Abstract
Two evolutionarily distant plant species, rice (Oryza sativa L.), a short-day (SD) plant, and Arabidopsis thaliana, a long-day plant, share a conserved genetic network controlling photoperiodic flowering. The orthologous floral regulators—rice Heading date 1 (Hd1) and Arabidopsis CONSTANS (CO)—integrate circadian clock and external light signals into mRNA expression of the FLOWERING LOCUS T (FT) group floral inducer. Here, we report that the rice Early heading date 1 (Ehd1) gene, which confers SD promotion of flowering in the absence of a functional allele of Hd1, encodes a B-type response regulator that might not have an ortholog in the Arabidopsis genome. Ehd1 mRNA was induced by 1-wk SD treatment, and Ehd1 may promote flowering by inducing FT-like gene expression only under SD conditions. Microarray analysis further revealed a few MADS box genes downstream of Ehd1. Our results indicate that a novel two-component signaling cascade is integrated into the conserved pathway in the photoperiodic control of flowering in rice.
Keywords: Photoperiodism, flowering time, two-component signaling
The gene network underlying photoperiodic flowering consists of photoreceptors, circadian clock systems, and floral integrator genes (Izawa et al. 2002; Mouradov et al. 2002; Simpson and Dean 2002; Yanovsky and Kay 2002; Hayama et al. 2003). Molecular genetic studies have revealed that orthologous genes in rice (Oryza sativa), a short-day (SD) plant, and Arabidopsis thaliana, a long-day (LD) plant, such as Heading date 1 (Hd1)/CONSTANS (CO) and Hd3a/FLOWERING LOCUS T (FT), are implicated in the regulation of photoperiodic flowering (Kardailsky et al. 1999; Kobayashi et al. 1999; Samach et al. 2000; Yano et al. 2000; Izawa et al. 2002; Kojima et al. 2002).
The molecular basis of day-length measurement in both species was recently explained according to a physiology-based model, termed the external coincidence model, which was first proposed in the 1930s (Izawa et al. 2002; Yanovsky and Kay 2002; Hayama and Coupland 2003). First, daily light/dark cycles set the phase of circadian clocks. Then, the circadian clocks express Hd1/CO mainly during the subjective night. In rice, Hd1 inhibits floral transition by suppressing FT-like genes, including FTL, Hd3a, and RFT1, on long days, possibly interacting with Pfr phytochromes produced during the daytime. On short days, the longer duration of darkness may lead to a gradual decrease in levels of Pfr phytochrome during the night, resulting in alteration of the function of Hd1 from a suppressor to an activator of FT-like genes at dawn (Izawa et al. 2002). Therefore, Hd1 promotes flowering only under SD conditions. In Arabidopsis, long days cause CO expression at dusk, although short days do not. Recently, it was revealed that the day-length-dependent CO expression was caused through interaction mediated by FKF1 (FLAVIN-BINDING, KELCH REPEAT, F-BOX 1) between blue-light signal transduction and circadian clocks (Imaizumi et al. 2003). CRYPTOCHROME 2 (or PHYTOCHROME A)-mediated light signaling makes the CO protein an activator of FT at dusk (Yanovsky and Kay 2002). Therefore, CO promotes flowering under LD conditions. Thus, the regulation of FT group genes by Hd1/CO plays a central role in photoperiodism of both SD and LD plants.
Regardless of the important role of Hd1, this model does not fully explain photoperiodic flowering in rice, because Hd1-deficient strains exhibit a significant delay of flowering under continuous-light conditions compared with SD conditions, and still preferentially express FT-like genes at daytime under SD conditions (Izawa et al. 2002). It has also been shown that Hd2, another flowering-time quantitative trait locus (QTL) of rice, contributes to flowering responses to photoperiod independently of Hd1 (Yano et al. 2001).
To better understand the molecular mechanisms of photoperiodic flowering in rice, we studied a japonica cultivar, Taichung 65 (T65), which exhibits a broad regional adaptability because of its reduced response to photoperiod (Nishida et al. 2001). In the present study, we show that T65 contains loss-of-function alleles of Hd1 and Ehd1 (Early heading date 1, formerly Ef(t); Doi and Yoshimura 1998), both of which play important roles in photoperiodic flowering of rice. We demonstrate that Ehd1 can function as a floral inducer independently of Hd1 and encodes a B-type response regulator. In addition, we analyzed four alleles of Ehd1 to determine a functional nucleotide polymorphism that is related to flowering phenotypes, because we earlier identified the Ehd1 locus as a QTL. We also reveal that Ehd1 induces mRNA expression of FT-like genes after SD treatment, indicating that a novel two-component signaling cascade is integrated in the conserved photoperiodic flowering pathway in rice. Microarray analysis and RT–PCR assays revealed several genes downstream of Ehd1, suggesting that Ehd1 signaling is specific to the control of photoperiodic flowering.
Results
Ehd1 can promote flowering independently of Hd1
The Ehd1 gene was originally identified as a flowering-time QTL on rice chromosome 10 by using a cross-combination between T65 and an accession of African rice (Oryza glaberrima Steud. accession no. IRGC104038; Doi et al. 1998). The O. glaberrima allele of Ehd1 (Ehd1-gla) is dominant and confers early flowering (Doi and Yoshimura 1998). This gene was detected as a QTL by using a cross-combination between T65 and Nipponbare. Single-point QTL analysis revealed that Nipponbare allele of Ehd1 (Ehd1-Nip) confers early flowering (Fig. 1A). In addition, no major QTL near the Ehd1 locus was found in a Nipponbare × Kasalath (an indica cultivar) cross; the only nearby QTL was a minor one, Hd14 (Yano et al. 2001). Therefore, T65 may carry a loss-of-function allele of Ehd1 (ehd1-T65), whereas O. glaberrima, Nipponbare, and Kasalath may contain functional Ehd1 alleles (Ehd1-gla, Ehd1-Nip, and Ehd1-Kas).
