Abstract
The differentiation of mesenchymal cells into chondrocytes and chondrocyte proliferation and maturation are fundamental steps in skeletal development. Runx2 is essential for osteoblast differentiation and is involved in chondrocyte maturation. Although chondrocyte maturation is delayed in Runx2-deficient (Runx2–/–) mice, terminal differentiation of chondrocytes does occur, indicating that additional factors are involved in chondrocyte maturation. We investigated the involvement of Runx3 in chondrocyte differentiation by generating Runx2-and-Runx3-deficient (Runx2–/–3–/–) mice. We found that chondrocyte differentiation was inhibited depending on the dosages of Runx2 and Runx3, and Runx2–/–3–/– mice showed a complete absence of chondrocyte maturation. Further, the length of the limbs was reduced depending on the dosages of Runx2 and Runx3, due to reduced and disorganized chondrocyte proliferation and reduced cell size in the diaphyses. Runx2–/–3–/– mice did not express Ihh, which regulates chondrocyte proliferation and maturation. Adenoviral introduction of Runx2 in Runx2–/– chondrocyte cultures strongly induced Ihh expression. Moreover, Runx2 directly bound to the promoter region of the Ihh gene and strongly induced expression of the reporter gene driven by the Ihh promoter. These findings demonstrate that Runx2 and Runx3 are essential for chondrocyte maturation and that Runx2 regulates limb growth by organizing chondrocyte maturation and proliferation through the induction of Ihh expression.
Keywords: Runx2, Runx3, chondrocyte proliferation, chondrocyte maturation, Ihh
Vertebrate skeletons are constructed through the formation of bone structures, a process that is achieved by intramembranous or endochondral ossification. Intramembranous bones, which are directly formed by osteoblasts, are restricted to the cranial vault, some facial bones, and parts of the mandible and clavicle, whereas the rest of the skeleton is composed of endochondral bones that are formed as a cartilaginous template which is then replaced by bone. In early skeletal development, mesenchymal cells condense and acquire the phenotypes of chondrocytes including the ability to produce Col2a1 and proteoglycan. In the process of endochondral ossification, immature chondrocytes proliferate, and chondrocytes at the center of the cartilaginous skeleton begin to mature to become prehypertrophic chondrocytes, which express parathyroid hormone/parathyroid hormone-related peptide (Pthlh) receptor (Pthr1) and Indian hedgehog (Ihh). The prehypertrophic chondrocytes further mature to hypertrophic chondrocytes, which express Col10a1. Upon the terminal differentiation of chondrocytes, the terminal hypertrophic chondrocytes express osteopontin, the matrix is mineralized, vascular vessels invade the calcified cartilage, and finally the cartilage is replaced by bone. Chondrocyte proliferation and differentiation occur in an organized manner and result in the formation of a growth plate that is composed of layers of chondrocytes at different stages of differentiation, including resting, proliferating, prehypertrophic, hypertrophic, and terminal hypertrophic chondrocytes (Gilbert 1997; Inada et al. 1999).
Ihh plays important roles in the formation of the growth plate. Ihh induces Pthlh expression at the epiphysial surface, probably through indirect pathways. The expression of Pthlh at the epiphysial surface results in a gradient of Pthlh with decreasing levels towards the diaphysis, and inhibits chondrocyte maturation through Pthr1 (Vortkamp et al. 1996). The limbs of Ihh–/– mice are severely diminished due to reduced chondrocyte proliferation, indicating that Ihh promotes chondrocyte proliferation (St-Jacques et al. 1999). Therefore, Ihh regulates both the maturation and proliferation of chondrocytes.
