Abstract
Myc proteins control cell proliferation, cell cycle progression, and apoptosis, and play important roles in cancer as well in establishment of pluripotency. Here we investigated the control of myc gene expression by the Pou5f1/Oct4 pluripotency factor in the early zebrafish embryo. We analyzed the expression of all known zebrafish Myc family members, myca, mycb, mych, mycl1a, mycl1b, and mycn, by whole mount in situ hybridization during blastula and gastrula stages in wildtype and maternal plus zygotic pou5f1 mutant (MZspg) embryos, as well as by quantitative PCR and in time series microarray data. We found that the broad blastula and gastrula stage mych expression, as well as late gastrula stage mycl1b expression, both depend on Pou5f1 activity. We analyzed ChIP-Seq data and found that both Pou5f1 and Sox2 bind to mych and mycl1b control regions. The regulation of mych by Pou5f1 appears to be direct transcriptional activation, as overexpression of a Pou5f1 activator fusion protein in MZspg embryos induced strong mych expression even when translation of zygotically expressed mRNAs was suppressed. We further showed that MZspg embryos develop enhanced apoptosis already during early gastrula stages, when apoptosis was not be detected in wildtype embryos. However, Mych knockdown alone did not induce early apoptosis, suggesting potentially redundant action of several early expressed myc genes, or combination of several pathways affected in MZspg. Experimental mych overexpression in MZspg embryos did significantly, but not completely suppress the apoptosis phenotype. Similarly, p53 knockdown only partially suppressed apoptosis in MZspg gastrula embryos. However, combined knockdown of p53 and overexpression of Mych completely rescued the MZspg apoptosis phenotype. These results reveal that Mych has anti-apoptotic activity in the early zebrafish embryo, and that p53-dependent and Myc pathways are likely to act in parallel to control apoptosis at these stages.
Introduction
Apoptosis plays a crucial role during development and maintenance of homeostasis in multicellular organisms by eliminating damaged or unneeded cells [1], [2]. Programmed cell death is intensely studied in model organisms, because its deregulation is involved in many diseases including cancer, Alzheimer's disease, or immune deficiencies [3], [4]. Today many components of the apoptosis pathway and their regulatory interactions are known [5]–[7]. In recent years, zebrafish have been increasingly used as model for studying apoptosis, because of the ease of experimental manipulation and the close homology of the apoptosis pathway core factors between fish and mammals [8]–[13].
Myc proteins are transcription factors involved in regulation of various cellular functions including cell-cycle progression [14], differentiation [15], [16], cell growth [17], and apoptosis [18]. Myc forms heterodimers with its transcriptional co-regulator Max to bind specific DNA sites in the promoter regions of its target genes [19], [20]. It was shown that Myc might be associated both with gene activation [21], [22] and repression [23], [24]. The myc proto-oncogene family consists of the three members c-myc [25], N-myc [26] and L-myc [27]. Myc deregulation is often linked to tumor formation in animals and human [28]–[30]. During normal mouse embryonic development c-myc is expressed in many different cell types [31]–[34]. C-Myc was shown to correlate with cell proliferation in several tissues, while it induces or sensitizes cells to apoptosis in others [35]. L-myc and N-myc expression are restricted to specific tissues in the course of cell differentiation [31]–[34]. The importance of c-Myc in promoting early development was shown by the fact that c-myc homozygous knock-out mice are embryonic lethal [36], and its role in stem cell pluripotency when mouse fibroblasts were reprogrammed by overexpressing c-Myc together with Oct4, Sox2 and Klf4 [37]. In embryonic stem cells, Myc makes complex contributions to pluripotency [38]–[40].
In zebrafish, homologous genes for all myc family members have been identified [41], [42]. As a result of the genome duplication that occurred in the evolution of teleosts [43] two fish paralogues each exist for c-myc (myca and mycb) and L-myc (mycl1a and mycl1b; Figure S1)[41], [42]. In contrast, there is only one homologous gene for N-myc known in zebrafish, mycn [42], [44]. Recently, a new member of the myc gene family, mych, was identified in zebrafish. Mych shows a high similarity to N-Myc and c-Myc in its C-terminal amino acid sequence, but has no direct orthologous gene in higher vertebrates [41], [45]. Zebrafish myc homologous genes are differentially expressed during embryonic development, and in specific adult tissues [41], [42], [44], [45]. The spatial distributions of the early gene expression patterns have been previously described for mych, mycn and mycl1a. All three genes show a broad expression at blastula and gastrula stages [44]–[46]. Meijer et al. [41] described the postgastrula expression patterns of all known zebrafish myc homologues in great detail (see also Figure S2). However, the functions of myc genes in early zebrafish development are poorly understood. The recently identified mych gene has been linked to cell survival and neural crest development [45].
In mammalian ES cells, myc genes are regulated by the master stem cell factors Pou5f1 (Oct4) and Nanog [38], [40]. Previous work in our lab revealed that the transcript levels of two zebrafish myc genes, mycl1b and mych, are reduced in pou5f1 (also named Pou2 or Pou5f3; www.zfin.org) maternal and zygotic (MZspg) mutant embryos as judged from microarray analysis [47]. Here we analyzed the early expression patterns of the zebrafish myc genes and their expression in Pou5f1 deficient embryos. We showed that the zebrafish mych and mycl1b genes are bound by Pou5f1 and are likely direct targets of Pou5f1. We further showed that MZspg gastrulae display enhanced apoptosis compared to wildtype (WT) controls. The apoptosis phenotype was partially rescued by mych overexpression, indicating a role of Mych in regulation of cell survival during gastrulation.
Results
Pou5f1 activity is required to induce zygotic expression of mych and mycl1b
Temporal expression profiles from time series microarray analyses (zygote to 8 hours post fertilization - hpf) of wildtype (WT) and Pou5f1 maternal and zygotic deficient embryos (MZspg) [47] have revealed myc genes potentially regulated by Pou5f1 in the early zebrafish embryo (Figure S3). For the zebrafish c-myc homologues myca and mycb, in MZspg embryos compared to WT, the mRNA levels derived from maternal expression have been found to be reduced before onset of zygotic transcription (midblastula transition - MBT), but elevated at post-MBT stages (Figure S3A and C). mycl1a expression was not altered in Pou5f1 mutants compared to WT (Figure S3E). mycl1b expression in MZspg has been found to be normal during blastula and early gastrula stages, but downregulated during late gastrulation (Figure S3G). The expression of mych and mycn has been shown to be strongly activated after MBT in WT embryos, but not in Pou5f1 mutants (Figure S3I and K). Whereas the induction of mycn expression only appeared to be delayed in MZspg embryos (Figure S3I), mych continued to be expressed at strongly reduced levels in MZspg throughout gastrulation (Figure S3K). These findings suggested that Pou5f1 function is crucial for a proper regulation of mycl1b and mych gene expression in early zebrafish development.
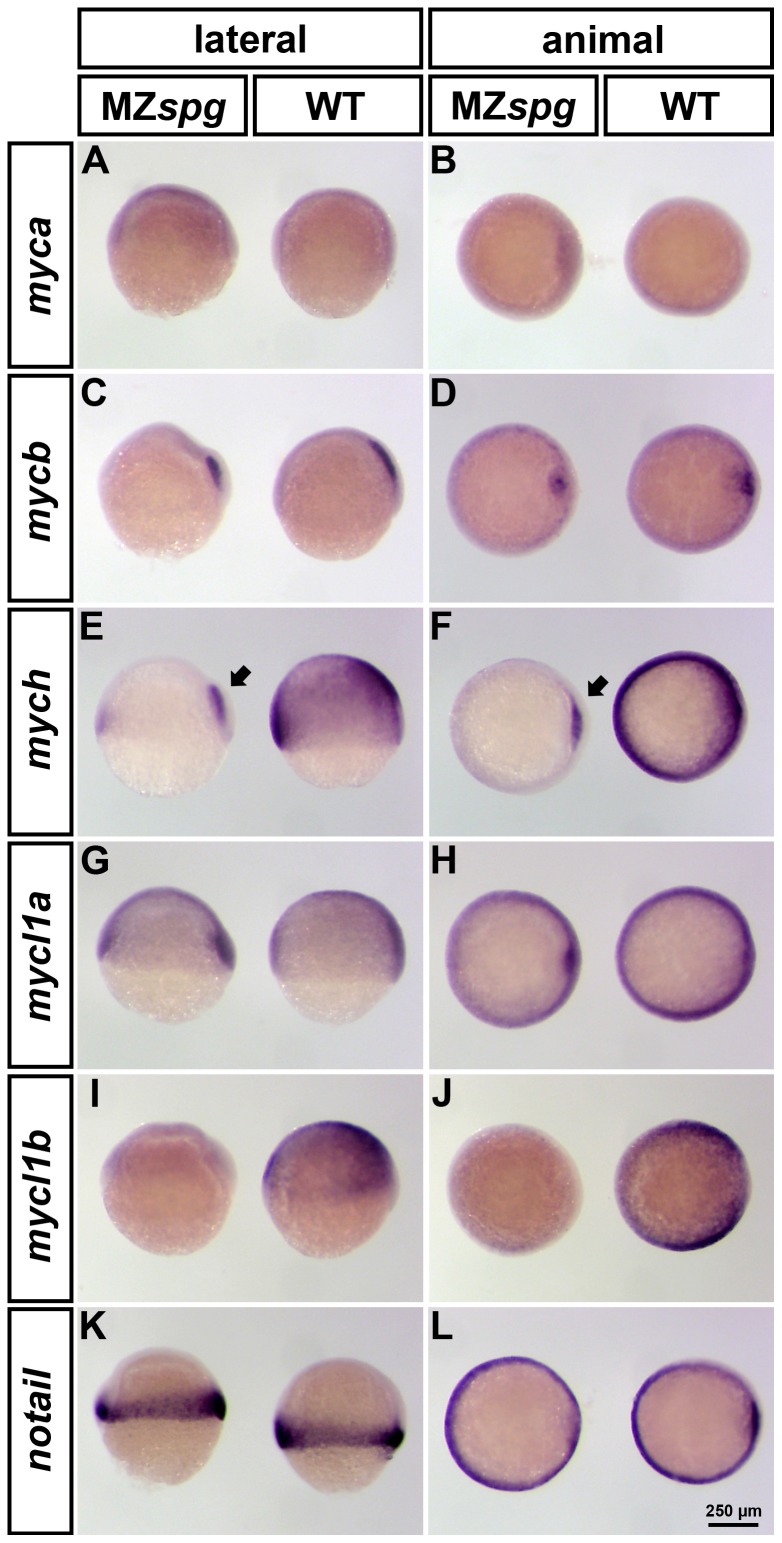
To confirm the microarray data and to obtain more information about the spatial distribution of gastrula stage myc gene expression patterns and their changes in MZspg embryos, we performed whole-mount in situ hybridization (WISH) expression analysis for myca, mycb, mych, mycl1a and mycl1b in WT and MZspg mutants at 60% epiboly (Figure 1). Early expression patterns of mych and mycn have been previously reported [44], [45]. The mesodermal marker no tail (ntl; [48] was used as a control for this experiment, because ntl is not regulated by Pou5f1 [47]. Only one of the two c-myc homologs, mycb, was detected with spatially restricted expression at mid-gastrulation (Figure 1A-D). At 60% epiboly mycb was specifically expressed in the region of the embryonic shield (Figure 1C-D). The expression patterns of both myca and mycb displayed no alterations in Pou5f1 deficient embryos compared to WT. The slightly higher microarray expression levels of myca and mycb at this stage in MZspg (Fig S3A,C) were not clearly detected by WISH except for a minor increase in stain intensity in MZspg. mych, mycl1a and mycl1b were found to be broadly expressed throughout the blastoderm at 60%-epiboly, while mycl1b, in contrast to mych and mycl1a, was excluded from the embryonic shield (Figure 1E-J). We found that in MZspg mutants mych and mycl1b expression were strongly downregulated (Figure 1E-F and I-J). Therefore, mych and mycl1b expression appeared to depend on the Pou5f1 transcription factor. Interestingly, expression of mych was found to depend on Pou5f1 activity in most early gastrula cells, except for the cells of the embryonic shield (Figure 1E-F).
