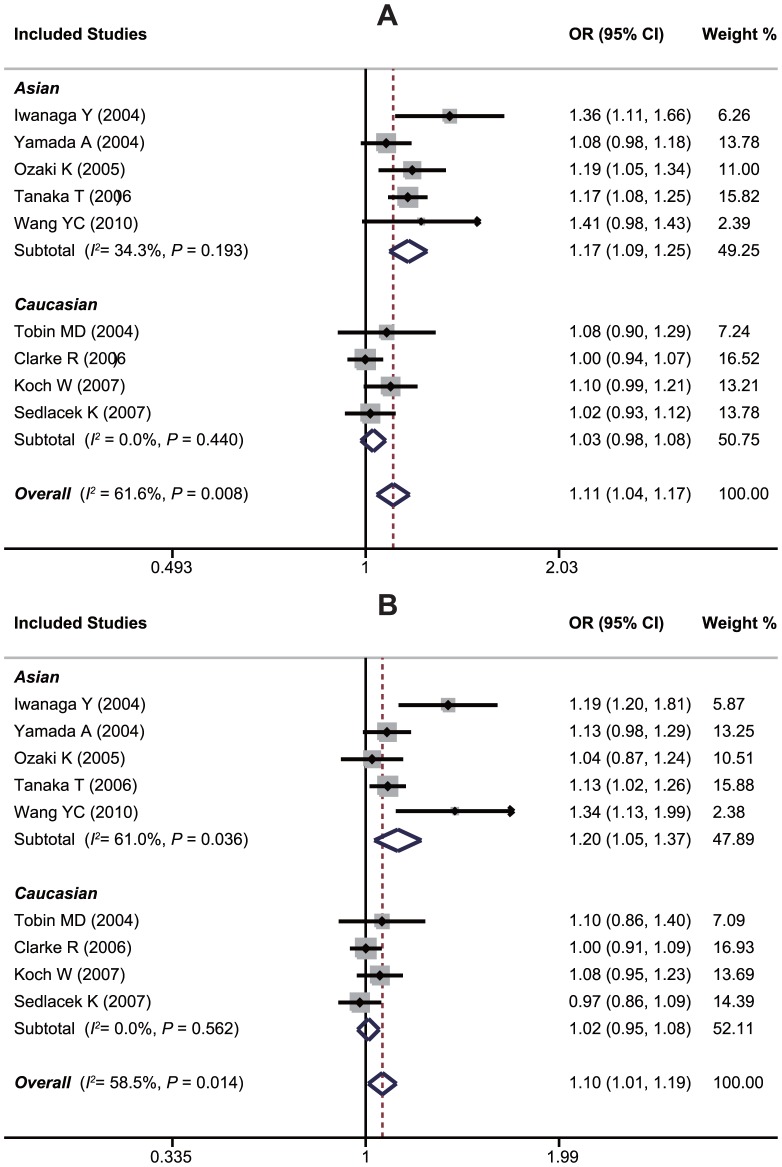
An evaluation of the association between the LTA A252G polymorphism and susceptibility to MI is summarized in Table 2. Twelve case-control studies investigated the relationship between A252G and susceptibility to MI with a total of 17,348 MI patients and 9,629 healthy controls. Since between-study heterogeneity obviously existed (P<0.10 and I2>50% under all genetic models), the random-effects model was used. As shown in Table 2, a borderline significant overall association was found, which indicates that GG genotype may confer an increased susceptibility to MI compared with AA and AG genotypes under recessive and homozygous genetic models (GG vs. AA+AG: OR = 1.20, 95%CI: 1.01–1.44, P = 0.040; GG vs. AA: OR = 1.21, 95%CI: 1.01–1.45, P = 0.039). However, exclusion of one hospital-based study by Yamada et al. made the summary ORs in population-based studies become insignificant, and after excluding of one non-HWE study by Ozaki et al., the pooled ORs in HWE studies also became insignificant, suggesting that the A252G polymorphism is unlikely to have a major role in the risk of MI. Interestingly, in a stratified analysis by ethnicity, we found that this polymorphism played different roles in Asian and Caucasian populations. In Asians, subjects harboring the 252G variant are approximately 20% more likely to have a MI when compared to subjects with the 252A allele (G allele vs. A allele: OR = 1.16, 95%CI: 1.07–1.26, P<0.001; AG+GG vs. AA: OR = 1.26, 95%CI: 1.04–1.54, P = 0.020; GG vs. AA+AG: OR = 1.28, 95%CI: 1.04–1.56, P = 0.018; GG vs. AA: OR = 1.35, 95%CI: 1.16–1.58, P<0.001). However, no significant effects were found for the Caucasians (all P>0.05) (Figure 3). Stratification based on genotyping method showed significant associations between LTA A252G and susceptibility to MI in the PCR-RFLP subgroup (GG vs. AA+AG: OR = 1.52, 95%CI: 1.30–1.77, P<0.001; GG vs. AA: OR = 1.33, 95%CI: 1.09–1.62, P = 0.005; GG vs. AA: OR = 1.69, 95%CI: 1.16–1.58, P<0.001), whereas no significant association was found in the non-PCR-RFLP subgroup (all P>0.05). Univariate and multivariate meta-analyses further confirmed the above results of subgroup analysis, which suggested that ethnicity might be the major source of between-study heterogeneity with 87% was explained by this covariate (Supplement S4).