Runx2 (runt-related transcription factor 2) is a transcription factor that belongs to the Runx family (Komori 2002) and acquires DNA binding activity by heterodimerizing with Cbfβ (Kundu et al. 2002; Miller et al. 2002; Yoshida et al. 2002). Runx2–/– mice die just after birth and completely lack bone formation due to the absence of osteoblast differentiation, demonstrating that Runx2 is essential for osteoblast differentiation (Komori et al. 1997; Otto et al. 1997). Further, chondrocyte maturation is delayed in Runx2–/– mice (Inada et al. 1999; Kim et al. 1999), Runx2 promotes chondrocyte maturation (Enomoto et al. 2000; Takeda et al. 2001; Ueta et al. 2001; Stricker et al. 2002), dominant-negative Runx2 inhibits chondrocyte maturation (Ueta et al. 2001; Stricker et al. 2002), and the overexpression of Runx2 in chondrocytes restores the chondrocyte maturation in Runx2–/– mice (Takeda et al. 2001). However, chondrocyte maturation to the terminal stage does eventually occur in Runx2–/– mice (Komori et al. 1997; Otto et al. 1997; Inada et al. 1999; Kim et al. 1999), indicating that Runx2 is not essential for chondrocyte maturation and that other factors are involved in chondrocyte maturation.
Runx3, which also belongs to the Runx family, plays important roles in the growth regulation of gastric epithelial cells and the development of dorsal root ganglion neurons (Inoue et al. 2002; Levanon et al. 2002; Li et al. 2002). As Runx3 is expressed in the cartilage (Levanon et al. 2001; Stricker et al. 2002), we investigated the involvement of Runx3 in chondrocyte differentiation by generating Runx2–/–3–/– mice. Chondrocyte maturation was mildly delayed in Runx3–/– mice during embryogenesis, but it was completely absent in Runx2–/–3–/– mice, demonstrating that Runx2 and Runx3 are functionally redundant in chondrocytes and play an essential role in chondrocyte maturation. Runx2–/–3–/– mice, which completely lacked Ihh expression, had severely shortened limbs due to reduced and disorganized chondrocyte proliferation and reduced cell size in the diaphyses. Further, the introduction of Runx2 to Runx2–/– chondrocytes induced Ihh expression, and Runx2 directly regulated the Ihh promoter. These findings indicate that Runx2 is also involved in chondrocyte proliferation, and that Runx2 coordinates chondrocyte maturation and proliferation by regulating Ihh expression.
Results
Runx3 expression in the skeleton
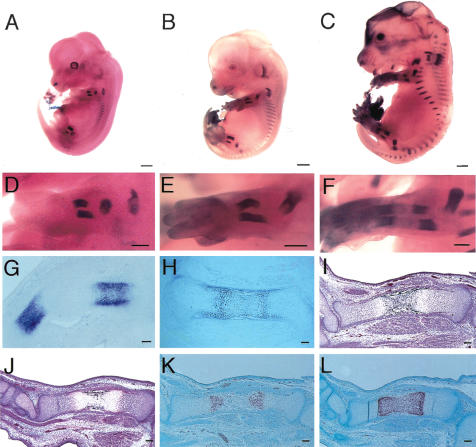
We examined Runx3 expression in the skeleton of Runx3+/– mice during embryogenesis using the β-galactosidase gene, which was integrated in exon 3 of Runx3 in Runx3+/– mice (Li et al. 2002), followed by detection of β-galactosidase activity (Fig. 1). Runx3 expression was first detected in the skeleton at embryonic day E12.5 (E12.5) in the scapulae, ribs, and skeletal parts of the limbs (Fig. 1A,D). Runx3 expression extended to the pelvic bones and vertebrae at E13.5 (Fig. 1B,E), and to most of the cartilaginous skeletons at E15.5 (Fig. 1C,F). The level of Runx3 expression increased at the stage of prehypertrophic chondrocytes and was maintained in hypertrophic chondrocytes, but it decreased in terminal hypertrophic chondrocytes (Fig. 1G,H). The expression patterns were compatible with the previous findings, which were obtained by in situ hybridization (Stricker et al. 2002).
Figure 1.