Figure 1. Spatial expression pattern of zebrafish myc genes in WT and MZspg embryos at 60% epiboly.
Whole mount in situ hybridization (WISH) analysis of myca, mycb, mych, mycl1a and mycl1b expression in WT (right embryo in each panel) and MZspg (left embryo in each panel). All embryos are shown in lateral (left column) and animal (right column) views with dorsal oriented to the right. All analyzed myc genes are broadly expressed in mid-gastrula embryos, except for c-myc homologues, myca and mycb. mycb is specifically expressed in the shield, whereas myca was not detectable at this stage (A-D). mych and mycl1a in addition have a strong expression domain in the involuting axial mesoderm (E-H). Only mych (E-F) and mycl1b (I-J) depend on the function of Pou5f1 and their expression is strongly decreased in MZspg mutants. However, the mych expression domain in the involuting axial mesoderm is less affected in Pou5f1 deficient embryos (E-F; arrows). We used notail as control, because its expression is not altered in MZspg mutants compared to WT (K-L).
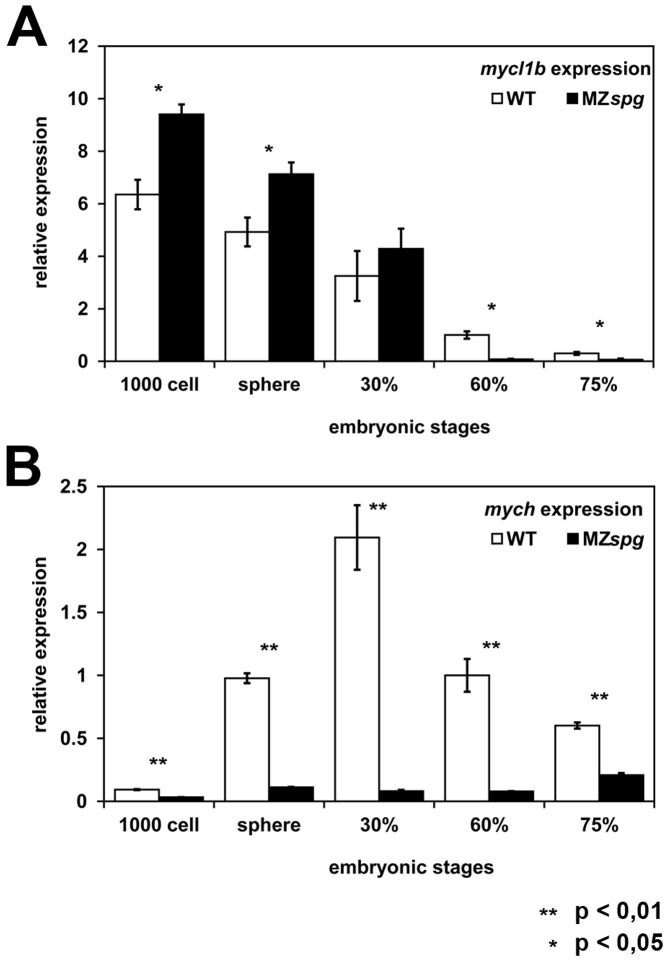
We next quantified the expression of mych and mycl1b in WT and MZspg embryos for five different developmental stages ranging from 1000-cell (MBT) to 75% epiboly by relative quantitative real-time PCR (qPCR) (Figure 2). We used the housekeeping gene translation elongation factor 1α (ef1α also named eef1a1l1) as reference gene to normalize the expression levels of mycl1b and mych in MZspg and WT. The developmental expression profiles, determined by qPCR, for both genes were in agreement with microarray results (Figure S3G,K) [47]. The developmental profile for mycl1b showed a strong signal at mid-blastula transition in WT and Pou5f1 mutants and expression decreased during gastrulation (Figure 2A). This suggested that most of the early mycl1b mRNA is maternally expressed. In comparison to WT, the expression level of mycl1b in MZspg was about 1.5 times higher at MBT, but found to decline rapidly in MZspg mutant embryos during gastrulation, resulting in a 12.5 fold downregulation compared to WT at 60%-epiboly (Figure 2A). This difference may be caused by two mechanisms: Pou5f1 may protect maternal mycl1b by direct or indirect mechanisms, or Pou5f1 may induce zygotic expression of mycl1b. In contrast to mycl1b, mych was not detected to be expressed maternally. mych expression was found to be activated immediately after MBT in WT embryos and to reach a high point at 30% epiboly before declining again (Figure 2B). In MZspg embryos mych expression was strongly reduced at all analyzed developmental stages (Figure 2B). Our results demonstrate that the Pou5f1 transcription factor is important for the early zygotic activation of mych expression, and for maintaining mycl1b RNA levels. In summary, our data indicate that Pou5f1 function is important for the activation of the early zygotic expression of mych, and mycl1b during the first 8 hours of development.
Figure 2. Temporal analysis of mych and mycl1b expression in WT and MZspg.
Relative expression levels of mych and mycl1b in WT and MZspg embryos for five distinct time points, between 1000-cell (MBT) and 75%-epiboly stage using quantitative real-time PCR. Bars show average values of three independent biological replicates each. The WT expression at 60%-epiboly stage was normalized to 1 for both genes. mycl1b is maternally expressed and declines as embryonic development progresses (A, white bars). In MZspg mutants, mycl1b expression is upregulated at MBT, likely reflecting higher maternal mRNA levels, but in comparison to WT it declines faster during gastrulation (A, black bars). In WT mych is activated at MBT, than its expression increases until 30%-epiboly before it starts to slowly fade (B, white bars). mych expression levels are up to 26-fold reduced in MZspg mutants compared to WT during early embryogenesis (B, black bars).
mych and mycl1b are directly activated by Pou5f1
To distinguish between direct and indirect Pou5f1 targets, Onichtchouk et al. [47] performed pou5f1 mRNA overexpression experiments in MZspg embryos, and inhibited translation of zygotic mRNAs after MBT by adding the translation elongation inhibitor cycloheximide (CHX) at the 64-cell stage. They compared by microarray analysis expression profiles at 30% epiboly of embryos injected with pou5f1 mRNA versus non-injected controls, both treated with CHX, such that only the expression of direct Pou5f1 targets should be differentially affected. We analyzed these published data to answer the question whether the myc genes may be regulated directly or indirectly by Pou5f1 (Figure S3). We found that expression of mych, mycl1b, and the c-myc homologous genes were more than 2-fold increased after the injection of pou5f1 mRNA and subsequent addition of CHX, which suggests direct regulation by Pou5f1 (Figure S3B, D, H and L).