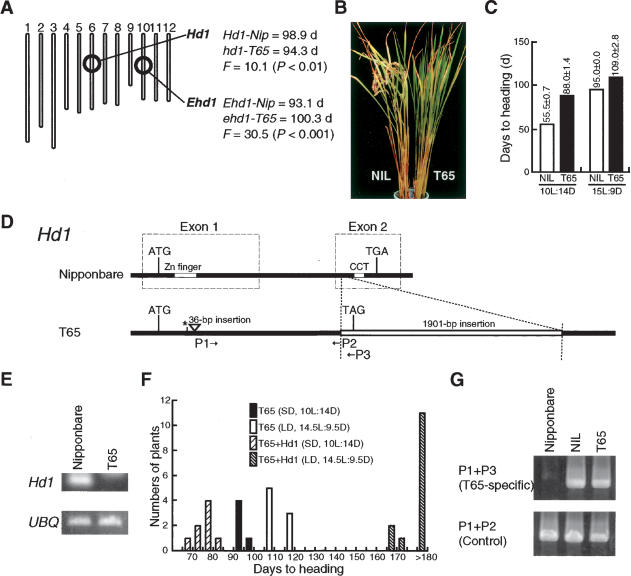
Figure 1.
Genetic analysis of Ehd1 and Hd1.(A) Major quantitative trait loci (QTLs) identified in 89 recombinant inbred lines derived from a cross between T65 and Nipponbare. Columns are chromosomes. Average of days to heading of each genotype and F values of single point QTL analyses at the Hd1 and Ehd1 loci are shown to the right of the rice linkage map. (B) Photograph of 80-day-old plants of NIL(Ehd1-gla) (left) and T65 (right) grown under SD (9L:15D) condition. (C) Days to heading of NIL(Ehd1-gla) and T65 under SD (10L:14D) and LD (15L:9D) conditions. (D) Allelic variation in Hd1. The genomic sequences of the functional Nipponbare and defective T65 alleles are shown schematically. The open triangle and asterisk indicate an insertion and a nucleotide substitution, respectively. T65 has a 1901-bp insertion in exon 2 that results in a premature stop codon (TAG) ahead of the CCT motif. P1, P2, and P3 are primers used in E and G. (E) mRNA levels of Hd1 in Nipponbare and T65. RT–PCR was performed with the combination of primers P1 and P2. UBQ (ubiquitin) is shown as a control. (F) Days to heading of T65 and a T2 transgenic line of T65 carrying the functional Hd1-Nip allele (T65 + Hd1). Similar results were obtained using independent transgenic lines. (G) NIL(Ehd1-gla) carries the defective hd1 allele from T65. T65-allele-specific bands were amplified with primers P1 and P3 from genomic DNA. Controls amplified with P1 and P2 are also shown.
To further characterize the function of Ehd1 in photoperiodic flowering of rice, we generated the nearly isogenic line NIL(Ehd1-gla), in which the Ehd1-gla allele was introgressed into T65 by six rounds of backcrossing. Comparison of flowering time between T65 and NIL(Ehd1-gla) demonstrated that Ehd1 confers early flowering, especially under SD condition (10 h light/14 h darkness, or 10L:14D; Fig. 1B,C).
We detected another QTL at the Hd1 locus in the T65 × Nipponbare cross (Fig. 1A), in which the Nipponbare allele of Hd1 was functional (Yano et al. 2000). This led us to analyze the genomic DNA sequence of Hd1 in T65. We found that a retroelement-like fragment was inserted into the second exon, and the T65 allele of Hd1 seemed to be a loss-of-function allele (Fig. 1D). Supporting this, Hd1 mRNA was severely reduced in T65, probably by nonsense-mediated decay (Fig. 1E). To further confirm the deficiency of Hd1 in T65, we transformed the functional Nipponbare allele of Hd1 (Hd1-Nip) into T65 to generate the T65 + Hd1 transgenic line. Hd1-Nip in T65 clearly promoted flowering under SD condition (10L:14D) and delayed flowering dramatically under LD condition (14.5L:9.5D; Fig. 1F). This is consistent with previous results (Yano et al. 2000; Izawa et al. 2002) in which Hd1 functioned as a floral inducer under SD and as a floral repressor under LD. We confirmed that NIL(Ehd1-gla) contains the defective T65 allele of Hd1 (Fig. 1G). Taken together, this genetic evidence clearly indicates that Ehd1 can function to promote flowering in rice, independently of Hd1.
Ehd1 encodes a B-type response regulator
To confirm the Hd1-independent photoperiodic flowering pathway in rice, we performed map-based cloning of Ehd1 (Fig. 2A–D). High-resolution mapping revealed that Ehd1 is in a 16-kb genomic region on the BAC clone OSJNBa0071K18 (Fig. 2B). Transformation of T65 with the corresponding Kasalath genomic fragments verified that Ehd1 encodes a B-type response regulator (RR; Hwang et al. 2002) of a 341 amino acid protein (Fig. 3A,B). It has a receiver domain at its N terminus and a GARP (Golden2, Arabidopsis RESPONSE REGULATOR [ARR], and Chlamydomonas regulatory protein of P-starvation acclimatization response [Psr1]) DNA-binding motif (Riechmann et al. 2000) in the middle portion (Fig. 3B–D).
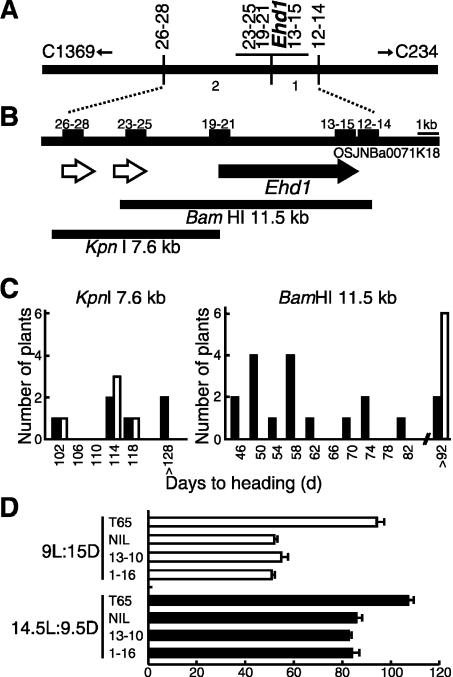
Figure 2.
Map-based cloning of Ehd1.(A) High-resolution linkage map of Ehd1 generated by using a mapping population of >2500 plants. Ehd1 is located on the Clemson BAC clone OSJNBa0071K18 (GenBank accession no. AC027038). (B) Physical map of Ehd1. The CAPS markers (12–14, 13–15, 19–21, 23–25, and 26–28) used for the linkage analysis are indicated by bars. Genes predicted by the GENSCAN program (Burge and Karlin 1997) are shown by arrows. BamHI (11.5 kb) and KpnI (7.6 kb) genomic fragments from Kasalath were introduced into T65. (C) Days to heading of T0 transformants under SD (10L:14D) condition. (Open bar) Empty vector; (solid bar) vector with the genome fragment. Only plants transformed with the 11.5-kb BamHI fragment showed promotion of flowering. (D) Days to heading of two independent T2 lines homozygous for the Ehd1-Kas transgene (1–16, 13–10), NIL(Ehd1-gla) and T65 under SD (9L:15D) and LD (14.5L:9.5D) conditions.