Runx3 expression in Runx3+/– mice and skeletal development in Runx3–/– mice during embryogenesis. (A–H), Runx3 expression in Runx3+/– mice at E12.5 (A,D,G), E13.5 (B,E), E15.5 (C,F) and E16.5 (H). We examined Runx3 expression by the activity of β-galactosidase, whose gene had been integrated in exon 3 of Runx3, in Runx3+/– mice. (A–F) Runx3 was expressed in cartilaginous skeletons. (G,H) Runx3 was specifically expressed in chondrocytes but not in osteoblasts in the skeleton, and its expression was up-regulated in prehypertrophic chondrocytes and down-regulated in terminal hypertrophic chondrocytes. (I–L) Skeletal development in Runx3–/– mice at E15.5. H&E and Kossa staining (I,J) and in situ hybridization using Col10a1 antisense probe (K,L) of wild-type (I,K) and Runx3–/– (J,L) tibiae. Calcification was stained black in I and J. We detected no signal using a sense probe of Col10a1 (data not shown). The diaphyses of wild-type tibiae were composed of calcified cartilage that was negative for Col10a1 expression (K), and blood vessels had invaded the diaphyses (I). (J,L) The diaphyses of Runx3–/– tibiae were composed of Col10a1-positive hypertrophic chondrocytes without vascular invasion. Bars: A–C, 1 mm; D–F, 500 μm; G–L, 100 μm.
Delayed endochondral ossification in Runx3–/– and Runx2+/–3–/– mice
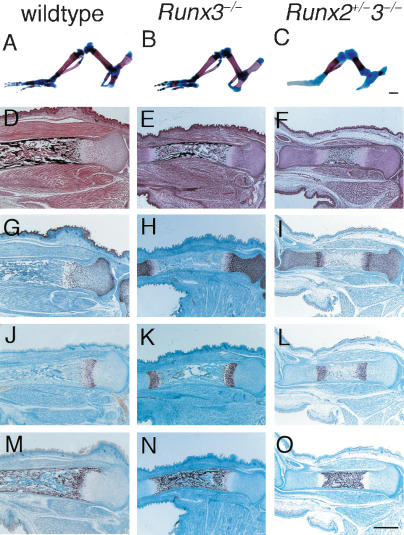
Although Runx3+/– mice develop normally, Runx3–/– mice of the C57BL/6 background die shortly after birth (Li et al. 2002). In Runx3–/– embryos, chondrocyte maturation and vascular invasion into the cartilage were slightly delayed at E15.5 (Fig. 1I–L), although the mice showed grossly normal skeletal development at the neonatal stage (Fig. 2B; data not shown). To further investigate the involvement of Runx3 in skeletal development, we mated Runx3+/– mice with Runx2+/– mice, and finally we generated Runx2–/–3–/– mice. Although Runx2+/– mice achieved grossly normal endochondral ossification at the newborn stage (Komori et al. 1997; Otto et al. 1997; data not shown), endochondral ossification was clearly delayed in the Runx2+/–3–/– mice, as shown by the smaller mineralized regions in the chondrocranium, vertebrae, ribs, and limbs at the newborn stage (Fig. 2C; data not shown). In the tibiae of wild-type and Runx3–/– mice at E18.5, the cartilage was largely replaced by bone (Fig. 2D,E). However, in the Runx2+/–3–/– tibiae, the diaphyses were composed of calcified cartilage that was covered with the bone collar, blood vessels had just started to invade the calcified cartilage, and no trabecular bone was observed (Fig. 2F). These features of the Runx2+/–3–/– tibiae were apparently caused by delayed chondrocyte maturation because the expression of Pthr1, Ihh, Col10a1, and osteopontin were restricted to the diaphyses (Fig. 2L,O; data not shown). These findings indicate that Runx3 is required for endochondral ossification if the Runx2 gene dosage is halved.
Figure 2.