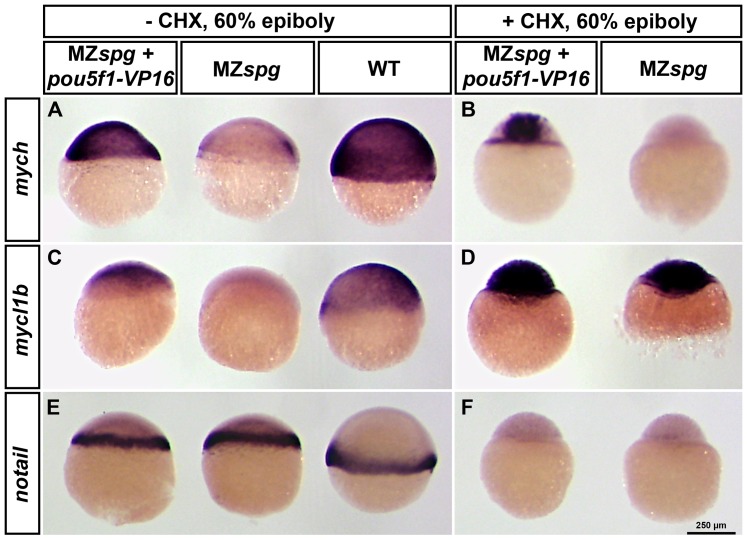
To determine the spatial extent of myc gene regulation by Pou5f1 in the embryo, and to confirm the microarray data, we addressed experimentally whether and to what level Pou5f1 may be able to directly activate mych and mycl1b expression. We microinjected mRNA encoding a fusion protein of Pou5f1 with the strong transcriptional activator domain VP16 [49] into MZspg embryos, and inhibited translation of zygotically expressed mRNAs by CHX. Therefore, the effect of Pou5f1-VP16 on mych and mycl1b should be direct, and not mediated by indirect effects of Pou5f1 targets on mych and mycl1b. The amount of pou5f1-VP16 mRNA injected was chosen such that the embryos showed a partial rescue of the morphological MZspg phenotype as described by Lunde et al. [49] (Figure S4B), which demonstrates the functionality of the used fusion mRNA. To reveal the specificity of the experiment, we used no tail as a control (Figure 3E), which has been shown not to be regulated by Pou5f1 [49], and is not affected by pou5f1-VP16 mRNA in our experiment. We analyzed the expression of mych and mycl1b by WISH at 60% epiboly. Injection of pou5f1-VP16 fusion mRNA into MZspg embryos induced strong mych and mycl1b expression (Figure 3A and C, Supplemental Table S1) also in the presence of CHX (Figure 3B and D). However, mycl1b RNA levels were not only increased by pou5f1-VP16 overexpression in CHX treated MZspg embryos, but also by the CHX treatment alone in control MZspg embryos (Figure3 D). Given the strong maternal expression (Figs. 2 and S3), CHX treatment may indirectly prevent degradation of maternal mycl1b RNA by mechanisms depending on translation of zygotic protein products.
Figure 3. mych and potentially also mycl1b are directly regulated by Pou5f1.
Pou5f1 overexpression in MZspg mutants by pou5f1-VP16 mRNA injection into one-cell stage embryos (A-F; left embryos in each panel) and non-injected controls (A, C and E; MZspg middle and WT right embryos in each panel; B, D and F; MZspg right embryo in each panel). The embryos in B, D and F are in addition treated with CHX starting at 1.5 hpf. The mych expression can be rescued by Pou5f1-VP16 in both, CHX treated and untreated embryos, and therefore the regulatory influence of Pou5f1 should be direct (A-B). The expression of mycl1b can also be rescued by Pou5f1-VP16 overexpression in CHX untreated MZspg embryos, but is also strongly upregulated in CHX treated MZspg embryos, even without the injection of pou5f1-VP16 mRNA (C-D). Thus the experiment cannot proove whether activation of mycl1b by Pou5f1 is direct or not. We used notail as negative control, because its expression is independent of Pou5f1 function (E), and depends on zygotic gene products.
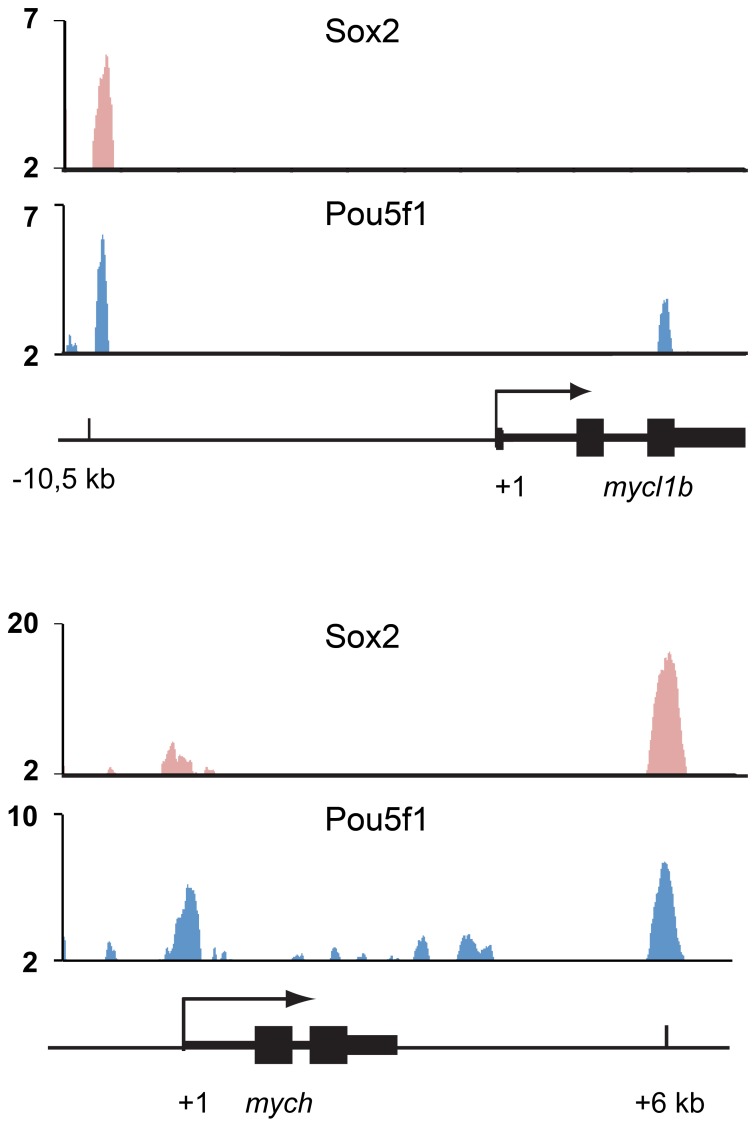
To investigate whether Pou5f1 may bind to the mych and mycl1b regulatory regions, we analyzed published genome-scale chromatin immunoprecipitation data (ChIP-seq) for Pou5f1 [50]. Figure 4 shows that at 50% epiboly stage Pou5f1 was detected bound in the proximity of both the mych and mycl1b genes (+6 kb and -10,5 kb from transcription start sites, respectively). In addition, smaller Pou5f1 binding peaks marked the basal promoter region of mych. Three Pou5f1 ChIP-Seq peaks also correlated with Sox2 ChIP-Seq peaks, suggesting that Pou5f1 and Sox2 act together in this regulation. In summary, our data reveal that zygotic transcription of mych and mycl1b are directly regulated by Pou5f1.
Figure 4. Pou5f1 binds to the promoters and regulatory regions of mych and mycl1b.
ChIP-seq reads for Sox2 (red) and Pou5f1 (blue) [50] mapped to the mycl1b (A) and mych (B) genetic loci are shown. Colocalisation of Pou5f1 (blue) and Sox2 (red) at the regulatory regions of mycl1b and mych were detected. Y-axis: ChIP-Seq reads per base, y-axis floor is set to 2.
Pou5f1 mutants show enhanced apoptosis
The most prominent early phenotype of MZspg embryos is delayed and ultimately arrested progression of epiboly (compare Figure 5B and J) [49], [51], [52]. Pou5f1-dependent regulation of cell adhesion and cell motility has been shown to contribute to the epiboly delay phenotype in MZspg [53], but does not appear to explain the full extent of phenotypic abnormalities in MZspg. Additional causes for the epiboly delay phenotype may include slowdown of cell proliferation or increase in apoptosis. Lachnit et al. [51] demonstrated that until 5 hpf stage there is no reduction in number of deep cells detectable in MZspg mutants compared to wildtype. However, cell death and proliferation have not been analyzed during mid to late gastrula stages in MZspg so far.
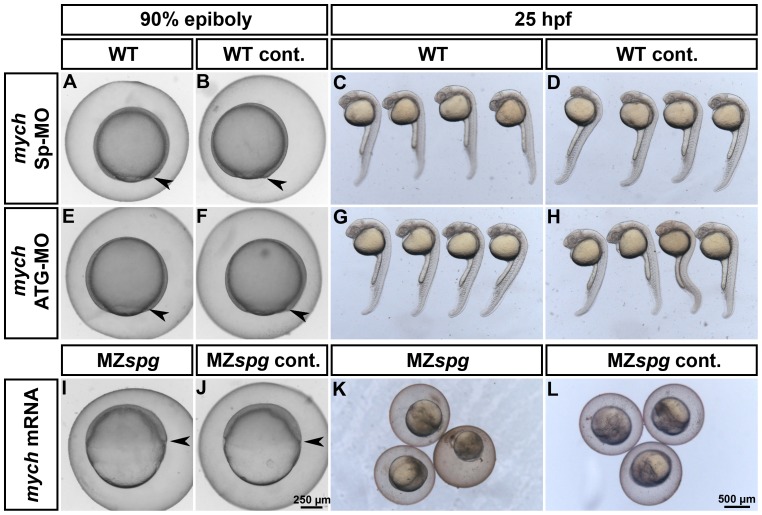
Figure 5. Analysis of the Mych contribution to the morphology of the MZspg mutant phenotype.
Knockdown of Mych function in WT by injecting splice-blocking (A-D) or translation-blocking (E-H) morpholinos. Morphological analysis of injected (A,C,E,G) and non-injected (B,D,F,H) embryos at 90%-epiboly and 25 hpf. Morphants show neither a delay in epiboly (A, E) nor obvious morphological defects by 25 hours development (C, G). Rescue of Mych activity in MZspg mutants by injecting mych mRNA into 1-cell stage embryos (I-L). Mych alone is not able to rescue the strong defects developed by Pou5f1 deficient embryos during gastrulation (I) and after 25 hpf (K).
We analyzed cell proliferation at 90% epiboly by calculating the mitotic index in MZspg mutants and WT embryos (Figure S5). We fixed embryos and stained chromatin using the Sytox Green fluorescent dye. We recorded animal view confocal stacks of the stained embryos and determined the total number of nuclei as well as the number of metaphase and anaphase nuclei, which characterize cells in mitosis. We calculated the ratio of mitotic nuclei and total nuclei (Figure S5). We found that even at 90% epiboly stage, when the MZspg epiboly phenotype is very pronounced, the mitotic index of the MZspg mutants did not differ significantly from WT embryos (Figure S5A, Supplemental Table S3). Thus, a reduction in cell number caused by decreased proliferation during late gastrulation may be excluded as cause for the MZspg epiboly phenotype.