Table 2. Meta-analysis of the association between LTA A252G and the risk of myocardial infarction (MI).
| Subgroups | No. of study | G allele vs. A allele | AG+GG vs. AA | GG vs. AA+AG | GG vs. AA | GG vs. AG | ||||||||||
| (Case/Control) | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | |
| Overall | 12 (17,348/9,629) | 1.08 | 0.99–1.18 | 0.082 | 1.08 | 0.93–1.26 | 0.306 | 1.20 | 1.01–1.44 | 0.040 | 1.21 | 1.01–1.45 | 0.039 | 1.23 | 0.98–1.55 | 0.069 |
| Ethnicity | ||||||||||||||||
| Caucasian | 6 (12,749/5,501) | 1.00 | 0.88–1.13 | 0.970 | 0.93 | 0.75–1.16 | 0.534 | 1.16 | 0.88–1.54 | 0.303 | 1.07 | 0.83–1.39 | 0.607 | 1.30 | 0.87–1.94 | 0.201 |
| Asian | 6 (4,599/4,128) | 1.16 | 1.07–1.26 | <0.001 | 1.26 | 1.04–1.54 | 0.020 | 1.28 | 1.04–1.56 | 0.018 | 1.35 | 1.16–1.58 | <0.001 | 1.21 | 0.94–1.55 | 0.142 |
| Source of control | ||||||||||||||||
| PB | 11 (15,457/7,831) | 1.09 | 0.98–1.20 | 0.113 | 1.09 | 0.91–1.29 | 0.358 | 1.22 | 0.99–1.51 | 0.056 | 1.23 | 0.99–1.51 | 0.058 | 1.26 | 0.97–1.64 | 0.088 |
| HB | 1 (1,891/1,798) | 1.08 | 0.98–1.19 | 0.103 | 1.13 | 0.98–1.29 | 0.083 | 1.08 | 0.91–1.28 | 0.409 | 1.15 | 0.95–1.39 | 0.155 | 1.03 | 0.86–1.23 | 0.789 |
| HWE status | ||||||||||||||||
| HWE | 11 (16,215/8,623) | 1.08 | 0.98–1.18 | 0.141 | 1.10 | 0.92–1.30 | 0.295 | 1.15 | 0.97–1.36 | 0.105 | 1.16 | 0.97–1.40 | 0.100 | 1.18 | 0.94–1.49 | 0.149 |
| Non-HWE | 1 (1,133/1,006) | 1.16 | 1.03–1.31 | 0.019 | 1.02 | 0.85–1.22 | 0.838 | 1.69 | 1.32–2.15 | <0.001 | 1.58 | 1.21–2.06 | 0.001 | 1.77 | 1.37–2.29 | <0.001 |
| Genotyping method | ||||||||||||||||
| PCR-RFLP | 5 (3,097/2,688) | 1.06 | 0.90–1.25 | 0.506 | 0.93 | 0.66–1.32 | 0.699 | 1.52 | 1.30–1.77 | <0.001 | 1.33 | 1.09–1.62 | 0.005 | 1.69 | 1.16–2.44 | 0.006 |
| Non-PCR-RFLP | 7 (14,251/6,941) | 1.10 | 0.98–1.23 | 0.105 | 1.18 | 1.00–1.40 | 0.049 | 1.05 | 0.87–1.27 | 0.639 | 1.14 | 0.90–1.45 | 0.289 | 0.99 | 0.85–1.16 | 0.915 |
OR, Odd ratio; 95%CI, 95% confidence interval; HB, Hospital-based; PB, Population-based; HWE, Hardy-Weinberg equilibrium; PCR-RFLP, Polymerase chain reaction-restriction fragment length polymorphism.
Figure 3. Forest plots of ORs for the association between the LTA A252G polymorphism and susceptibility to myocardial infarction in subgroup analysis based on ethnicity under the recessive model (A) and the homozygous model (B).
Association between the LTA G10A Polymorphism and Susceptibility to MI
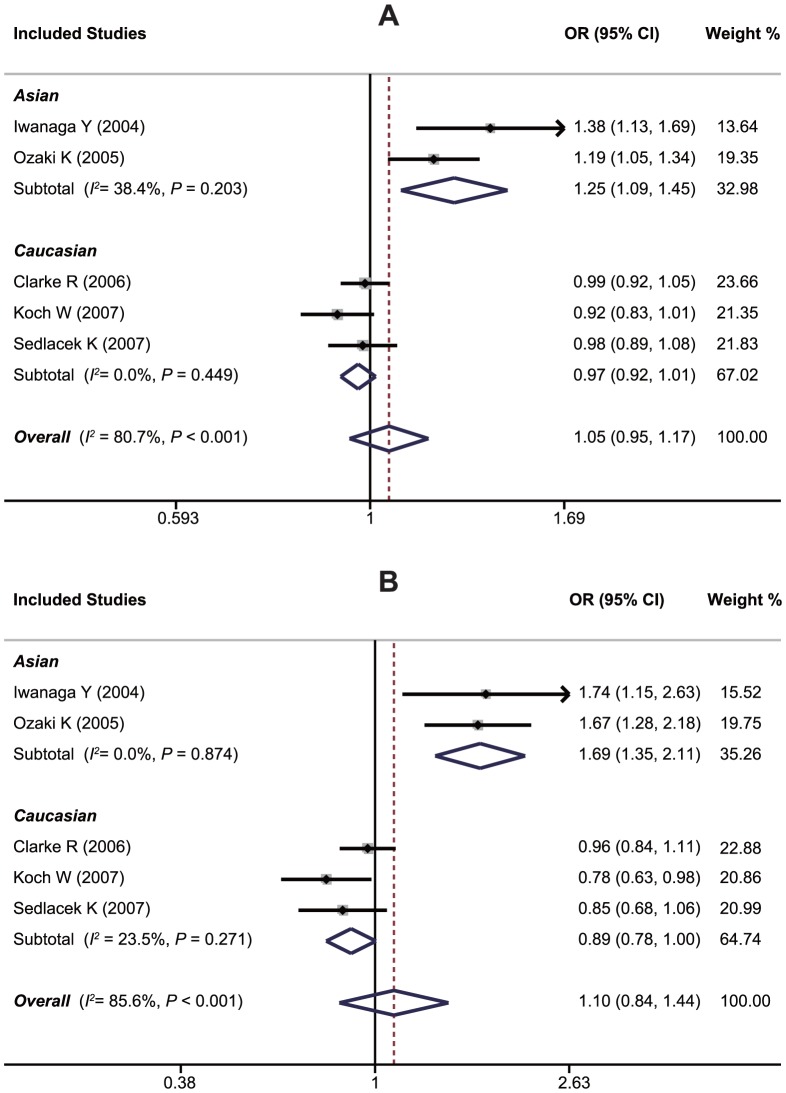
A summary of findings on the relationship between the LTA G10A polymorphism and susceptibility to MI is provided in Table 3. Data from five studies, in total compromised of 14,653 MI cases and 7,873 healthy controls, were pooled together for analysis. The random-effects model was conducted since heterogeneity obviously existed (P<0.10 and I2>50% under all genetic models). The overall analysis showed that LTA G10A may not be associated with the risk of MI under any of the five genetic models (A allele vs. G allele: OR = 1.05, 95%CI: 0.95–1.17, P = 0.349; GA+AA vs. GG: OR = 1.01, 95%CI: 0.96–1.07, P = 0.651; AA vs. GG+GA: OR = 1.07, 95%CI: 