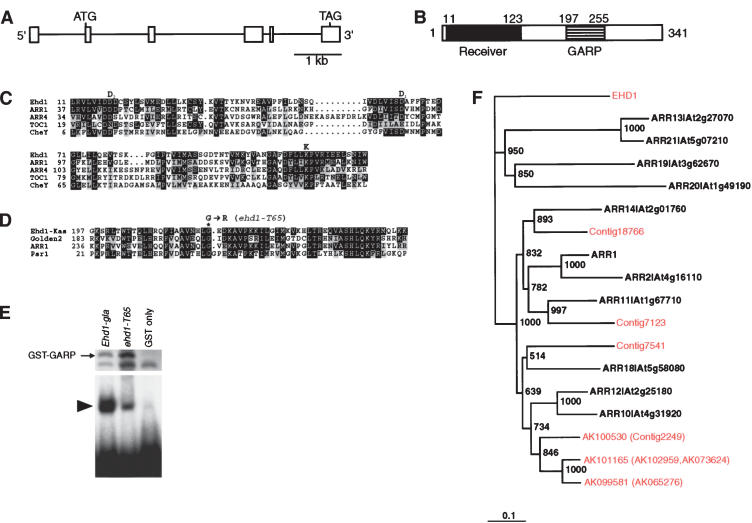
Figure 3.
Structure of Ehd1.(A) Gene organization of Ehd1. White boxes indicate exons. (B) Protein structure of Ehd1. (C) Comparison of receiver domains of Ehd1-Kas and representative Arabidopsis response regulators ARR1 (B-type; GenBank accession no. T51246), ARR4 (A-type; BAA34726), TOC1 (Pseudo-RR; AAF86252), and CheY (BAA15698). Residues identical to those in Ehd1 are in black boxes; similar ones are in gray. Asp (D1 and D2) and Lys (K) residues crucial for the phosphorelay function are conserved in Ehd1. (D) Alignment of GARP domains of Ehd1-Kas, Golden2 (AAG32325), ARR1 (T51246), and Psr1 (AAD55941). The position of amino acid variation in T65 (G→R) is indicated by an asterisk. Sequences are highlighted as in C. (E) Comparison of the DNA-binding ability of the GARP domains of Ehd1-gla and ehd1-T65. Amounts of fusion proteins in Escherichia coli extracts are shown at the top. Extra bands in the GST–GARP lanes may be partly digested products. The same extracts were used for gel-mobility shift assay (bottom). The arrowhead indicates the band corresponding to the retarded probe. (F) Phylogenetic comparison of B-type RRs between rice and Arabidopsis. ARR clones with MIPS (Munich Information Center for Protein Sequences) codes correspond to Arabidopsis B-type RRs. The others are EHD1 and B-type RRs from rice database information. B-type RRs corresponding to rice full-length cDNAs are indicated by AK accession numbers. Sequences prefixed with Contig are derived from the 93-11 rice genome sequences of the Beijing Genomics Institute.
To identify functional nucleotide variations corresponding to flowering phenotypes, we determined the genomic DNA sequences of Ehd1 from T65, Kasalath, and O. glaberrima and obtained that of Nipponbare from GenBank (accession no. AC027038). Comparison of the deduced amino acid sequences of these four Ehd1 alleles revealed that one amino acid was changed to Arg in the conserved residue of the GARP domain in the defective T65 allele, whereas the other three functional alleles had Gly at the corresponding position (Fig. 3D). The other amino acid changes are not restricted to T65 (Supplementary Fig. 1). We next used gel mobility-shift assays to compare the in vitro DNA-binding ability of the GARP domains of Ehd1-gla and ehd1-T65. Because the GARP domain of Ehd1 shares significant homology with that of ARR1, the same probes for ARR1 were used for this binding assay (Sakai et al. 2000). The results demonstrated that the DNA-binding ability of the ehd1-T65 allele was greatly reduced (Fig. 3E). Therefore, this Gly-to-Arg mutation may be crucial for the late-flowering phenotype of T65.
It is noteworthy that we could not define any Arabidopsis B-type RR as an Ehd1 ortholog when comparing the receiver and GARP domains of Ehd1 and of all rice and Arabidopsis B-type RR proteins found in the public databases (Fig. 3F). Other parts of Ehd1 than the receiver and GARP DNA-binding domains lacked significant homology with any database-registered protein. Furthermore, we found several pseudo-RRs and A-type RRs in the rice genome (Izawa et al. 2003; Murakami et al. 2003; data not shown). Therefore, Ehd1 seems to have evolved originally as a B-type RR since the separation of monocot (rice) from dicot (Arabidopsis). It is very likely that the Ehd1-signaling cascade exists in rice but not in Arabidopsis.
Diurnal expression of Ehd1
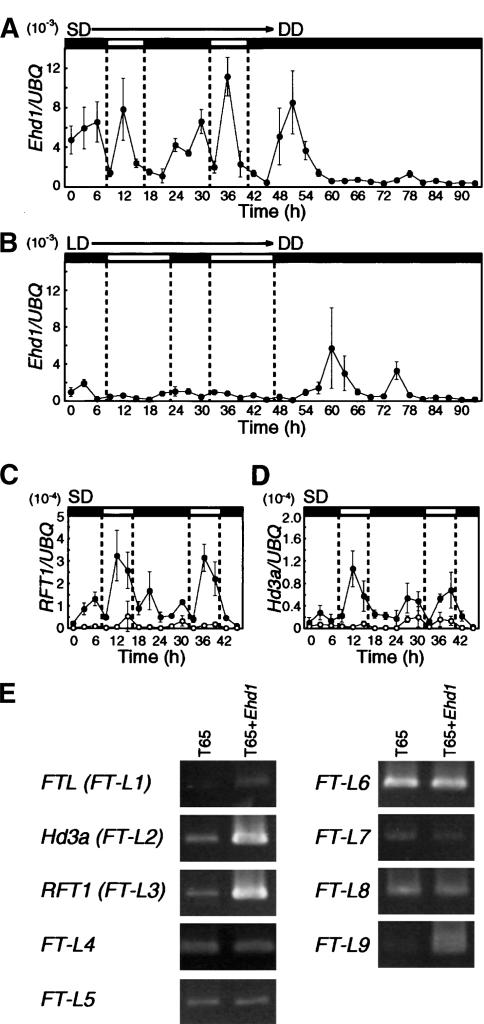
In the T65 background, Ehd1-gla exhibited a diurnal expression pattern under SD (9L:15D) condition and a rapid damping of circadian oscillation in subsequent continuous darkness (Fig. 4A). Two peaks, one at 2 h before dawn and the other at 4 h after dawn, occurred under SD. The second peak is probably an acute response to the light-on signals, because no such second peak was observed during continuous darkness. The damping of the rhythm in continuous darkness suggests that Ehd1 is controlled also by circadian clocks. Therefore, both circadian clocks and acute responses to light may cause the diurnal rhythm of Ehd1 mRNA levels. Under LD (15L:9D) conditions, the levels of Ehd1-gla mRNA were consistently low, whereas one peak was seen in subsequent continuous darkness 14 h after the onset of darkness, and a second peak occurred 15 h after the first peak (Fig. 4B). This suggests that Ehd1 mRNA may be induced by a long-night treatment. In addition, the induction of Ehd1 mRNA under SD strongly supports the idea that Ehd1 functions as a floral inducer in the SD-promotion pathway.