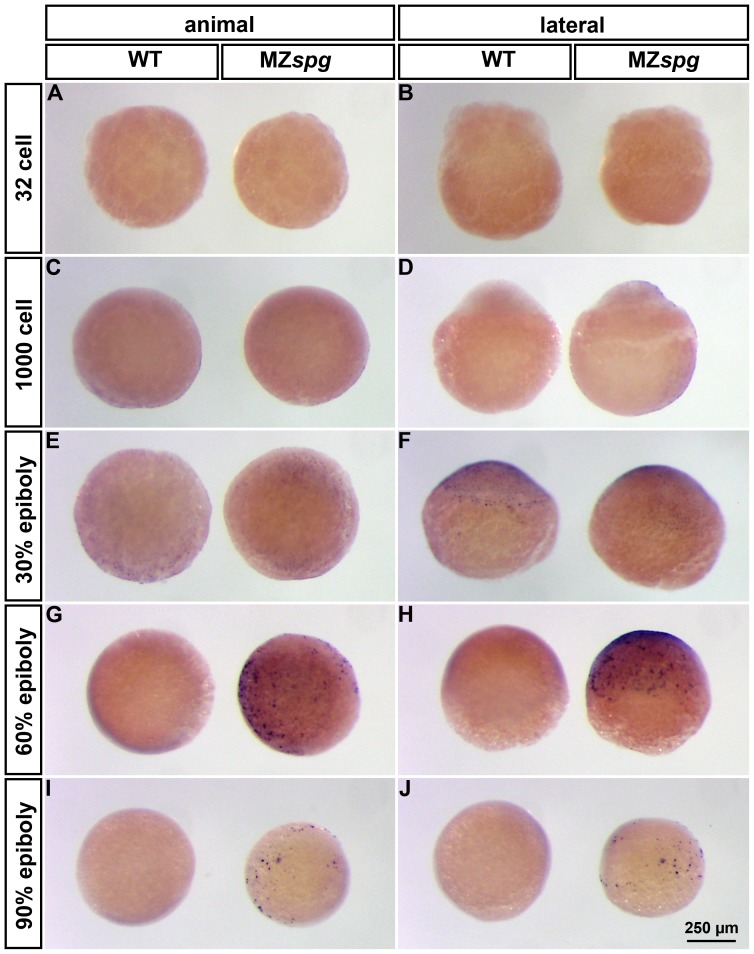
Controlled cell death, and specifically apoptosis, contributes to embryonic development across the animal kingdom [54]. Cole and Ross [55] described the temporal and spatial distribution of apoptotic cells during normal zebrafish development. The earliest apoptotic cells in wildtype development were reported around 12 hpf in the dorsal midline and the segmental plate. To investigate whether MZspg embryos have enhanced apoptosis before this time stage, we analyzed apoptosis in MZspg mutant and WT embryos using TUNEL assay staining from 32-cell stage up to bud stage (Figs. 6 and 7). We could not detect any apoptotic cells in both MZspg and WT embryos until early gastrula stage (Figure 6A-F). Later in development, starting at 60% epiboly, we found enhanced apoptosis in MZspg mutants, whereas WT embryos showed no TUNEL staining (Figure 6G-J and Figure 7A-D).
Figure 6. MZspg mutants have enhanced apoptosis during gastrulation.
TUNEL staining of WT (left embryo in each panel) and MZspg (right embryo in each panel) embryos at distinct embryonic stages between 32 cells and 90%-epiboly. Embryos are shown in animal (left column) and lateral (right column) view. Pou5f1 deficient embryos show enhanced apoptosis, starting at 60%-epiboly, compared to WT (G-J).
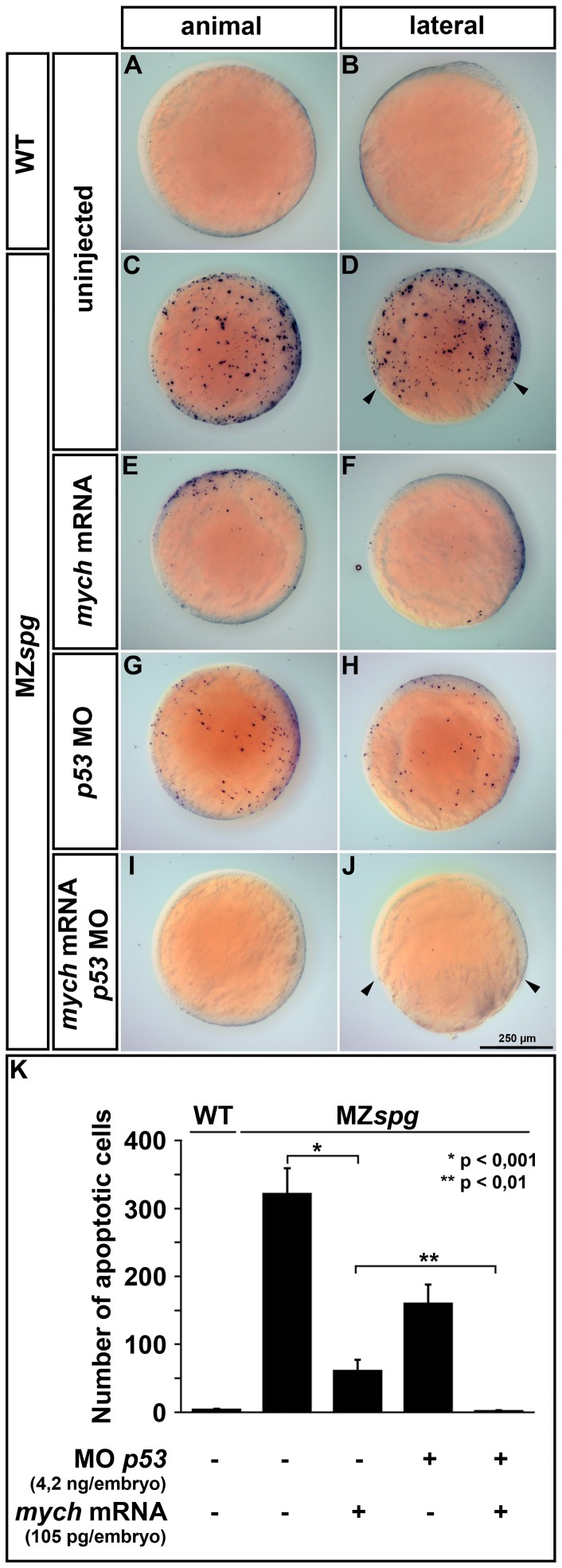
Figure 7. Mych overexpression and p53 knockdown suppress cell death in MZspg gastrulae.
TUNEL staining to detect apoptosis at bud stage (A-J) and subsequent computational image analysis (K) for quantification of the number of apoptotic cells. The images show maximum intensity projections of z-stacks taken from single embryos with dorsal to the right. WT embryos display almost no apoptosis, whereas MZspg mutants show an increase in cell death throughout the embryo (A-D). This mutant apoptosis phenotype was partially repressed either by mych overexpression (E,F) or p53 morpholino knockdown (G,H). The co-injection of mych mRNA and p53-morpholino could completely suppress cell death in MZspg mutants, but did not rescue the delay in epiboly movement (I-J; arrowheads). The quantification of cell death (K) revealed that the number of apoptotic cells is decreased by a factor of six in MZspg embryos after mych mRNA injection. By combined knockdown of p53 and Mych overexpression, apoptosis in MZspg embryos was reduced to WT levels.
Mych controls apoptosis in zebrafish
To investigate a potential contribution of loss of Mych to the MZspg phenotype we analyzed the morphology after mych overexpression in MZspg mutants and Mych knockdown in WT embryos at 90% epiboly and 25 hpf (Figure 5). The knockdown of Mych activity using a splice blocking (Sp-MO) morpholino, which blocks splicing of the second intron of the zygotic mych pre-mRNA, or a translation blocking (ATG-MO) morpholino, which blocks the translation of both maternal and zygotic mych mRNAs, did not cause a delay of epiboly movement (Figure 5A-B and E-F). To demonstrate the functionality of our mych ATG-MO, we knocked down GFP in embryos injected with gfp mRNA fused to the mych ATG-MO binding site (Figure S6A-H). The effectiveness of our mych Sp-MO was shown by mych RT-PCR after injecting different concentrations of the splice blocking morpholino (Figure S6I). Also, broad overexpression of mych in MZspg mutants by mRNA injection could not rescue the MZspg phenotype (Figure 5I-J). One-day-old manipulated embryos developed morphologically similar to WT or MZspg control embryos, respectively (Figure 5 C-D, G-H and K-L; Supplemental Table S2). A more detailed analysis of the late phenotypes of Mych morphants was reported by Hong et al. [45].
Next, we analyzed whether Mych activity may contribute to control of apoptosis during gastrula stages. Following mych knockdown and analysis by TUNEL assay, we found enhanced apoptosis in Mych morphants at 24 hpf (data not shown), comparable to the results reported by Hong et al. [45]. To analyze whether mych overexpression can rescue the apoptosis phenotype, we injected mych mRNA into one-cell stage MZspg embryos, and compared the numbers of apoptotic cells at late gastrula stage between injected and non-injected embryos. We found that mych mRNA injection led to a reduced number of apoptotic cells in the MZspg mutants at bud stage (Figure 7E-F). In parallel we performed p53 knockdown in MZspg embryos, using a p53 specific morpholino, which has been reported to block apoptosis [56]. MZspg p53 MO injected embryos showed a strong reduction in apoptosis compared to non-injected MZspg controls (Figure 7G-H). The co-injection of mych mRNA and p53-MO completely rescued the MZspg apoptosis phenotype (Figure 7I-J). We quantified the numbers of apoptotic cells in these experiments (Figure 7K, Figure S7, Supplemental Table S4, Methods). When MZspg embryos injected with mych mRNA were compared to control MZspg embryos, a statistically significant six-fold reduction of apoptotic cells in mych overexpressing MZspg was observed. p53 knockdown caused a less pronounced reduction of the apoptotic phenotype in MZspg, and the combined overexpression of mych and knockdown of p53 rescued the apoptosis essentially back to low wildtype levels (Figure 7K). While we microinjected well-established amounts of p53 MO for knockdown, and a relatively large amount of mych mRNA for rescue, we cannot completely exclude that incomplete rescue or knockdown may contribute to the apparent partial rescue of apoptosis only. We conclude that Mych and p53 dependent pathways are likely to contribute in parallel to the MZspg apoptotic phenotype.