0.82–1.41, P = 0.618; AA vs. GG: OR = 1.10, 95%CI: 0.84–1.44, P = 0.498; AA vs. GA: OR = 1.00, 95%CI: 0.91–1.10, P = 0.975). Exclusion of one non-HWE study by Ozaki et al. did not make the summary ORs significant in HWE studies. However, stratified analyses based on ethnicity suggested that the 10A allele may confer a significantly increased susceptibility to MI in Asians (A allele vs. G allele: OR = 1.25, 95%CI: 1.09–1.45, P = 0.002; GA + AA vs. GG: OR = 1.17, 95%CI: 1.01–1.36, P = 0.034; AA vs. GG + GA: OR = 1.63, 95%CI: 1.27–2.08, P<0.001; AA vs. GG: OR = 1.69, 95%CI: 1.35–2.11, P<0.001; AA vs. GA: OR = 1.61, 95%CI: 1.30–2.00, P<0.001), but not in Caucasians, as shown in Figure 4. Due to insufficient observations, meta-regression analyses were not conducted for this SNP.
Table 3. Meta-analysis of the association between LTA G10A and myocardial infarction risk.
| Genetic model | Subgroup | No. of study (CA/CO) | OR [95% CI] | POR | Ph | Method |
| A allele vs. G allele | Overall | 5 (14,653/7,873) | 1.05 [0.95, 1.17] | 0.349 | <0.001 | RE |
| (Allele model) | Asian | 2 (1,610/1,378) | 1.25 [1.09, 1.45] | 0.002 | 0.203 | FE |
| Caucasian | 3 (12,406/6,495) | 0.97 [0.92, 1.02] | 0.179 | 0.449 | FE | |
| HWE | 4 (12,883/6,867) | 1.02 [0.92, 1.13] | 0.749 | 0.004 | RE | |
| Non-HWE | 1 (1,133/1,006) | 1.19 [1.05, 1.34] | 0.007 | - | FE | |
| GA + AA vs. GG | Overall | 5 (14,653/7,873) | 1.01 [0.96, 1.07] | 0.651 | 0.014 | RE |
| (Dominant model) | Asian | 2 (1,610/1,378) | 1.17 [1.01, 1.36] | 0.034 | 0.010 | RE |
| Caucasian | 3 (12,406/6,495) | 0.99 [0.93, 1.05] | 0.682 | 0.520 | FE | |
| HWE | 4 (12,883/6,867) | 1.01 [0.95, 1.08] | 0.737 | 0.006 | RE | |
| Non-HWE | 1 (1,133/1,006) | 1.04 [0.87, 1.24] | 0.682 | - | FE | |
| AA vs. GG + GA | Overall | 5 (14,653/7,873) | 1.07 [0.82, 1.41] | 0.618 | <0.001 | RE |
| (Recessive model) | Asian | 2 (1,610/1,378) | 1.63 [1.27, 2.08] | <0.001 | 0.255 | FE |
| Caucasian | 3 (12,406/6,495) | 0.88 [0.78, 1.00] | 0.051 | 0.222 | FE | |
| HWE | 4 (12,883/6,867) | 0.92 [0.78, 1.09] | 0.350 | 0.062 | RE | |
| Non-HWE | 1 (1,133/1,006) | 1.78 [1.39, 2.27] | <0.001 | - | FE | |
| AA vs. GG | Overall | 5 (14,653/7,873) | 1.10 [0.84, 1.44] | 0.498 | <0.001 | RE |
| (Homozygous model) | Asian | 2 (1,610/1,378) | 1.69 [1.35, 2.11] | <0.001 | 0.874 | FE |
| Caucasian | 3 (12,406/6,495) | 0.89 [0.78, 1.00] | 0.059 | 0.271 | FE | |
| HWE | 4 (12,883/6,867) | 0.97 [0.78, 1.22] | 0.799 | 0.008 | RE | |
| Non-HWE | 1 (1,133/1,006) | 1.67 [1.28, 2.18] | <0.001 | - | FE | |
| AA vs. GA | Overall | 5 (14,653/7,873) | 1.00 [0.91, 1.10] | 0.975 | <0.001 | RE |
| (Heterozygous model) | Asian | 2 (1,610/1,378) | 1.61 [1.30, 2.00] | <0.001 | 0.035 | RE |
| Caucasian | 3 (12,406/6,495) | 0.89 [0.80, 0.99] | 0.030 | 0.219 | FE | |
| HWE | 4 (12,883/6,867) | 0.90 [0.79, 1.02] | 0.087 | 0.252 | FE | |
| Non-HWE | 1 (1,133/1,006) | 1.87 [1.44, 2.41] | <0.001 | - | FE |
CA, Case group; CO, Control group; OR, Odd ratio; 95%CI, 95% confidence interval; Ph, P value of heterogeneity; HWE, Hardy-Weinberg equilibrium; RE, Random-effect model; FE, Fix-effect model.