Figure 4.
Expression analyses of Ehd1, RFT1, and Hd3a. (A,B) Ehd1 mRNA expression in NIL(Ehd1-gla) under SD (A) and LD (B) condition. (C,D) Expression of RFT1 (C) and Hd3a (D) in NIL(Ehd1-gla) (solid symbols) and T65 (open symbols) under SD condition. The ratios of the average expression levels of Ehd1, RFT1, and Hd3a to that of UBQ (ubiquitin) are plotted on the graphs. The average values were obtained from at least three real-time RT–PCR assays. Periods of light and darkness are indicated with white and black bars, respectively. (E) FT-like gene expression in T65 and T65 + Ehd1 (Fig. 2D, lines 1–16).
mRNA levels of ehd1-T65 did not show any clear diurnal or circadian patterns or any significant differences between SD and LD conditions, although irreproducible peaks were observed under LD (Supplementary Fig. 2). Whether Ehd1-gla mRNA oscillation is regulated by its own gene product in some feedback mechanism remains to be examined.
Ehd1 promotes flowering by inducing FT-like genes
FT-like genes are floral inducers of rice (Izawa et al. 2002; Kojima et al. 2002) and the possible targets of regulation by Ehd1. Therefore, we assessed the mRNA expression of FT-like genes in NIL(Ehd1-gla) and T65. After SD treatment, mRNA expression of RFT1 and Hd3a increased in NIL(Ehd1-gla) and showed a diurnal expression with two peaks— one weak peak before dawn, and the other peak during daytime (Fig. 4C,D). In contrast, levels of expression were clearly lower in T65. Expression of RFT1, and Hd3a under LD was induced in neither T65 nor NIL(Ehd1-gla; data not shown). In addition, other FT-like gene expression was also examined by using a transgenic line that contains Ehd1-Kas in a T65 background (T65 + Ehd1), because at least nine FT-like genes exist in the rice genome (Izawa et al. 2002). In addition to Hd3a and RFT1, FTL and FT-L9 were induced by Ehd1 in the transgenic line (Fig. 4E). These results indicate that a novel two-component signaling cascade is integrated into the conserved photoperiod pathway in rice.
Other genes downstream of Ehd1
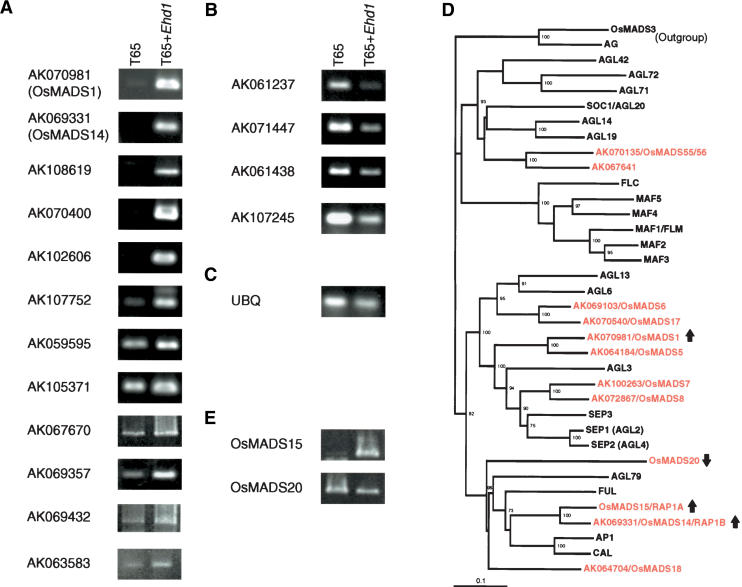
To further identify genes downstream of Ehd1, we performed a microarray analysis using custom 60-bp oligonucleotide arrays with 22,000 probes designed with full-length cDNA sequences of rice (Kikuchi et al. 2003). To minimize the effect of genetic background, we used T65 and T65 + Ehd1 for the microarray analysis. We collected leaves from 1-month-old plants of T65 and T65 + Ehd1 grown under SD, when floral transition could occur only in T6 5+ Ehd1, and prepared both total RNA and mRNA. Two independent color-swap experiments using both total RNA and mRNA provided us with 29 candidate genes, 21 up-regulated and 8 down-regulated, in T65 + Ehd1 (Supplementary Table 1). Six of the 21 up-regulated genes were detected in common between the experiments using total RNA and mRNA. Subsequent RT–PCR analysis verified that at least 12 genes were induced and four were reduced by Ehd1 upon floral transition (Fig. 5A–C; Table 1). We observed no difference in eight other genes by RT–PCR and could not detect any expression of the remaining five candidates. Some discrepancy between the results of microarray analysis and RT–PCR could be partly due to the low threshold of the log ratio, 0.25, when the candidate clones were selected in the microarray analysis.
Figure 5.
Genes downstream of Ehd1 in RT–PCR assays. The same cDNA samples as used in Figure 4E were used as templates. (A) Clones up-regulated in T65 + Ehd1. (B) Clones down-regulated in T65 + Ehd1. (C) UBQ (ubiquitin) as a control. (D) Phylogenetic tree of MADS box genes in rice and Arabidopsis. All rice and Arabidopsis MADS box proteins belonging to AP1/CAL/FUL, SEP, and SOC1 clades in public databases plus AG and OsMADS3 as an outgroup are included in the tree. Accession numbers corresponding to full-length rice cDNAs are also shown. (E) RT–PCR of the remaining two rice MADS box genes, OsMADS15 and OsMADS20.