Discussion
Myc proteins contribute to the control of cell proliferation by regulating cell cycle progression and apoptosis, and play important roles in cancer [28] as well in establishment of pluripotency [37]. Here we investigated the control of myc gene expression by the Pou5f1/Oct4 pluripotency factor in the early zebrafish embryo. We found that early zygotic mych expression as well as late gastrula stage mycl1b expression both directly depend on Pou5f1 activity. We further showed that Pou5f1 deficient MZspg embryos developed enhanced apoptosis already during early gastrula stages, and that mych overexpression in MZspg embryos was able to significantly suppress the apoptosis phenotype. Combined knockdown of p53 and overexpression of Mych completely rescued the MZspg apoptosis phenotype. Both results together reveal that Mych has anti-apoptotic activity in the early zebrafish embryo, and that p53-dependent and Myc pathways act in parallel to control apoptosis at these stages.
Zebrafish myc genes are broadly expressed during the first hours of development
Our and others previous studies have shown that zebrafish L-myc and c-myc homologous genes are maternally expressed and mRNAs deposited in the egg, which remain stable until blastula stages [42], [44], [45], whereas mycn and mych expression are first detectable after mid-blastula transition [44]. Zebrafish mycl1a, mycl1b, and mych genes are broadly expressed in blastoderm cells after MBT. Broad myc gene expression during early development was also shown in other vertebrate species. The Xenopus c-myc, L-myc and N-myc homologous genes are all maternally expressed, but their expression levels decrease during early embryogenesis [57], [58]. xc-myc expression is maintained at a low level during blastula and gastrula stages before it increases again during neurulation. During mouse gastrulation c-myc and N-myc are widely expressed in embryonic and extraembryonic tissues [32]. Thus, the broad myc gene expression during blastula and gastrula stages is evolutionary conserved throughout the vertebrate subphylum. However, not much is known about potentially conserved myc gene functions during blastula and gastrula stages.
Pou5f1 activity is required for proper mych, mycl1b and mycn expression
We found that the transcription factor Pou5f1 is required for proper transcription of mych after MBT from the 1000-cell stage on. While Pou5f1 appears to directly activate the broad early zygotic expression of mych after MBT, Pou5f1 activity is not required for the initiation of the later mych expression domain in the involuted axial mesoderm on the dorsal side of the embryo. Pou5f1 protein is also involved in proper maternal regulation of mycl1b, as enhanced levels of mycl1b were detected at pre-MBT and blastula stages in MZspg. Further, Pou5f1 is required for proper maintenance of mycl1b expression level during mid- to late gastrula stages, when mycl1b drops significantly below wildtype levels in MZspg embryos. In addition, Pou5f1 is required for proper early post-MBT activation of mycn expression, which is delayed in MZspg mutants.
The regulation of c-myc expression has been intensively studied in several systems. A number of regulatory cis-elements have been identified in mammals and it was shown that many transcription factors can bind to the c-myc regulatory elements in vivo [59]. Chen et al. [60] analyzed the core transcriptional network of embryonic stem cells using chromatin immunoprecipitation coupled with ultra-high-throughput DNA sequencing (ChIP-seq) to map the locations of key pluripotency transcription factors like Oct4, Sox2, Nanog, c-Myc and n-Myc. They could show that Oct4 and Sox2 directly bind to the n-myc promoter region, but do not interact with c-myc. This reported regulation of myc family members is consistent with our results in early zebrafish embryogenesis, where mych and mycl1b genes are directly induced by Pou5f1 and potentially also bound by Sox2. Thus, regulation of myc gene expression by Pou5f1 and Sox2 during early embryogenesis, respectively in ES cells, may be an evolutionary conserved feature in vertebrate development.
Maternal and zygotic pou5f1 mutants show normal proliferation, but enhanced apoptosis at gastrulation stages
When analyzing cell division rates and cell survival at blastula to gastrula stages, we found no differences in the mitotic rates between MZspg and WT embryos, while apoptosis rates were markedly increased in MZspg embryos starting from 60% epiboly. The relationship of Oct4 to apoptosis in mammalian ES cells is not well understood. Oct4 is a central factor in the ES cell transcriptional network, controlling many parameters of ES cell biology [38], [40]. This role is compatible with both promoting proliferation and inhibiting apoptosis, as ES cells are immortal cell population with fast self-replication rates. Oct4 deprivation results in global expression changes, cessation of rapid cell proliferation and finally differentiation of ES cells to trophectoderm. However, Oct4 has also been linked to survival and anti-apoptotic pathways in ES cells by several different potential mechanisms: by a STAT3/Survivin route [61], by Trp53 regulation [62], or by a miR-125b pathway [63]. Next to its role in ES cells, Oct4 has also been suggested to promote survival of cancer stem cells by inhibiting apoptosis [63], [64].
In zebrafish, like in mammals, Pou5f1 controls expression of a large network of developmentally important signaling molecules and transcription factors [47], [50]. The majority of these factors is expressed in embryonic tissue-specific patterns, starting already from mid-gastrulation stages [47], [49], [50], [65]. Therefore, it is possible, that the absence of some of these factors may not be compatible with cell survival within a specific tissue, which may lead to the activation of safeguarding apoptotic mechanism. However, we did not detect any obvious tissue-specific pattern of apoptosis in MZspg mutants, which suggests that Pou5f1 may be required throughout the whole embryo to prevent the activation of apoptotic cascades.
Mechanisms of anti-apoptotic action of Pou5f1: separate Myc and p53 branches
The expression of multiple myc genes, mych, mycl1b, and to a lesser degree mycn, is reduced in MZspg embryos. Since myc genes are known anti-apoptotic factors acting through different routes including p19ARF, BIM and BCL2 (reviewed in [66]) and MDM2 [67], they may convey the anti-apoptotic action of Pou5f1. Enhanced early apoptosis in MZspg may be caused by reduced Myc activity, where individual myc genes may act partially redundantly. Indeed, we found that mych mRNA overexpression is able to rescue most of the ectopic apoptosis in MZspg embryos. However, the knockdown of mych in wildtype embryos did not induce apoptosis during gastrulation stages (10 hpf stage; data not shown), strengthening the notion that mych, mycl1b and mycn gene activities, which are all broadly expressed in the blastula and gastrula embryo [44], [45], may be required redundantly downstream of Pou5f1 to prevent early activation of the apoptotic programs.
p53, together with co-factors and depending on the type of stress a cell is subjected to, is a universal activator of apoptosis [68], acting in response to various stimuli. In many cell systems, myc genes contribute to control of apoptosis by regulating p53, e.g. in a p19ARF dependent manner [66]. We determined whether Pou5f1 may regulate levels of tp53 mRNA by reanalyzing published time-series microarray data for MZspg and WT embryos [47], and found no significant differences in tp53 mRNA levels from zygote to end of gastrulation. Thus, Pou5f1 does not regulate p53 expression, and an involvement of p53 may be through indirect pathways controlling p53 activity. If all apoptosis in MZspg would be p53-dependent, apoptosis should be completely abolished by knockdown of p53 through injection of p53-morpholino. However, in p53-MO injected MZspg embryos, as in mych mRNA injected embryos, apoptosis was found to be only partially suppressed. Complete suppression of apoptosis in MZspg embryos was achieved only by simultaneous increase of Mych activity and suppression of p53. This suggests that myc genes, specifically mych, in the early zebrafish embryo are able to suppress apoptosis through a p53-independent pathway. A similar p53-independent mechanism may potentially be involved in the suppression of apoptosis in the neural plate by Mych at later developmental stages [45]. Our study suggests that the zebrafish embryo may be a suitable model system to dissect p53-dependent and independent anti-apoptotic activities of Myc proteins during embryonic development. However, the coexpression of several myc genes throughout the early embryonic stages will likely require combined inactivation of each of these genes, which so far has hindered progress towards analysis of molecular mechanisms.
Materials and Methods
Ethics statement
This study was performed with the approval of the State of Baden-Württemberg Regierungspraesidium Freiburg Animal Protection Authorities in accordance with the German Animal Protection Act under permission number 35-9185.81/G-12/40.
Fish and embryo care
We used WT embryos of AB x TÜB strain crosses (http://www.ZFIN.org) and MZspg embryos carrying the m793 allele of the spg mutation [69](ZFIN ID: ZDBGENE-980526-485, ZDB-GENO-081023-1). Fish were raised, maintained and crossed under standard conditions as described [70]. Embryos were incubated or raised in egg water or in 0.3× Danieau's solution at 28.5°C. Developmental age is reported as hours post fertilization (hpf) when incubated at 28.5°C. Developmental stages of MZspg embryos were indirectly determined by observation of WT embryos born at the same time and incubated under identical conditions.
Morpholinos
Morpholino oligos mych-Sp-MO: 5′-GTAGCAAAAGACTCACCAGAATCGC-3′, mych-ATG-MO: 5′-GCAGCATCTTGACGGAACCTTTTTC-3′, and standard control morpholino SCMO: 5′-CCTCTTACCTCAGTTACAATTTATA-3′ were ordered from Gene Tools (Philomath, USA). The mych-ATG-MO blocks the translation of the mych mRNA into protein by binding to the translation start site. The mych-Sp-MO prevents splicing of the second intron resulting in a non-functional truncated protein missing the DNA binding domain. To test the specificity of mych-ATG-MO, the sequence -4 to +21 bp from the mych translation start site ATG was cloned upstream of GFP ORF in the CS2+ vector, to obtain the mych-GFP construct. 50 pg/embryo of in vitro transcribed mych-GFP mRNA was injected into one-cell stage embryos together with 1.4 ng, 4.1 ng or 8.6 ng per embryo of the mych-ATG-MO or without Morpholino (Figure S6). The efficiency of mych-Sp-MO was tested by injecting 1.4 ng, 4.1 ng or 8.6 ng into single cell WT embryos and subsequent RT-PCR analysis at 60% epiboly stage. The ratio of correctly spliced mych mRNA (162 bp) and mRNA containing the second intron (396 bp) shows that the injection of 4.1 ng Morpholino is sufficient to nearly completely inhibit splicing (Figure S6). To address the contribution of mych to epiboly delay phenotype in MZspg mutant embryos we injected 4.6 ng of mych-Sp-MO and 1.4 ng mych-ATG-MO into 1-cell stage embryos respectively. As control we injected the same amount of SCMO.