Figure 4. Forest plots of ORs for the association between the LTA G10A polymorphism and susceptibility to myocardial infarction in subgroup analysis based on ethnicity under the allele model (A) and the homozygous model (B).
Association between the LTA C804A Polymorphism and Susceptibility to MI
Table 4 summarizes the association between the LTA C804A polymorphism and susceptibility to MI. Nine case-control studies had investigated the relationship between C804A and susceptibility to MI with a total of 19,404 MI patients and 13,684 healthy controls. Since between-study heterogeneity obviously existed (P<0.10 and I2>50% under all genetic models), the random-effects model was used. Overall, the LTA C804A polymorphism was found to be associated with a significant increased susceptibility to MI (A allele vs. C allele: OR = 1.11, 95%CI: 1.04–1.17, P = 0.001; CA+AA vs. CC: OR = 1.10, 95%CI: 1.01–1.19, P = 0.020; AA vs. CC+CA: OR = 1.24, 95%CI: 1.08–1.41, P = 0.002; AA vs. CC: OR = 1.21, 95%CI: 1.01–1.45, P<0.001; AA vs. CA: OR = 1.21, 95%CI: 1.04–1.39, P = 0.012). As shown in Table 4, neither the exclusion of one hospital-based study by Yamada et al. nor of one non-HWE study by Ozaki et al. changed the summary ORs significantly in population-based studies and HWE studies. Like the other two variants of the LTA gene involved in the current study, stratified analysis based on ethnicity revealed a significant association in Asians, but not in Caucasians (Figure 5). Stratification based on genotyping method showed a significant association between LTA C804A and susceptibility to MI in the PCR-RFLP subgroup (A allele vs. C allele: OR = 1.16, 95%CI: 1.10–1.23, P<0.001; CA+AA vs. CC: OR = 1.11, 95%CI: 1.02–1.20, P = 0.018; AA vs. CC+CA: OR = 1.45, 95%CI: 1.17–1.79, P = 0.001; AA vs. CC: OR = 1.46, 95%CI: 1.26–1.69, P<0.001; AA vs. CA: OR = 1.44, 95%CI: 1.11–1.86, P = 0.006), whereas no significant association was found in the other subgroups (all P>0.05). Similar to A252G, univariate and multivariate meta-analyses also suggested that ethnicity might be the major source of between-study heterogeneity for C804A with 83% was explained by this covariate (Supplement S4).
Table 4. Meta-analysis of the association between LTA C804A and the risk of myocardial infarction (MI).
| Subgroups | No. of study | A allele vs. C allele | CA+AA vs. CC | AA vs. CC+CA | AA vs. CC | AA vs. CA | ||||||||||
| (Case/Control) | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | |
| Overall | 9 (19,404/13,684) | 1.11 | 1.04–1.17 | 0.001 | 1.10 | 1.01–1.19 | 0.020 | 1.24 | 1.08–1.41 | 0.002 | 1.21 | 1.01–1.45 | <0.001 | 1.21 | 1.04–1.39 | 0.012 |
| Ethnicity | ||||||||||||||||
| Caucasian | 4 (12,953/7,000) | 1.03 | 0.98–1.08 | 0.234 | 1.02 | 0.95–1.08 | 0.628 | 1.11 | 0.98–1.26 | 0.097 | 1.27 | 1.11–1.45 | 0.099 | 1.11 | 0.98–1.26 | 0.113 |
| Asian | 5 (6,451/6,684) | 1.17 | 1.09–1.25 | <0.001 | 1.20 | 1.05 –1.37 | 0.007 | 1.35 | 1.10–1.66 | 0.004 | 1.41 | 1.20–1.66 | <0.001 | 1.28 | 1.00–1.65 | 0.052 |
| Source of control | ||||||||||||||||
| PB | 8 (17,513/11,886) | 1.11 | 1.04–1.19 | 0.003 | 1.10 | 1.00–1.20 | 0.043 | 1.27 | 1.09–1.48 | 0.002 | 1.29 | 1.11–1.51 | 0.001 | 1.24 | 1.06–1.46 | 0.009 |
| HB | 1 (1,891/1,798) | 1.08 | 0.98–1.18 | 0.113 | 1.13 | 0.98–1.29 | 0.083 | 1.07 | 0.90–1.27 | 0.459 | 1.14 | 0.94–1.38 | 0.174 | 1.02 | 0.85–1.22 | 0.857 |
| HWE status | ||||||||||||||||
| HWE | 8 (18,271/12,678) | 1.10 | 1.03–1.17 | 0.005 | 1.11 | 1.02–1.21 | 0.021 | 1.18 | 1.05–1.32 | 0.006 | 1.22 | 1.07–1.39 | 0.002 | 1.14 | 1.02–1.28 | 0.024 |
| Non-HWE | 1 (1,133/1,006) | 1.19 | 1.05–1.34 | 0.007 | 1.04 | 0.87–1.24 | 0.689 | 1.78 | 1.39–2.27 | <0.001 | 1.67 | 1.28–2.18 | <0.001 | 1.87 | 1.44–2.41 | <0.001 |
| Genotyping method | ||||||||||||||||
| PCR-RFLP | 3 (4,513/4,910) | 1.16 | 1.10–1.23 | <0.001 | 1.11 | 1.02–1.20 | 0.018 | 1.45 | 1.17–1.79 | 0.001 | 1.46 | 1.26–1.69 | <0.001 | 1.44 | 1.11–1.86 | 0.006 |
| TaqMan | 4 (12,883/6,867) | 1.07 | 0.98–1.18 | 0.132 | 1.08 | 0.95–1.23 | 0.255 | 1.14 | 0.99–1.30 | 0.072 | 1.18 | 0.99–1.42 | 0.069 | 1.11 | 0.97–1.26 | 0.120 |
| AS-PCR | 2 (2,008/1,907) | 1.16 | 0.92–1.47 | 0.215 | 1.36 | 0.85–2.16 | 0.196 | 1.07 | 0.90–1.26 | 0.455 | 1.15 | 0.95–1.39 | 0.151 | 1.01 | 0.85–1.21 | 0.891 |
OR, Odd ratio; 95%CI, 95% confidence interval; HB, Hospital-based; PB, Population-based; HWE, Hardy-Weinberg equilibrium; PCR-RFLP, Polymerase chain reaction-restriction fragment length polymorphism.