Table 1.
List of genes whose mRNA expression was affected by Ehd1
| Up- or down-regulated | GenBank accession no. | Corresponding rice genes | Most similar database protein excluding Oryza sativa, [species], score (bits), E valuea |
|---|---|---|---|
| Up | AK059595 | gi|15236625 polygalacturonase, putative [Arabidopsis thaliana], 569, 1e-161 | |
| Up | AK105371 | gi|7433295 ω-6 desaturase [Gossypium hirsutum], 510, 1e-143 | |
| Up | AK069357 | gi|15810385 unknown protein [Arabidopsis thaliana], 375, 1e-127 | |
| Up | AK070981 | OsMADS1 (S53306) | gi|3892652 putative MADS-domain transcription factor [Zea mays], 320, 2e-86 |
| Up | AK069331 | OsMADS14 (AAF19047)/RAP1B (BAA94342) | gi|29372748 putative MADS-domain transcription factor [Zea mays], 264, 2e-69 |
| Up | AK067670 | LIRI (Q03200) | gi|7448057 cytokinin-repressed protein CR9 [Cucumis sativus], 87, 4e-16 |
| Up | AK069432 | gi|30698182 senescence-associated protein [Arabidopsis thaliana], 78, 5e-13 | |
| Up | AK070400 | gi|15241489 replication protein, putative [Arabidopsis thaliana], 75, 4e-12 | |
| Up | AK107752 | gi|7488303 ribonucleoprotein homolog T419.1 [Arabidopsis thaliana], 46, 1e-04 | |
| Up | AK063583 | gi|34530688 unnamed protein product [Homo sapiens], 41, 0.015 | |
| Up | AK102606 | gi|37546363 hypothetical protein [Homo sapiens], 36, 1.9 | |
| Up | AK108619 | No significant similarity found | |
| Down | AK061438 | Drought-induced S-like ribonuclease (AAL33776) | gi|20271131 S-like RNase [Triticum aestivum], 419, 1e-116 |
| Down | AK071447 | gi|8389329 (+)-delta-cadinene synthase [Gossypium hirsutum], 343, 5e-93 | |
| Down | AK061237 | gi|21554074 putative acid phosphatase [Arabidopsis thaliana], 297, 3e-79 | |
| Down | AK107245 | gi|15241275 ids4-related protein [Arabidopsis thaliana], 226, 9e-58 |
The results of NCBI BlastX search against “nr” database (2003.10.16) excluding Oryza sativa clones.
Microarray clones verified by RT—PCR are summarized.
Two of the twelve up-regulated genes are MIKC-type MADS box genes in rice. One of them is identical to OsMADS14/RAP1B, an AP1 ortholog of rice, and the other is OsMADS1, which is related to the SEP family of Arabidopsis (Fig. 5D) and is involved in flower development in rice (Jeon et al. 2000). These results are consistent with the view that Ehd1 is a floral inducer. The relatively small number of genes regulated by Ehd1 suggests that Ehd1 signaling is specific to floral induction in rice. The biological functions of those downstream genes remain to be analyzed, because there are no apparent links to floral induction among downstream genes other than the OsMADS genes.
Identification of the two MADS box genes as genes downstream of Ehd1 further led us to examine whether other MADS box genes possibly related to floral induction are affected by Ehd1. Phylogenetic analysis of MADS box genes using genomic and cDNA sequences of rice and Arabidopsis in public databases revealed that 12 rice MADS box genes belong to the AP1/CAL/FUL, SEP, and SOC1 clades (Fig. 5D, in red). Note that no rice MADS box gene has been found in the FLC clade (Izawa et al. 2003). Among those MADS box genes, 10 correspond to full-length cDNA clones in the microarrays (AK clones in Fig. 5D). We confirmed that mRNA expression of eight MADS box clones other than OsMADS14 and OsMADS1 was not affected by Ehd1 in the microarray analyses (data not shown). The remaining two MADS-box genes, OsMADS15 (another AP1 ortholog) and OsMADS20, were further examined by RT–PCR. The results indicate that OsMADS15 was induced in T65 + Ehd1, whereas OsMADS20 was slightly reduced (Fig. 5E). Therefore, three of the 12 rice MADS box genes—OsMADS14, OsMADS1, and OsMADS15—were induced by Ehd1 upon floral transition.
Discussion
Hd1-independent short-day promotion pathway in rice
We have demonstrated that a B-type RR gene that may not have an ortholog in Arabidopsis plays an important role in photoperiodic control of flowering in the SD plant rice. We could conclude that Ehd1 may not have an ortholog in Arabidopsis, as both rice and Arabidopsis genome sequences have been sequenced. However, it is possible that Arabidopsis B-type RR not-related to rice Ehd1 might be involved in floral pathways. The fact that Ehd1 can induce FT-like genes (Izawa et al. 2002; Kojima et al. 2002) in an Hd1-deficient background reveals that a novel two-component signaling cascade is integrated into the floral pathway. Floral regulation by Ehd1 would be assigned as an SD-promotion pathway in rice. Genetic analyses indicated that Ehd1-gla, Ehd1-Nip, and Ehd1-Kas alleles were functional, and ehd1-T65 was a recessive and loss-of-function allele. It is interesting that T65 still responds to photoperiod, although it has the loss-of-function alleles in both Hd1 and Ehd1. There may be another unidentified photoperiodic floral pathway in rice, which is not genetically identified yet in T65. Alternatively, this may be due to the remained Ehd1 activity of ehd1-T65 with its low DNA-binding ability, although ehd1-T65 mRNA expression pattern does not support this idea (Supplementary Fig. 2).
The expression pattern of Ehd1 is clearly distinct from that of Hd1, which functions as a circadian clock mediator. Hd1 mRNA is expressed during subjective night, and the expression patterns of Hd1 mRNA are similar under both LD and SD conditions (Izawa et al. 2002). In contrast, Ehd1 mRNA is induced only under SD conditions. This induction of Ehd1 mRNA was observed in the presence of the defective allele of Hd1, although the expression of Ehd1 is likely to be controlled by circadian clocks (Fig. 4A). Therefore, an unidentified circadian clock mediator other than Hd1 may be involved in Ehd1 mRNA expression.