Cycloheximide experiment
MZspg embryos were injected with 10 pg pou5f1-VP16 mRNA at the 1-cell stage or left non-injected as controls. Embryos were treated with 15 mg/ml of cycloheximide (CHX, Calbiochem) dissolved in egg water. CHX was added at 1.5 hpf to allow for translation of injected mRNAs, but to block translation of the earliest zygotic transcripts. In the presence of CHX, direct Pou5f1 targets are transcribed after MBT, but these mRNAs are not translated, avoiding indirect downstream regulatory effects. Loss of ntl expression in CHX embryos was used as control for efficient inhibition of translation [71].
Plasmids used in this study
The zebrafish expression construct CS2+ Pou5f1-VP16 has been described [49]. The following myc gene EST clones were ordered from RZPD (Deutsches Ressourcenzentrum für Genomforschung GmbH): IMAGp998E208991Q1 (mych), IRBOp991F125D2 (myca), IRBOp991G0414D2 (mycb), IRBOp991E0238D2 (mycl1a), IRAKp961I20283Q2 (mycl1b).
Whole-mount in situ hybridization and in situ detection of apoptosis
Whole-mount in situ hybridization was performed as described [72].
The ApopTag Peroxidase In Situ Apoptosis Detection Kit (Chemicon/Merck Millipore) was used to detect apoptotic cells in early embryonic stages. To investigate whether MZspg embryos develop enhanced apoptosis, MZspg and WT embryos were fixed in 4% PFA at several developmental stages between 32-cell and bud stage. The fixed embryos were incubated in 100% methanol to make them permeable for the TUNEL staining. To analyze the influence of Mych and p53 on this phenotype, 105 pg mych mRNA and/or 4.2 ng p53-MO were injected at one-cell stage into MZspg embryos. Non-injected WT and MZspg embryos were used as controls. Embryos were fixed at 10 hpf. To quantify apoptosis image z-stacks of 5-14 stained embryos were taken for each experiment using transmitted light microscopy. Thereafter, maximum intensity projections of z-stacks were calculated for each embryo using ImageJ (Rasband, W.S., ImageJ, U.S. National Institutes of Health, Bethesda, Maryland, USA, http://imagj.nih.gov/ij/, 1997–2012) and apoptotic nuclei were automatically detected using the following set of parameters in Volocity Image Analysis Software (PerkinElmer): (1) Find Objects by Intensity; (2) exclude objects smaller than 10 μm2; (3) separate touching objects greater than 50 μm2 (Figure S7).
Cell proliferation analysis
To determine a potential role of Mych in proliferation, 105 pg mych mRNA and/or 4.2 ng p53-MO were injected into one-cell stage MZspg embryos. Non-injected WT and MZspg embryos were used as controls. Embryos were fixed in 4% PFA at 10 hpf. The embryos were incubated in 100% methanol to make them permeable for the Sytox-Green DNA stain. Embryos were stained in 2 μM Sytox-Green (Invitrogen) at room temperature (20 to 25°C) for 2 hours. Mitotic nuclei (prometa-, meta- and anaphase nuclei) were manually detected by their characteristic chromosome arrangement and dense DNA structure, resulting in higher Sytox-Green intensity. The mitotic index is the proportion of mitotic nuclei to the total number of nuclei. For comparisons WT mitotic index was normalized to 1.
Microinjections
mRNAs were synthesized using the mMessage mMachine kit (Invitrogen) according to manufacturer's instructions. mRNA or morpholino were injected into the yolk of freshly fertilized zygotes (younger than 15 minutes) mounted on 1% (w/v) agarose ramps, using microinjection pipettes connected to an air pressure driven microinjector. A volume of 0.5–1 nl, containing mRNA or morpholino and 0.5% (v/v) phenol red in water, was injected into each zygote.
Quantitative RT-PCR
60–100 embryos per sample were snap-frozen in liquid nitrogen, and total RNA was isolated using the RNA Easy kit (Qiagen). cDNA was synthesized using Superscript III kit (Invitrogen). cDNA was amplified using gene-specific primers and ABsolute SYBR Green Fluorescein (ABgene, Thermo Scientific) according to manufacturer's instructions on an Bio-Rad i-Cycler. Results were calculated using the ddCT method and zebrafish ef1α as a normalization control. Primers used: mych forward 5′-CCAGGCACTGGAGAGCGAAGAC-3′, reverse 5′-AGAGCGTGTCAGGGTGCTGGAG-3′; mycl1b forward 5′-GGTCAGAATCTCGCACCGACTC-3′, reverse 5′-ATGTGGAAACGCTTCATGCAGG-3′; and ef1α forward 5′-CCTGGGAGTGAAACGCTGATC-3′, reverse 5′-CCGATCTTCTTGATGTATGCGCTG-3′. Calculations are based on Pfaffl [73].
Statistical analysis
Error bars in Figures show Standard Error of the Mean (SEM). Significance was evaluated using Student's T-test (Microsoft Excel).
Supporting Information
Phylogenetic analysis of the six zebrafish myc genes. Phylogenetic tree of myc family genes. In zebrafish two paralogous genes each exist for L-myc, mycl1a and mycl1b, and c-myc, myca and mycb. In addition, there is a single copy each for the mycn and mych genes. The later one is closely related to the N-myc and c-myc genes, but has no known homologues in other vertebrate species. Trees were built using phylip proml, and 100 datasets for bootstrapping. The alignment was done with clustalw. Sequences used are: hs-nmyc NP_005369.2, mm-nmyc NP_032735.2, ha-myc NP_002458.2, mm-myc NP_034979.3, hs-lmyc NP_001028253.1, mm-lmyc NP_032532.1, hs-smyc E10909, mm-smyc NP_034980.1, mm-bmyc NP_075815.2, dr-myca NP_571487.2, dr-mycb NP_956466.1, dr-mych XP_005166306.1, dr-nmyc NP_997779.1, dr-mycl1a NP_998102.1, dr-mycl1b NP_001038607.1.
(TIF)
WISH analysis of myc gene expression at 24 hpf. WISH analysis of myca, mycb, mych, mycl1a and mycl1b expression in WT (A-J). All embryos are shown in lateral (left column) and dorsal (right column) view. notail expression was used as control to evaluate stain background levels in the head, where notail is not expressed (K-L). All myc genes show a gene specific expression pattern and are mainly expressed in proliferating and neural tissues. The expression of the c-Myc orthologous genes, myca and mycb (A-D), and of the L-Myc orthologous genes, mycl1a and mycl1b (G-J), show partially complementary patterns.
(TIF)
myc gene expression profiles and transcriptional regulation by Pou5f1 and Sox2. (A, C, E, G, I, K) Microarray time series data [47] of myc gene expression profiles in WT (white squares) and MZspg (black triangles) within the first 8–hours of development. The highest expression value for each gene was normalized to 100. (B, D, F, H, J, L) Microarray analysis of transcriptomes of MZspg embryos injected with mRNA encoding Pou5f1, and developed in presence of CHX from 64-cell stage on. Data are from [47]. Non-injected MZspg control was normalized to 1.
(TIF)
Morphological analysis of CHX treated and pou5f1-VP16 mRNA injected control embryos. (A) Morphological phenotype of MZspg embryos treated with CHX from 64-cell stage on and developed until WT control embryos reached 60% epiboly. Treated embryos are arrested before sphere stage, but do not degenerate until 60% epiboly equivalent age. (B) The injection of 10 pg pou5f1-VP16 mRNA into 1-cell MZspg embryos is sufficient to rescue the MZspg phenotype, but it also may ventralize the embryo as Pou5f1 overexpression in WT would do [74]. The experiment demonstrates that pou5f1-VP16 was injected in our experiments at concentrations that could be considered physiological for embryonic development.
(TIF)
Analysis of the mitotic index at 90%-epiboly. Quantification of the proportion of cells undergoing cell division in WT, MZspg and MZspg injected with mych mRNA and/or p53 morpholinos by calculating the mitotic index (ratio between the total number of nuclei and nuclei undergoing cell division). (A). The calculated mitotic indices are not significantly different between the different genotypes and experimental conditions. Mitotic index of WT embryos was set to 1. Confocal microscopy Z-stacks were taken from the animal region of 90%-epiboly stage embryos, whose nuclei are stained by Sytox fluorescent DNA dye (B). Chromatin is highly condensed during meta- and anaphase of the cell division, which leads to an increase in Sytox stain intensity (B; arrows).
(TIF)
Testing of mych morpholino functionality. (A-H) The functionality of the mych translation-blocking morpholino (ATG-MO) was tested by injecting a fusion mRNA, where the MO target sequence was fused to the gfp ORF at the start ATG, together with different concentrations of the ATG-MO into one-cell stage embryos. The GFP signal was analyzed using fluorescence microscopy (left panel) and the normal morphology of the embryos after morpholino injection was documented using transmitted light microscopy (right panel). The translation of gfp was completely blocked by injecting as little as 1.4 ng of the ATG-MO (C). For the splice-blocking morpholino (Sp-MO) the functionality was tested by RT-PCR using a pair of primers overlapping the second intron (I), whose splicing sites are targeted by the mych-Sp-MO. In WT the 162 bp fragment reflects the proper splicing of the pre-mRNA, whereas after the injection of 4.1 ng or more of mych-Sp-MO the detected fragment contains the intron and its size increased to 396 bp (I).