Figure 5. Forest plots of ORs for the association between the LTA C804A polymorphism and susceptibility to myocardial infarction in subgroup analysis based on ethnicity under the allele model (A) and the dominant model (B).
Sensitivity Analyses and Publication Bias
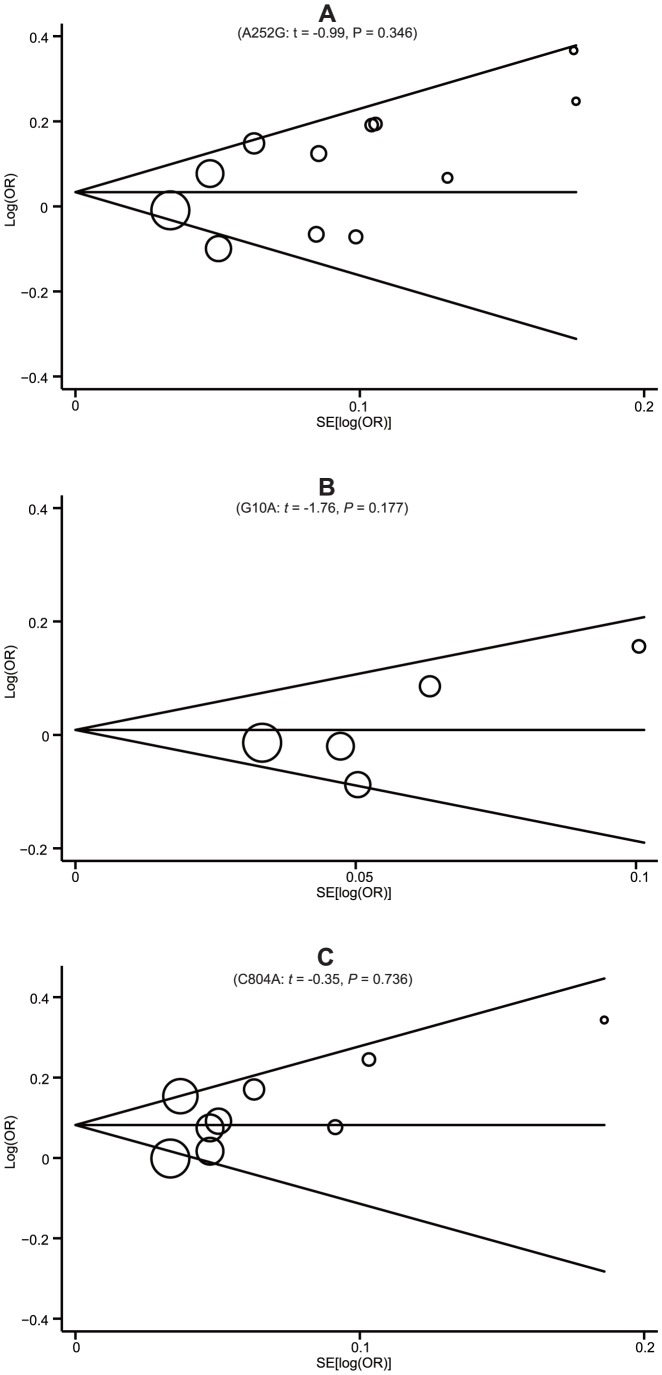
Sensitivity analyses for LTA A252G, G10A and C804A were conducted to determine the influence of individual datasets on the pooled ORs by sequential removal of each eligible study. Omission of one study at a time reveals that the pooled estimates remain virtually the same with each study excluded, indicating that no single study heavily influenced summary ORs in this meta-analysis (data not shown). Begg's funnel plots and Egger's linear regression test were used to assess the potential publication bias of included studies under the allele model. The shapes of the funnel plots did not reveal any evidence of obvious asymmetry (Figure 6). In addition, Egger's test did not show any statistical evidence of publication bias (A252G: t = −0.99, P = 0.346; G10A: t = −1.76, P = 0.177; C804A: t = −0.35, P = 0.736). The above tests indicated a promising level of robustness and accuracy for the results of this meta-analysis.
Figure 6. Begg's funnel plots of publication bias for the associations the LTA A252G (A), G10A (B) and C804A (C) polymorphisms and susceptibility to myocardial infarction under the allele model.
Discussion
MI is a multifactorial disease significantly associated with certain genetic factors [38]. Many genome association and candidate gene studies have been conducted to identify MI-susceptible genes, discover novel molecular pathways and help susceptible individuals prevent the development of MI [16], [39]–[41]. To date, several genetic association studies (GAS) have found that SNPs in the LTA gene and the LTA protein are associated with MI onset. LTA is a proinflammatory cytokine with homology to inflammatory cytokine tumor necrosis factor alpha (TNF-α) and is related with the development of atherosclerotic lesions in coronary arteries [42]. Common SNPs of the LTA gene can modify the function of the LTA protein qualitatively and quantitatively, thereby conferring a risk for MI, as shown in Figure 1. Amino-acid substitutions in the LTA gene, such as A-to-C in intron 1, G-to-A in exon 1, and C-to-A in exon 3, may influence the inflammatory biological activities through causing functional mutations in LTA protein, which in turn affect the degree of inflammation and confer a higher susceptibility to MI than normal LTA protein. Based on this fact, several investigations have been conducted to assess the association between common LTA genetic polymorphisms and susceptibility to MI. Previously, Padovani et al. first investigated the association between LTA A252G and susceptibility to MI in a Brazilian population, but failed to find a significant association [32]. Koch et al. conducted a similar study in a Germany population, and again found no significant results [33]. However, in subsequent studies on Asians, both Iwanaga et al. and Ozaki et al. reported strong evidence for the influence of LTA genetic polymorphisms on susceptibility to MI in Japanese populations [34], [37]. Furthermore, Wang et al. observed a significant effect of LTA A252G and C804A on susceptibility to MI in a Taiwanese population [19], whereas Kimura et al. did not find any effect in a Korean population [20]. The discrepancy of these studies is partly due to their limited sample sizes and insufficient statistical power to demonstrate significant associations. In addition, these studies included different populations and sampling strategies, making their results difficult to interpret. Thus, in the present study, we performed a meta-analysis to derive a relatively comprehensive assessment of the relationships between the LTA A252G, G10A and C804A polymorphisms and susceptibility to MI.