Ehd1 as a B-type response regulator
It has been shown so far that plant two-component (or His-to-Asp phosphorelay) signaling cascades, which include histidine kinases (HKs), His-containing phosphotransfer proteins, and RRs, play important roles in phytohormone signaling in Arabidopsis (Stock et al. 2000; Lohrmann and Harter 2002). Ethylene receptors and cytokinin receptors encode HKs, although no molecular link between ethylene receptors and RRs was reported in Arabidopsis (Chang and Stadler 2001; Inoue et al. 2001). Recently, it was reported that an Arabidopsis HK, CKI1, which was believed to be a cytokinin receptor, plays a role in megagametogenesis (Pischke et al. 2002).
Phylogenetic analysis indicates that there are three major clades of RR in Arabidopsis, two clades of canonical RRs (A-type and B-type) and one atypical group (pseudo-RRs; Hwang et al. 2002). The rice Ehd1 belongs to the B-type RRs, although none of the 12 B-type RRs of Arabidopsis seem to be orthologous to Ehd1 (Fig. 3E). In Arabidopsis, ARR1, a B-type RR, functions in cytokinin signaling during Arabidopsis development (Sakai et al. 2001). In transient assays, the ARR1, ARR2, and ARR10 B-type RRs are involved in cytokinin signaling and regulate the mRNA expression of A-type RRs, such as ARR6 (Hwang and Sheen 2001). Recent work has revealed that the ARR11 B-type RR is also involved in cytokinin signaling (Imamura et al. 2003). In addition, a cytokinin-inducible A-type RR, ARR4, can interact directly with phytochrome B and may control stability of the Pfr form (Sweere et al. 2001). However, the biological functions of other Arabidopsis B-type RRs have not yet been identified.
In contrast, pseudo-RRs have been shown to regulate flowering time in Arabidopsis. TOC1/APRR1, an Arabidopsis pseudo-RR, is believed to be a plant circadian clock component (Matsushika et al. 2000; Strayer et al. 2000). It affects flowering time through the phase setting of CO expression (Yanovsky and Kay 2002). TOC1/APRR1 belongs to a small gene family in Arabidopsis (Matsushika et al. 2000). Recently, at least five rice pseudo-RR genes orthologous to the TOC1/APRR family were analyzed to show circadian mRNA expression (Murakami et al. 2003). Pseudo-RRs may have lost their phosphorelay activity and are thus constitutively active because they lack highly conserved Asp residues. Because Ehd1 is a B-type RR with a canonical receiver domain and a functional GARP DNA-binding domain, it is likely to be evolutionarily distant from the pseudo-RR type and not to be a circadian clock component. Preferential expression of Ehd1 mRNA only after SD treatment supports this idea. We examined the expression of a possible clock component in rice, OsLHY (Izawa et al. 2002), by a quantitative RT–PCR method, but did not find a significant difference in OsLHY expression by Ehd1 (T. Izawa, K. Doi, A. Yoshimura, and M. Yano, unpubl.).
FT-like and MADS box genes lie downstream of Ehd1
The expression patterns of Hd3a and RFT1 induced by Ehd1 seem to parallel that of Ehd1 mRNA expression under SD (Fig. 4A,C,D). Because the GARP DNA-binding domain can specifically recognize only 5-bp nucleotides in vitro (Sakai et al. 2000) and Ehd1 probably functions as a transcription factor, it is possible that Ehd1 directly regulates the mRNA expression of these FT-like genes. Several possible binding sites of Ehd1 in the Hd3a and RFT1 promoter regions were found (data not shown), although the in vivo function of these candidate binding sites remains to be tested.
Previous studies have revealed that the expression of FT-like genes starts before dawn and continues until dusk in cultivars of rice that contain functional alleles of both Hd1 and Ehd1 (Izawa et al. 2002; Kojima et al. 2002). The expression of FT-like genes around dawn can be attributed to Hd1, but it does not explain the daytime expression of FT-like genes well (Izawa et al. 2002). However, the present study has provided a good explanation; Ehd1 causes the expression of FT-like genes during day-time (Fig. 4). The FT-like genes are more highly expressed during daytime-induced peaks of Ehd1 expression than the circadian peaks before down. In contrast, Ehd1 mRNA exhibits bimodal expression patterns more clearly (Fig. 4C,D). Therefore, this daytime peak of FT-like genes may be affected by the external coincidence of light with Ehd1 gene product.
On the other hand, we have identified three MADS box genes regulated downstream of Ehd1. Phylogenetic analysis tells that two of them, OsMADS14 and OsMADS15, are orthologs of Arabidopsis AP1, and the other, OsMADS1, is related to the Arabidopsis SEP family (Fig. 5D). In Arabidopsis floral pathways, AP1 lies downstream of floral integrators such as FT, SOC1, and LEAFY (Simpson and Dean 2002). Recently, wheat VRN1 in the floral vernalization pathway was revealed to encode an AP1 ortholog and to be induced by vernalization treatment (Yan et al. 2003). Meanwhile, Arabidopsis SEP genes are required for flower development (Pelaz et al. 2000), and the expression of SEP1 and SEP2 starts in stage-2 flower primordia upon floral transition (Flanagan and Ma 1994; Savidge et al. 1995). OsMADS1 mRNA expression was detected in floral meristem regions upon floral transition (Chung et al. 1994). Furthermore, mutations in OsMADS1 resulted in florets with elongated leafy paleas and lemmas, and overexpression of OsMADS1 caused early flowering (Jeon et al. 2000). This evidence clearly indicates that Ehd1 activates downstream genes such as FT-like genes and MADS box genes, which are tightly related to floral transition.
Two-component Ehd1 signaling cascade in short-day promotion pathway
Because a phytochrome deficiency derepresses mRNA expression of FT-like genes in rice (Izawa et al. 2002), the possibility that the Ehd1 product is phosphorylated by phytochromes remains to be tested, although phosphorylation of B-type RRs has not been proved yet, even in Arabidopsis. Although no study has reported the HK activity of a phytochrome in higher plants, HK activity has been detected in an ancestral phytochrome, Cph1, in cyanobacteria, and relevant RRs have been identified (Yeh et al. 1997). Alternatively, the acute expression of Ehd1 in response to light-on signals suggests that Ehd1 mRNA expression is regulated by phytochromes. It has been shown that cytokinin-inducible ARR4 (an A-type RR; Brandstatter and Kieber 1998) can directly interact with phytochrome B in Arabidopsis (Sweere et al. 2001). Therefore, it is also possible that some A-type RRs transcriptionally regulated by Ehd1 are involved in the control of the stability of Pfr phytochromes, and so, control FT-like genes in rice. However, downstream genes identified by microarray analysis alone do not support this idea at this moment.