(TIF)
Quantification of apoptosis in WT and MZ spg embryos at bud stage. Detection of apoptotic cells by TUNEL staining (A) and subsequent computational image analysis (B). The images show a lateral maximum intensity projection of a z-stack taken from a single embryo. (B) The same z-stack after automatic object recognition using Volocity software (Perkin-Elmer), where most of the apoptotic cells are marked in red (B).
(TIF)
mych and potentially also mycl1b are directly regulated by Pou5f1. (Referring to: Figure 3)
(PDF)
Analysis of the Mych contribution to the morphology of the MZ spg mutant phenotype. (Referring to: Figure 5)
(PDF)
Analysis of the mitotic index at 90%-epiboly by Sytox nuclear stain. (Referring to: Figure S5)
(PDF)
Mych overexpression and p53 knockdown suppress cell death in MZ spg gastrulae. (Referring to: Figure 7)
(PDF)
Acknowledgments
We thank Karen Lunde for discussion and initial analysis of the MZspg cell death phenotype, Manuel Leichsenring for work on ChIP-Seq and advice on qPCR, and Rebecca Moessner for technical assistance.
Funding Statement
This work was supported by BMBF FORSYS center FRISYS, the Excellence Initiative of the German Federal and State Governments (BIOSS Centre for Biological Signalling Studies EXC 294), and German Research Foundation (DFG-SFB592-A3 & SFB850-A1, WD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jacobson MD, Weil M, Raff MC (1997) Programmed cell death in animal development. Cell 88: 347–354. [DOI] [PubMed] [Google Scholar]
- 2. Kerr JF, Wyllie AH, Currie AR (1972) Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26: 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Metzstein MM, Stanfield GM, Horvitz HR (1998) Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet 14: 410–416. [DOI] [PubMed] [Google Scholar]
- 4. Abrams JM (1999) An emerging blueprint for apoptosis in Drosophila. Trends Cell Biol 9: 435–440. [DOI] [PubMed] [Google Scholar]
- 5. Taylor RC, Cullen SP, Martin SJ (2008) Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol 9: 231–241. [DOI] [PubMed] [Google Scholar]
- 6. Danial NN, Korsmeyer SJ (2004) Cell death: critical control points. Cell 116: 205–219. [DOI] [PubMed] [Google Scholar]
- 7. Fuchs Y, Steller H (2011) Programmed cell death in animal development and disease. Cell 147: 742–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krumschnabel G, Podrabsky JE (2009) Fish as model systems for the study of vertebrate apoptosis. Apoptosis 14: 1–21. [DOI] [PubMed] [Google Scholar]
- 9. Eimon PM, Kratz E, Varfolomeev E, Hymowitz SG, Stern H, et al. (2006) Delineation of the cell-extrinsic apoptosis pathway in the zebrafish. Cell Death Differ 13: 1619–1630. [DOI] [PubMed] [Google Scholar]
- 10. Inohara N, Nunez G (2000) Genes with homology to mammalian apoptosis regulators identified in zebrafish. Cell Death Differ 7: 509–510. [DOI] [PubMed] [Google Scholar]
- 11. Kratz E, Eimon PM, Mukhyala K, Stern H, Zha J, et al. (2006) Functional characterization of the Bcl-2 gene family in the zebrafish. Cell Death Differ 13: 1631–1640. [DOI] [PubMed] [Google Scholar]
- 12. Eimon PM, Ashkenazi A (2010) The zebrafish as a model organism for the study of apoptosis. Apoptosis 15: 331–349. [DOI] [PubMed] [Google Scholar]
- 13. Storer NY, Zon LI (2010) Zebrafish models of p53 functions. Cold Spring Harb Perspect Biol 2: a001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eilers M, Schirm S, Bishop JM (1991) The MYC protein activates transcription of the alpha-prothymosin gene. Embo J 10: 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miner JH, Wold BJ (1991) c-myc inhibition of MyoD and myogenin-initiated myogenic differentiation. Mol Cell Biol 11: 2842–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coppola JA, Cole MD (1986) Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature 320: 760–763. [DOI] [PubMed] [Google Scholar]
- 17. van Riggelen J, Yetil A, Felsher DW (2010) MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10: 301–309. [DOI] [PubMed] [Google Scholar]
- 18. Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, et al. (1992) Induction of apoptosis in fibroblasts by c-myc protein. Cell 69: 119–128. [DOI] [PubMed] [Google Scholar]
- 19. Blackwood EM, Eisenman RN (1991) Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 251: 1211–1217. [DOI] [PubMed] [Google Scholar]
- 20. Prendergast GC, Lawe D, Ziff EB (1991) Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell 65: 395–407. [DOI] [PubMed] [Google Scholar]
- 21. Amati B, Dalton S, Brooks MW, Littlewood TD, Evan GI, et al. (1992) Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature 359: 423–426. [DOI] [PubMed] [Google Scholar]
- 22. Amin C, Wagner AJ, Hay N (1993) Sequence-specific transcriptional activation by Myc and repression by Max. Mol Cell Biol 13: 383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herkert B, Eilers M (2010) Transcriptional repression: the dark side of myc. Genes Cancer 1: 580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li LH, Nerlov C, Prendergast G, MacGregor D, Ziff EB (1994) c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. Embo J 13: 4070–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vennstrom B, Sheiness D, Zabielski J, Bishop JM (1982) Isolation and characterization of c-myc, a cellular homolog of the oncogene (v-myc) of avian myelocytomatosis virus strain 29. J Virol 42: 773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, et al. (1983) Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature 305: 245–248. [DOI] [PubMed] [Google Scholar]
- 27. Nau MM, Brooks BJ, Battey J, Sausville E, Gazdar AF, et al. (1985) L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature 318: 69–73. [DOI] [PubMed] [Google Scholar]
- 28. Dang CV (2012) MYC on the path to cancer. Cell 149: 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Swartling FJ (2012) Myc proteins in brain tumor development and maintenance. Ups J Med Sci 117: 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wolfer A, Ramaswamy S (2011) MYC and metastasis. Cancer Res 71: 2034–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Domashenko AD, Latham KE, Hatton KS (1997) Expression of myc-family, myc-interacting, and myc-target genes during preimplantation mouse development. Mol Reprod Dev 47: 57–65. [DOI] [PubMed] [Google Scholar]
- 32. Downs KM, Martin GR, Bishop JM (1989) Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev 3: 860–869. [DOI] [PubMed] [Google Scholar]
- 33. Mugrauer G, Alt FW, Ekblom P (1988) N-myc proto-oncogene expression during organogenesis in the developing mouse as revealed by in situ hybridization. J Cell Biol 107: 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zimmerman KA, Yancopoulos GD, Collum RG, Smith RK, Kohl NE, et al. (1986) Differential expression of myc family genes during murine development. Nature 319: 780–783. [DOI] [PubMed] [Google Scholar]
- 35. Pelengaris S, Khan M (2003) The many faces of c-MYC. Arch Biochem Biophys 416: 129–136. [DOI] [PubMed] [Google Scholar]
- 36. Davis AC, Wims M, Spotts GD, Hann SR, Bradley A (1993) A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev 7: 671–682. [DOI] [PubMed] [Google Scholar]
- 37. Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. [DOI] [PubMed] [Google Scholar]
- 38. Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, et al. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chappell J, Sun Y, Singh A, Dalton S (2013) MYC/MAX control ERK signaling and pluripotency by regulation of dual-specificity phosphatases 2 and 7. Genes Dev 27: 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, et al. (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440. [DOI] [PubMed] [Google Scholar]
- 41. Meijer AH, van dSAM, Cunha C, Lamers GE, Laplante MA, et al. (2008) Identification and real-time imaging of a myc-expressing neutrophil population involved in inflammation and mycobacterial granuloma formation in zebrafish. Dev Comp Immunol 32: 36–49. [DOI] [PubMed] [Google Scholar]
- 42. Schreiber-Agus N, Horner J, Torres R, Chiu FC, DePinho RA (1993) Zebra fish myc family and max genes: differential expression and oncogenic activity throughout vertebrate evolution. Mol Cell Biol 13: 2765–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Amores A, Force A, Yan YL, Joly L, Amemiya C, et al. (1998) Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711–1714. [DOI] [PubMed] [Google Scholar]
- 44. Loeb-Hennard C, Kremmer E, Bally-Cuif L (2005) Prominent transcription of zebrafish N-myc (nmyc1) in tectal and retinal growth zones during embryonic and early larval development. Gene Expr Patterns 5: 341–347. [DOI] [PubMed] [Google Scholar]
- 45. Hong SK, Tsang M, Dawid IB (2008) The mych gene is required for neural crest survival during zebrafish development. PLoS One 3: e2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thisse B, Thisse C (2004) Fast Release Clones: A High Throughput Expression Analysis.