Meta-analysis has the advantage of synthesizing data from published GAS to obtain greater statistical power for detecting significant associations than available in an individual GAS, especially in the absence of large heterogeneity between studies [43]. A large number of meta-analyses have been conducted to investigate the association between the LTA gene and various diseases, including gastric cancer [44], [45], breast cancer [46], asthma [47], sepsis [48], migraine [49], and lymphoma [50]. To the best of our knowledge, our study is the first meta-analysis to describe the associations of the LTA genetic polymorphisms with susceptibility to MI. This systematic review provides a more comprehensive summary of the currently available evidence on the associations between the LTA A252G, G10A and C804A polymorphisms and susceptibility to MI. In this meta-analysis, the LTA C804A polymorphisms seem to be associated with an increased susceptibility to MI, whereas LTA A252G and G10A may confer a significant increased susceptibility to MI only in Asians.
Heterogeneity is a major problem when interpreting the results of meta-analyses. As the results of meta-regression shows, ethnicity was an important source of this heterogeneity as individuals of different ethnicities may have diverse genetic backgrounds and environmental factors, and as a result the same polymorphism may play different roles in different populations. Interestingly, our subgroup analysis by ethnicity showed that the LTA genetic polymorphisms played different roles in Asian and Caucasian populations. All these LTA genetic polymorphism had significant increased effects on the risk of MI in Asians, whereas no significant effects were found in Caucasians. These conflicting results may be due to the different genetic backgrounds of these populations, subsequently leading to different genetic susceptibility to the same disease. In addition, the source of controls was another factor that contributed to heterogeneity. The genotype distribution in population-based controls may be similar to normal and thus population-based controls could be more reliable than hospital-based controls. This might partially explain why the results of the stratified analysis by the source of the controls showed differences between the two subgroups for LTA A252G. Thus, further studies with larger sample size using population-based controls are warranted. Moreover, subgroup analysis based on genotyping method showed a significant association between LTA A252G and C804A and susceptibility to MI in the PCR-RFLP subgroup, whereas no significant association was found in the non-PCR-RFLP subgroups. This result may be due to the overlapping effect of ethnicity, since the significant association was also observed in Asians and most included studies on Asian subjects used the PCR-RFLP genotyping method. In addition, this association may also be due to chance especially for C804A, for which only three studies were involved in the PCR-RFLP subgroups. Hence, further investigations are also warranted to assess the effect of different genotyping methods on the significance of the association between LTA genetic polymorphisms and susceptibility to MI.
In interpreting the results of this meta-analysis, some specific issues need to be addressed. First, the significant associations found for the LTA A252G and G10A polymorphisms should be interpreted in caution. For A252G, exclusion of hospital-based and non-HWE studies made the summary ORs become insignificant, suggesting that this polymorphism is unlikely to have a major role in susceptibility to MI. For G10A, the significant association in Asians should also be treated carefully, since only two studies on Asians were involved and thus statistical power was limited due to the small sample size. Hence, the significant associations of LTA A252G and G10A with susceptibility to MI should be interpreted in caution and more large-scale studies are warranted for confirming these relationships. Second, as with other complex traits, susceptibility to MI may be modulated by other genetic markers besides the LTA gene. Thus, fully elucidating the pathogenesis of MI would demand an investigation into the association and combined interaction of many gene variants with susceptibility to MI. Third, the included studies only focused on the Asian and Caucasian populations and thus further studies on a wider spectrum of subjects should be carried out to investigate the role of these variants in different ethnicities. Last, this meta-analysis was based on unadjusted ORs and possible effect modifiers, such as sex, age, BMI, diabetes mellitus, and smoking status, may influence the estimates of associations. Unfortunately, the calculation of adjusted pooled ORs and further subgroup analyses based on these factors could not be performed because of limited data. Thus, further well-designed GAS need to focus on exploring these sources of heterogeneity. Despite these limitations, our study is the first comprehensive meta-analysis of all eligible studies on the association between the LTA genetic polymorphisms and MI risk.
In summary, the current meta-analysis indicates that the LTA A252G and C804A polymorphisms may be associated with an increased risk of MI, whereas LTA G10A may confer a significant increased risk of MI only in Asian populations. Thus, these polymorphisms of the LTA gene could possibly be used with other genetic markers together to identify individuals at high risk for MI. However, due to the limitations of this study, these results should be interpreted with caution and still require future large-scale studies to confirm their accuracy. Moreover, considering that MI is a complex disease with a multifactorial etiology, the development of MI might be associated with gene-gene and gene-environment interactions, whose effects should be considered in future GAS and subsequent meta-analyses that may provide more conclusive evidence regarding the genetic susceptibility to MI.
Supporting Information
The Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) Checklist.
(DOC)
The Strengthening the Reporting of Genetic Association Studies (STREGA) quality score systems.
(DOC)
The Newcastle-Ottawa Scale (NOS) criteria.
(DOC)
Univariate and multivariate meta-analyses of potential source of heterogeneity.