Possible developmental or environmental signals mediated by Ehd1 may include not only light signals, but also phytohormones such as cytokinins or ethylene, which may affect floral induction in some plant species (Bernier et al. 1993; Samach et al. 2000). All reported functions of B-type RRs were involved in cytokinin signaling in Arabidopsis (Hwang et al. 2002). However, profiles of genes downstream of Ehd1 do not strongly support this idea, because all identified downstream genes seem not to be related to cytokinin signaling except for AK067670, which has some homology with the cytokinin-repressed CR9 gene of Cucumis sativus, but is identical to the gene for lir1, a rice light-inducible protein (Table 1; Reimmann and Dudler 1993). Whether Ehd1 can mediate cytokinin signaling is now under investigation. Whatever the signal is, Ehd1 would lie in the latter part of the photoperiodic pathway of rice and function mainly as a floral inducer, because not many genes were found downstream of Ehd1. This floral inducer unique to rice will provide a clue to elucidating the diversity in the photoperiodic control of flowering of higher plants.
Materials and methods
Genetic analysis of Ehd1 and Hd1
For QTL analysis, 89 recombinant inbred (RI) lines derived from a cross between Nipponbare and T65 were planted at the experimental farm of Kyushu University. This experiment was conducted under natural field conditions. We analyzed the days-to-heading for 12 plants from each line to obtain the average of the line. The genotypes of the RI lines at the Hd1 and Ehd1 loci were determined by using PCR and cleaved amplified polymorphic (CAPS) markers.
For testing how Ehd1 acts in photoperiodic flowering, the BC6F2 near-isogenic line NIL(Ehd1-gla), containing the functional allele of Ehd1-gla from O. glaberrima, was developed in the T65 background. T65 and NIL(Ehd1-gla) were grown under SD (10L:14D) and LD (15L:9D) conditions in the daylength control facility at Kyushu University. Three primers (P1, P2, and P3) were used for RT–PCR and genotyping (Fig. 1D,E,G).
Map-based cloning of Ehd1
The initial mapping of Ehd1 by using a BC3F4 segregating population of 147 plants was reported previously (Doi and Yoshimura 1998). Progeny (>2500 plants) of heterozygotes of the initial mapping population were used for the high-resolution mapping. Linkage analysis using restriction-fragment-length polymorphism (RFLP) and CAPS markers revealed that Ehd1 was restricted in a 16-kb genomic region on the BAC clone OSJNBa0071K18 (GenBank accession no. AC027038). To test the complementation of Ehd1, we cloned two Kasalath genomic fragments, the 11.5-kb BamHI fragment and the 7.6-kb KpnI fragment, into the pPZP2H-lac binary vector (Fuse et al. 2001) and introduced them into T65 by Agrobacterium-mediated transformation. The heading date of T0 regenerated plants was examined under SD (10L:14D) condition after they were transplanted into soil in pots (Fig. 2C). Homozygous T2 progeny derived from single-copy T0 transformants were grown under SD (9L:15D) condition, and their heading dates were recorded. Only the 11.5-kb BamHI fragment could complement the Ehd1 phenotype. The structure of the mRNA of Ehd1-gla was determined by rapid amplification of cDNA ends (RACE) with a Marathon cDNA amplification kit (Clontech). Gel-mobility shift assays of GARP domains were performed essentially as previously described (Sakai et al. 2000).
Quantitative RT–PCR analysis of gene expression
For SD samples, plants were grown for 3 wk under LD (15L:9D) condition and transferred to SD (9L:15D) condition for 1 wk. Then, leaves were harvested every 3 h for 4 d, 2 d under SD, followed by 2 d under continuous darkness. For LD samples, plants were grown under LD for 4 wk and the sampling was done for 4 d, 2 d under LD followed by 2 d under continuous darkness. Real-time quantitative RT–PCR analysis was performed as previously described (Kojima et al. 2002). cDNA corresponding to 100 ng total RNA was used as the template for each TaqMan PCR reaction (Applied Biosystems). At least three PCR reactions using the same templates were performed to get average values of expression levels. The PCR conditions were 2 min at 50°C, then 10 min at 95°C, followed by 40 cycles of 15 sec at 95°C, and 1 min, 30 sec at 60°C. For Ehd1-gla mRNA expression, the specific primers 5′-GCGCTTCTGATTTCC TGC-3′ and 5′-CGGAATATGTGCTGCCAG-3′ and the probe 5′-GTGAGGATCGAAGAGCTGAGCAACA-3′ were used. RFT1 mRNA was quantified using the primer pair 5′-CGTCCATG GTGACCCAACA-3′ and 5′-CCGGGTCTACCATCACGAGT-3′ and the probe 5′-CGGTGGCAATGACATGAGGACGTTC -3′. Hd3a mRNA was quantified using primers and probes previously described (Kojima et al. 2002). For copy-number standards, quantified fragments of cloned cDNA were used.
Microarray analysis and RT–PCR
Agilent Technologies custom microarrays were hybridized according to the manufacturer's instructions. Because floral transition in T65 + Ehd1 (Fig. 2D, lines 1–16) occurred ∼25–30 d after sowing and Ehd1 is expressed at daytime under SD condition, sample leaves of T65 and T65 + Ehd1 were collected for RNA analysis at noon from 1-month-old plants grown under SD condition.
cDNA was synthesized from both total RNA and mRNA and labeled with the fluorescent dyes Cy5 and Cy3. Color-swap experiments were performed for both cDNA from total RNA and mRNA. Results were filtered to pick up candidate clones with an average log ratio >0.25 and a P value log ratio <0.01. The expression of candidate clones was verified by RT–PCR with specific primers under PCR conditions described in Supplementary Table 2. Independent RNA samples derived from different plants grown under the same conditions were used for the RT–PCR verification.
Acknowledgments
We are grateful for the financial support by the Program for Promotion of Basic Research Activities for Innovative Biosciences. This work was supported in part by Grants-in-Aid for Scientific Research in Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology to T.I. We thank Dr. Y. Takahashi for support with the TaqMan RT–PCR and Dr. T. Aoyama for advice on gel-mobility shift assays of B-type RRs. We also thank the Rice Genome Resource Center at NIAS for the use of the rice microarray analysis system and the technical support provided by Dr. Nagamura and Ms. Hashimoto.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1189604.