- 47. Onichtchouk D, Geier F, Polok B, Messerschmidt DM, Mossner R, et al. (2010) Zebrafish Pou5f1-dependent transcriptional networks in temporal control of early development. Mol Syst Biol 6: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schulte-Merker S, van EFJ, Halpern ME, Kimmel CB, Nusslein-Volhard C (1994) no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development 120: 1009–1015. [DOI] [PubMed] [Google Scholar]
- 49. Lunde K, Belting HG, Driever W (2004) Zebrafish pou5f1/pou2, homolog of mammalian Oct4, functions in the endoderm specification cascade. Curr Biol 14: 48–55. [DOI] [PubMed] [Google Scholar]
- 50. Leichsenring M, Maes J, Mossner R, Driever W, Onichtchouk D (2013) Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science 341: 1005–1009. [DOI] [PubMed] [Google Scholar]
- 51. Lachnit M, Kur E, Driever W (2008) Alterations of the cytoskeleton in all three embryonic lineages contribute to the epiboly defect of Pou5f1/Oct4 deficient MZspg zebrafish embryos. Dev Biol 315: 1–17. [DOI] [PubMed] [Google Scholar]
- 52. Reim G, Brand M (2006) Maternal control of vertebrate dorsoventral axis formation and epiboly by the POU domain protein Spg/Pou2/Oct4. Development 133: 2757–2770. [DOI] [PubMed] [Google Scholar]
- 53. Song S, Eckerle S, Onichtchouk D, Marrs JA, Nitschke R, et al. (2013) Pou5f1-dependent EGF expression controls E-cadherin endocytosis, cell adhesion, and zebrafish epiboly movements. Dev Cell 24: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Penaloza C, Lin L, Lockshin RA, Zakeri Z (2006) Cell death in development: shaping the embryo. Histochem Cell Biol 126: 149–158. [DOI] [PubMed] [Google Scholar]
- 55. Cole LK, Ross LS (2001) Apoptosis in the developing zebrafish embryo. Dev Biol 240: 123–142. [DOI] [PubMed] [Google Scholar]
- 56. Langheinrich U, Hennen E, Stott G, Vacun G (2002) Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signaling. Curr Biol 12: 2023–2028. [DOI] [PubMed] [Google Scholar]
- 57. Hourdry J, Brulfert A, Gusse M, Schoevaert D, Taylor MV, et al. (1988) Localization of c-myc expression during oogenesis and embryonic development in Xenopus laevis. Development 104: 631–641. [DOI] [PubMed] [Google Scholar]
- 58. Vize PD, Vaughan A, Krieg P (1990) Expression of the N-myc proto-oncogene during the early development of Xenopus laevis. Development 110: 885–896. [DOI] [PubMed] [Google Scholar]
- 59. Wierstra I, Alves J (2008) The c-myc promoter: still MysterY and challenge. Adv Cancer Res 99: 113–333. [DOI] [PubMed] [Google Scholar]
- 60. Chen X, Vega VB, Ng HH (2008) Transcriptional regulatory networks in embryonic stem cells. Cold Spring Harb Symp Quant Biol 73: 203–209. [DOI] [PubMed] [Google Scholar]
- 61. Guo Y, Mantel C, Hromas RA, Broxmeyer HE (2008) Oct-4 is critical for survival/antiapoptosis of murine embryonic stem cells subjected to stress: effects associated with Stat3/survivin. Stem Cells 26: 30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen T, Du J, Lu G (2012) Cell growth arrest and apoptosis induced by Oct4 or Nanog knockdown in mouse embryonic stem cells: a possible role of Trp53. Mol Biol Rep 39: 1855–1861. [DOI] [PubMed] [Google Scholar]
- 63. Wang YD, Cai N, Wu XL, Cao HZ, Xie LL, et al. (2013) OCT4 promotes tumorigenesis and inhibits apoptosis of cervical cancer cells by miR-125b/BAK1 pathway. Cell Death Dis 4: e760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lin Y, Yang Y, Li W, Chen Q, Li J, et al. (2012) Reciprocal regulation of Akt and Oct4 promotes the self-renewal and survival of embryonal carcinoma cells. Mol Cell 48: 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kotkamp K, Mössner R, Allen A, Onichtchouk D, Driever W (2013) A Pou5f1/Oct4 dependent Klf2a, Klf2b, and Klf17 regulatory sub-network contributes to EVL and ectoderm development during zebrafish embryogenesis. http://dx.doi.org/10.1016/j.ydbio.2013.10.025i. [DOI] [PubMed]
- 66. Adhikary S, Eilers M (2005) Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645. [DOI] [PubMed] [Google Scholar]
- 67. Slack A, Chen Z, Tonelli R, Pule M, Hunt L, et al. (2005) The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci U S A 102: 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Haupt S, Berger M, Goldberg Z, Haupt Y (2003) Apoptosis - the p53 network. J Cell Sci 116: 4077–4085. [DOI] [PubMed] [Google Scholar]
- 69. Belting HG, Hauptmann G, Meyer D, Abdelilah-Seyfried S, Chitnis A, et al. (2001) spiel ohne grenzen/pou2 is required during establishment of the zebrafish midbrain-hindbrain boundary organizer. Development 128: 4165–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Westerfield M (2000) The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). Eugene: Univ. of Oregon Press.
- 71. Leung T, Bischof J, Soll I, Niessing D, Zhang D, et al. (2003) bozozok directly represses bmp2b transcription and mediates the earliest dorsoventral asymmetry of bmp2b expression in zebrafish. Development 130: 3639–3649. [DOI] [PubMed] [Google Scholar]
- 72. Hauptmann G, Gerster T (2000) Multicolor whole-mount in situ hybridization. Methods Mol Biol 137: 139–148. [DOI] [PubMed] [Google Scholar]
- 73. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Belting HG, Wendik B, Lunde K, Leichsenring M, Mossner R, et al. (2011) Pou5f1 contributes to dorsoventral patterning by positive regulation of vox and modulation of fgf8a expression. Dev Biol 356: 323–336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic analysis of the six zebrafish myc genes. Phylogenetic tree of myc family genes. In zebrafish two paralogous genes each exist for L-myc, mycl1a and mycl1b, and c-myc, myca and mycb. In addition, there is a single copy each for the mycn and mych genes. The later one is closely related to the N-myc and c-myc genes, but has no known homologues in other vertebrate species. Trees were built using phylip proml, and 100 datasets for bootstrapping. The alignment was done with clustalw. Sequences used are: hs-nmyc NP_005369.2, mm-nmyc NP_032735.2, ha-myc NP_002458.2, mm-myc NP_034979.3, hs-lmyc NP_001028253.1, mm-lmyc NP_032532.1, hs-smyc E10909, mm-smyc NP_034980.1, mm-bmyc NP_075815.2, dr-myca NP_571487.2, dr-mycb NP_956466.1, dr-mych XP_005166306.1, dr-nmyc NP_997779.1, dr-mycl1a NP_998102.1, dr-mycl1b NP_001038607.1.
(TIF)
WISH analysis of myc gene expression at 24 hpf. WISH analysis of myca, mycb, mych, mycl1a and mycl1b expression in WT (A-J). All embryos are shown in lateral (left column) and dorsal (right column) view. notail expression was used as control to evaluate stain background levels in the head, where notail is not expressed (K-L). All myc genes show a gene specific expression pattern and are mainly expressed in proliferating and neural tissues. The expression of the c-Myc orthologous genes, myca and mycb (A-D), and of the L-Myc orthologous genes, mycl1a and mycl1b (G-J), show partially complementary patterns.
(TIF)
myc gene expression profiles and transcriptional regulation by Pou5f1 and Sox2. (A, C, E, G, I, K) Microarray time series data [47] of myc gene expression profiles in WT (white squares) and MZspg (black triangles) within the first 8–hours of development. The highest expression value for each gene was normalized to 100. (B, D, F, H, J, L) Microarray analysis of transcriptomes of MZspg embryos injected with mRNA encoding Pou5f1, and developed in presence of CHX from 64-cell stage on. Data are from [47]. Non-injected MZspg control was normalized to 1.
(TIF)
Morphological analysis of CHX treated and pou5f1-VP16 mRNA injected control embryos. (A) Morphological phenotype of MZspg embryos treated with CHX from 64-cell stage on and developed until WT control embryos reached 60% epiboly. Treated embryos are arrested before sphere stage, but do not degenerate until 60% epiboly equivalent age. (B) The injection of 10 pg pou5f1-VP16 mRNA into 1-cell MZspg embryos is sufficient to rescue the MZspg phenotype, but it also may ventralize the embryo as Pou5f1 overexpression in WT would do [74]. The experiment demonstrates that pou5f1-VP16 was injected in our experiments at concentrations that could be considered physiological for embryonic development.
(TIF)
Analysis of the mitotic index at 90%-epiboly. Quantification of the proportion of cells undergoing cell division in WT, MZspg and MZspg injected with mych mRNA and/or p53 morpholinos by calculating the mitotic index (ratio between the total number of nuclei and nuclei undergoing cell division). (A). The calculated mitotic indices are not significantly different between the different genotypes and experimental conditions. Mitotic index of WT embryos was set to 1. Confocal microscopy Z-stacks were taken from the animal region of 90%-epiboly stage embryos, whose nuclei are stained by Sytox fluorescent DNA dye (B). Chromatin is highly condensed during meta- and anaphase of the cell division, which leads to an increase in Sytox stain intensity (B; arrows).
(TIF)
Testing of mych morpholino functionality. (A-H) The functionality of the mych translation-blocking morpholino (ATG-MO) was tested by injecting a fusion mRNA, where the MO target sequence was fused to the gfp ORF at the start ATG, together with different concentrations of the ATG-MO into one-cell stage embryos. The GFP signal was analyzed using fluorescence microscopy (left panel) and the normal morphology of the embryos after morpholino injection was documented using transmitted light microscopy (right panel). The translation of gfp was completely blocked by injecting as little as 1.4 ng of the ATG-MO (C). For the splice-blocking morpholino (Sp-MO) the functionality was tested by RT-PCR using a pair of primers overlapping the second intron (I), whose splicing sites are targeted by the mych-Sp-MO. In WT the 162 bp fragment reflects the proper splicing of the pre-mRNA, whereas after the injection of 4.1 ng or more of mych-Sp-MO the detected fragment contains the intron and its size increased to 396 bp (I).
(TIF)
Quantification of apoptosis in WT and MZ spg embryos at bud stage. Detection of apoptotic cells by TUNEL staining (A) and subsequent computational image analysis (B). The images show a lateral maximum intensity projection of a z-stack taken from a single embryo. (B) The same z-stack after automatic object recognition using Volocity software (Perkin-Elmer), where most of the apoptotic cells are marked in red (B).
(TIF)
mych and potentially also mycl1b are directly regulated by Pou5f1. (Referring to: Figure 3)
(PDF)
Analysis of the Mych contribution to the morphology of the MZ spg mutant phenotype. (Referring to: Figure 5)
(PDF)
Analysis of the mitotic index at 90%-epiboly by Sytox nuclear stain. (Referring to: Figure S5)
(PDF)
Mych overexpression and p53 knockdown suppress cell death in MZ spg gastrulae. (Referring to: Figure 7)
(PDF)