(DOC)
Funding Statement
The authors have no support or funding to report.
References
- 1. Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, Dai S, et al. (2010) Heart disease and stroke statistics–2010 update: a report from the American Heart Association. Circulation 121: e46–e215. [DOI] [PubMed] [Google Scholar]
- 2. Schiller JS, Lucas JW, Ward BW, Peregoy JA (2012) Summary health statistics for U.S. adults: National Health Interview Survey, 2010. Vital Health Stat 10: 1–207. [PubMed] [Google Scholar]
- 3. Kung HC, Hoyert DL, Xu J, Murphy SL (2008) Deaths: final data for 2005. Natl Vital Stat Rep 56: 1–120. [PubMed] [Google Scholar]
- 4. Thygesen K, Alpert JS, Jaffe AS, Simoons ML, Chaitman BR, et al. (2012) Third universal definition of myocardial infarction. J Am Coll Cardiol 60: 1581–1598. [DOI] [PubMed] [Google Scholar]
- 5. Finn AV, Nakano M, Narula J, Kolodgie FD, Virmani R (2010) Concept of vulnerable/unstable plaque. Arterioscler Thromb Vasc Biol 30: 1282–1292. [DOI] [PubMed] [Google Scholar]
- 6. Dobaczewski M, Gonzalez-Quesada C, Frangogiannis NG (2010) The extracellular matrix as a modulator of the inflammatory and reparative response following myocardial infarction. J Mol Cell Cardiol 48: 504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carpeggiani C, Coceani M, Landi P, Michelassi C, L'Abbate A (2010) ABO blood group alleles: A risk factor for coronary artery disease. An angiographic study. Atherosclerosis 211: 461–466. [DOI] [PubMed] [Google Scholar]
- 8. Gigante B, Bennet AM, Leander K, Vikstrom M, de Faire U (2010) The interaction between coagulation factor 2 receptor and interleukin 6 haplotypes increases the risk of myocardial infarction in men. PLoS One 5: e11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ianni M, Callegari S, Rizzo A, Pastori P, Moruzzi P, et al. (2012) Pro-inflammatory genetic profile and familiarity of acute myocardial infarction. Immun Ageing 9: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nabel EG, Braunwald E (2012) A tale of coronary artery disease and myocardial infarction. N Engl J Med 366: 54–63. [DOI] [PubMed] [Google Scholar]
- 11. Asselbergs FW, Pai JK, Rexrode KM, Hunter DJ, Rimm EB (2007) Effects of lymphotoxin-alpha gene and galectin-2 gene polymorphisms on inflammatory biomarkers, cellular adhesion molecules and risk of coronary heart disease. Clin Sci (Lond) 112: 291–298. [DOI] [PubMed] [Google Scholar]
- 12. Watkins H, Farrall M (2006) Genetic susceptibility to coronary artery disease: from promise to progress. Nat Rev Genet 7: 163–173. [DOI] [PubMed] [Google Scholar]
- 13. Libby P, Ridker PM, Maseri A (2002) Inflammation and atherosclerosis. Circulation 105: 1135–1143. [DOI] [PubMed] [Google Scholar]
- 14. Clarke R, Xu P, Bennett D, Lewington S, Zondervan K, et al. (2006) Lymphotoxin-alpha gene and risk of myocardial infarction in 6,928 cases and 2,712 controls in the ISIS case-control study. PLoS Genet 2: e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Q (2005) Molecular genetics of coronary artery disease. Curr Opin Cardiol 20: 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, et al. (2002) Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet 32: 650–654. [DOI] [PubMed] [Google Scholar]
- 17. Tanaka T, Ozaki K (2006) Inflammation as a risk factor for myocardial infarction. J Hum Genet 51: 595–604. [DOI] [PubMed] [Google Scholar]
- 18. Panoulas VF, Nikas SN, Smith JP, Douglas KM, Nightingale P, et al. (2008) Lymphotoxin 252A>G polymorphism is common and associates with myocardial infarction in patients with rheumatoid arthritis. Ann Rheum Dis 67: 1550–1556. [DOI] [PubMed] [Google Scholar]
- 19. Wang YC, Chen CC, Zhang WD, Zhang SK, Chang FH, et al. (2010) The 252A/G and 804C/A polymorphisms of Lymphotoxin-alpha is associated to onset of acute myocardial infarction in Taiwan. Lab Medicine 41: 220–225. [Google Scholar]
- 20. Kimura A, Takahashi M, Choi BY, Bae SW, Hohta S, et al. (2007) Lack of association between LTA and LGALS2 polymorphisms and myocardial infarction in Japanese and Korean populations. Tissue Antigens 69: 265–269. [DOI] [PubMed] [Google Scholar]
- 21. Koch W, Hoppmann P, Michou E, Jung V, Pfeufer A, et al. (2007) Association of variants in the BAT1-NFKBIL1-LTA genomic region with protection against myocardial infarction in Europeans. Hum Mol Genet 16: 1821–1827. [DOI] [PubMed] [Google Scholar]
- 22. Sedlacek K, Neureuther K, Mueller JC, Stark K, Fischer M, et al. (2007) Lymphotoxin-alpha and galectin-2 SNPs are not associated with myocardial infarction in two different German populations. J Mol Med (Berl) 85: 997–1004. [DOI] [PubMed] [Google Scholar]
- 23. Ryan AW, O'Brien E, Shields D, McManus R (2008) Lack of association between NFKBIL1/LTA polymorphisms and hypertension, myocardial infarct, unstable angina and stable angina in a large Irish population sample. Atherosclerosis 197: 465–466. [DOI] [PubMed] [Google Scholar]
- 24. Moher D, Liberati A, Tetzlaff J, Altman DG (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 339: b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, et al. (2009) Strengthening the reporting of genetic association studies (STREGA): an extension of the STROBE Statement. Hum Genet 125: 131–151. [DOI] [PubMed] [Google Scholar]