References
- Bernier, G., Havelange, A., Houssa, C., Petitjean, A., and Lejeune, P. 1993. Physiological signals that induce flowering. Plant Cell 5: 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstatter, I. and Kieber, J.J. 1998. Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10: 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Chang, C. and Stadler, R. 2001. Ethylene hormone receptor action in Arabidopsis. BioEssays 23: 619–627. [DOI] [PubMed] [Google Scholar]
- Chung, Y.Y., Kim, S.R., Finkel, D., Yanofsky, M.F., and An, G. 1994. Early flowering and reduced apical dominance result from extopic expression of a rice MADS box gene. Plant Mol. Biol. 26: 657–665. [DOI] [PubMed] [Google Scholar]
- Doi, K. and Yoshimura, A. 1998. RFLP mapping of a gene for heading date in an African rice. Rice Genet. Newslett. 15: 148–149. [Google Scholar]
- Doi, K., Yoshimura, A., and Iwata, N. 1998. RFLP mapping and QTL analysis of heading date and pollen sterility using backcross populations between Oryza sativa L. and Oryza glaberrima Steud. Breed. Sci. 48: 395–399. [Google Scholar]
- Flangan, C.A. and Ma, H. 1994. Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant Arabidopsis flowers. Plant Mol. Biol. 26: 581–595. [DOI] [PubMed] [Google Scholar]
- Fuse, T., Sasaki, T., and Yano, M. 2001. Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 18: 219–222. [Google Scholar]
- Hayama, R. and Coupland, G. 2003. Shedding light on the circadian clock and the photoperiodic control of flowering. Curr. Opin. Plant Biol. 6: 13–19. [DOI] [PubMed] [Google Scholar]
- Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. 2003. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722. [DOI] [PubMed] [Google Scholar]
- Hwang, I. and Sheen, J. 2001. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389. [DOI] [PubMed] [Google Scholar]
- Hwang, I., Chen, H.C., and Sheen, J. 2002. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129: 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi, T., Tran, H.G., Swartz, T.E., Briggs, W.R., and Kay, S.A. 2003. FKF1 is essential for photoperiodic-specific light signaling in Arabidopsis. Nature 426: 302–306. [DOI] [PubMed] [Google Scholar]
- Imamura, A., Kiba, T., Tajima, Y., Yamashino, T., and Mizuno, T. 2003. In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 44: 122–131. [DOI] [PubMed] [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. 2001. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063. [DOI] [PubMed] [Google Scholar]
- Izawa, T., Oikawa, T., Sugiyama, N., Tanisaka, T., Yano, M., and Shimamoto, K. 2002. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes & Dev. 16: 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa, T., Takahashi, Y., and Yano, M. 2003. Comparative biology comes into bloom: Genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr. Opin. Plant Biol. 6: 113–120. [DOI] [PubMed] [Google Scholar]
- Jeon, J.S., Jang, S., Lee, S., Nam, J., Kim, C., Lee, S.H., Chung, Y.Y., Kim, S.R., Lee, Y.H., Cho, Y.G., et al. 2000. leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. 1999. Activation tagging of the floral inducer FT. Science 286: 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kikuchi, S., Satoh, K., Nagata, T., Kawagashira, N., Doi, K., Kishimoto, N., Yazaki, J., Ishikawa, M., Yamada, H., Ooka, H., et al. 2003. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. 1999. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., and Yano, M. 2002. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43: 1096–1105. [DOI] [PubMed] [Google Scholar]
- Lohrmann, J. and Harter, K. 2002. Plant two-component signaling systems and the role of response regulators. Plant Physiol. 128: 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika, A., Makino, S., Kojima, M., and Mizuno, T. 2000. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 41: 1002–1012. [DOI] [PubMed] [Google Scholar]
- Mouradov, A., Cremer, F., and Coupland, G. 2002. Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 14: S111–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, M., Ashikari, M., Miura, K., Yamashiro T., and Mizuno, T. 2003. The evolutionarily conserved OsPRR quintet: Rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 44: 1229–1236. [DOI] [PubMed] [Google Scholar]
- Nishida, H., Okumoto, Y., Nakagawa, H., Ichitani, K., Inoue, H., and Tanisaka, T. 2001. Analysis of tester lines for rice (Oryza sativa L.) heading-time genes using reciprocal photoperiodic transfer treatments. Ann. Bot. 88: 527–536. [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. 2000. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203. [DOI] [PubMed] [Google Scholar]
- Pischke, M.S., Jones, L.G., Otsuga, D., Fernandez, D.E., Drews, G.N., and Sussman, M.R. 2002. An Arabidopsis histidine kinase is essential for megagametogenesis. Proc. Natl. Acad.Sci. 99: 15800–15805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimmann, C. and Dudler, R. 1993. Circadian rhythmicity in the expression of a novel light-regulated rice gene. Plant Mol. Biol. 22: 165–170. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., Adam, L., Pineda, O., Ratcliffe, O.J., Samaha, R.R., et al. 2000. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Aoyama, T., and Oka, A. 2000. Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 24: 703–711. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Honma, T., Aoyama, T., Sato, S., Kato, T., Tabata, S., and Oka, A. 2001. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521. [DOI] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanovsky, M.F., and Coupland, G. 2000. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616. [DOI] [PubMed] [Google Scholar]
- Savidge, B., Rounsley, S.D., and Yanofsky, M.F. 1995. Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell 7: 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G.G. and Dean, C. 2002. Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289. [DOI] [PubMed] [Google Scholar]
- Stock, A.M., Robinson, V.L., and Goudreau, P.N. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69: 183–215. [DOI] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Mas, P., Panda, S., Kreps, J.A., and Kay, S.A. 2000. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771. [DOI] [PubMed] [Google Scholar]
- Sweere, U., Eichenberg, K., Lohrmann, J., Mira-Rodado, V., Baurle, I., Kudla, J., Nagy, F., Schafer, E., and Harter, K. 2001. Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294: 1108–1111. [DOI] [PubMed] [Google Scholar]
- Yan, L., Loukoinov, A., Tranquilli, G., Helguera, M., Fahima, T., and Dubcovsky, J. 2003. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. 100: 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., Baba, T., Yamamoto, K., Umehara, Y., Nagamura, Y., et al. 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M., Kojima, S., Takahashi Y., Lin, H.X., and Sasaki, T. 2001. Genetic control of flowering time in rice, a short-day plant. Plant Physiol. 127: 1425–1429. [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J. and Kay, S.A. 2002. Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312. [DOI] [PubMed] [Google Scholar]
- Yeh, K.C., Wu, S.H., Murphy, J.T., and Lagarias, J.C. 1997. A cyanobacterial phytochrome two-component light sensory system. Science 277: 1505–1508. [DOI] [PubMed] [Google Scholar]