- 26. Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25: 603–605. [DOI] [PubMed] [Google Scholar]
- 27. Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 28. Jackson D, White IR, Riley RD (2012) Quantifying the impact of between-study heterogeneity in multivariate meta-analyses. Stat Med 31: 3805–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sacks HS, Berrier J, Reitman D, Ancona-Berk VA, Chalmers TC (1987) Meta-analyses of randomized controlled trials. N Engl J Med 316: 450–455. [DOI] [PubMed] [Google Scholar]
- 30. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L (2006) Comparison of two methods to detect publication bias in meta-analysis. JAMA 295: 676–680. [DOI] [PubMed] [Google Scholar]
- 32. Padovani JC, Pazin-Filho A, Simoes MV, Marin-Neto JA, Zago MA, et al. (2000) Gene polymorphisms in the TNF locus and the risk of myocardial infarction. Thromb Res 100: 263–269. [DOI] [PubMed] [Google Scholar]
- 33. Koch W, Kastrati A, Bottiger C, Mehilli J, von Beckerath N, et al. (2001) Interleukin-10 and tumor necrosis factor gene polymorphisms and risk of coronary artery disease and myocardial infarction. Atherosclerosis 159: 137–144. [DOI] [PubMed] [Google Scholar]
- 34. Iwanaga Y, Ono K, Takagi S, Terashima M, Tsutsumi Y, et al. (2004) Association analysis between polymorphisms of the lymphotoxin-alpha gene and myocardial infarction in a Japanese population. Atherosclerosis 172: 197–198. [DOI] [PubMed] [Google Scholar]
- 35. Tobin MD, Braund PS, Burton PR, Thompson JR, Steeds R, et al. (2004) Genotypes and haplotypes predisposing to myocardial infarction: a multilocus case-control study. Eur Heart J 25: 459–467. [DOI] [PubMed] [Google Scholar]
- 36. Yamada A, Ichihara S, Murase Y, Kato T, Izawa H, et al. (2004) Lack of association of polymorphisms of the lymphotoxin alpha gene with myocardial infarction in Japanese. J Mol Med (Berl) 82: 477–483. [DOI] [PubMed] [Google Scholar]
- 37. Ozaki K, Tanaka T (2005) Genome-wide association study to identify SNPs conferring risk of myocardial infarction and their functional analyses. Cell Mol Life Sci 62: 1804–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Broeckel U, Hengstenberg C, Mayer B, Holmer S, Martin LJ, et al. (2002) A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat Genet 30: 210–214. [DOI] [PubMed] [Google Scholar]
- 39. Stanton LW, Garrard LJ, Damm D, Garrick BL, Lam A, et al. (2000) Altered patterns of gene expression in response to myocardial infarction. Circ Res 86: 939–945. [DOI] [PubMed] [Google Scholar]
- 40. Helgadottir A, Manolescu A, Thorleifsson G, Gretarsdottir S, Jonsdottir H, et al. (2004) The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet 36: 233–239. [DOI] [PubMed] [Google Scholar]
- 41. Helgadottir A, Manolescu A, Helgason A, Thorleifsson G, Thorsteinsdottir U, et al. (2006) A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat Genet 38: 68–74. [DOI] [PubMed] [Google Scholar]
- 42. Stoll LL, Denning GM, Li WG, Rice JB, Harrelson AL, et al. (2004) Regulation of endotoxin-induced proinflammatory activation in human coronary artery cells: expression of functional membrane-bound CD14 by human coronary artery smooth muscle cells. J Immunol 173: 1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Munafo MR, Flint J (2004) Meta-analysis of genetic association studies. Trends Genet 20: 439–444. [DOI] [PubMed] [Google Scholar]
- 44. Lu R, Dou X, Gao X, Zhang J, Ni J, et al. (2012) A functional polymorphism of lymphotoxin-alpha (LTA) gene rs909253 is associated with gastric cancer risk in an Asian population. Cancer Epidemiol 36: e380–386. [DOI] [PubMed] [Google Scholar]
- 45.Xu Z, Shi R, Zhang R, Zhang D, Wang L (2013) Association between tumor necrosis factor beta 252 A/G polymorphism and risk of gastric cancer: a meta-analysis. Tumour Biol. [DOI] [PubMed]
- 46. Zhou P, Huang W, Chu X, Du LF, Li JP, et al. (2012) The lymphotoxin-alpha 252A>G polymorphism and breast cancer: a meta-analysis. Asian Pac J Cancer Prev 13: 1949–1952. [DOI] [PubMed] [Google Scholar]
- 47. Yang M, Fu X, Zhang Y, Zhang J, He J, et al. (2012) The +252A/G polymorphism in the lymphotoxin-alpha gene increases the risk of asthma: a meta-analysis. Respirology 17: 1229–1236. [DOI] [PubMed] [Google Scholar]
- 48. Tiancha H, Huiqin W, Jiyong J, Jingfen J, Wei C (2011) Association between lymphotoxin-alpha intron +252 polymorphism and sepsis: a meta-analysis. Scand J Infect Dis 43: 436–447. [DOI] [PubMed] [Google Scholar]
- 49. Schurks M, Rist PM, Zee RY, Chasman DI, Kurth T (2011) Tumour necrosis factor gene polymorphisms and migraine: a systematic review and meta-analysis. Cephalalgia 31: 1381–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Skibola CF, Bracci PM, Nieters A, Brooks-Wilson A, de Sanjose S, et al. (2010) Tumor necrosis factor (TNF) and lymphotoxin-alpha (LTA) polymorphisms and risk of non-Hodgkin lymphoma in the InterLymph Consortium. Am J Epidemiol 171: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) Checklist.
(DOC)
The Strengthening the Reporting of Genetic Association Studies (STREGA) quality score systems.
(DOC)
The Newcastle-Ottawa Scale (NOS) criteria.
(DOC)
Univariate and multivariate meta-analyses of potential source of heterogeneity.
(DOC)