Skeletal development in Runx3–/– and Runx2+/–3–/– mice. (A–C) Hind limb skeletons of wild-type (A), Runx3–/– (B), and Runx2+/–3–/– (C) newborns. Calcified tissues were stained red with Alizarin red, and cartilage was stained blue with Alcian blue. Wild-type and Runx3–/– limb skeletons showed a similar pattern of mineralization (A,B), whereas the mineralized regions of Runx2+/–3–/– limbs were much smaller (C). (D–F) Histological analysis of the tibiae of wild-type (D), Runx3–/– (E), and Runx2+/–3–/– (F) embryos at E18.5. The sections were stained with H&E and Kossa. (G–O) In situ hybridization using the antisense probes of Col2a1 (G–I), Col10a1 (J–L), and osteopontin (M–O). We detected no signal using sense probes of Col2a1, Col10a1, or osteopontin (data not shown). In the tibiae of wild-type and Runx3–/– embryos, the cartilage was largely replaced by bone (D,E), osteopontin-positive osteoblasts occupied the bone marrow (M,N), and Col2a1-positive (G,H) or Col10a1-positive (J,K) chondrocytes were observed in the epiphyses. Runx2+/–3–/– tibiae were still cartilaginous (F), and the chondrocytes expressed Col2a1 (I) or Col10a1 (L) except for those in the mineralized diaphyses, which were occupied by osteopontin-positive terminal hypertrophic chondrocytes (O). Bars: A–C, 1 mm; D–O, 500 μm.
Mildly delayed early skeletal development in Runx2–/–3–/– embryos
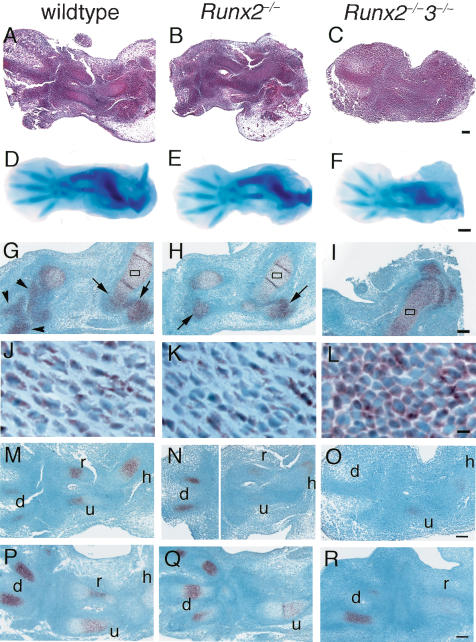
In the early skeletal development in Runx2–/–3–/– embryos, chondrocyte differentiation and proteoglycan production seemed to be mildly delayed (Fig. 3A–F). Cartilaginous anlagen was clearly seen in the wild-type and Runx2–/– embryos but not in the Runx2–/–3–/– embryos at E12.5, indicating that chondrocyte differentiation was mildly delayed in Runx2–/–3–/– embryos (Fig. 3A–C). Alcian blue staining of E13.5 Runx2–/–3–/– embryos was weaker than that of the wild-type and Runx2–/– embryos, indicating less proteoglycan production in Runx2–/–3–/– cartilage, which was probably due to delayed chondrocyte differentiation (Fig. 3D–F). As to Sox9 expression, which is essential for mesenchymal condensation (Bi et al. 1999), within the limb skeletons of E13.5 wild-type and Runx2–/– embryos, there was strong expression in the skeletal parts with immature chondrocytes, but weak expression in the skeletal parts with differentiating chondrocytes (Fig. 3G,H,J,K). In the limb skeletons of Runx2–/–3–/– embryos at E13.5, however, all chondrocytes strongly expressed Sox9 (Fig. 3I,L), indicating that all of the chondrocytes were immature. As Ihh is expressed in chondrocytes at an early stage of skeletal development (Bitgood and McMahon 1995), we examined Ihh expression at E12.5 and E13.5. Ihh was expressed in immature chondrocytes in Runx2–/–3–/– mice as well as in wild-type and Runx2–/– mice, although the appearance of Ihh expression was slightly delayed in Runx2–/–3–/– mice (Fig. 3M–R). This indicates that neither Runx2 nor Runx3 is essential for the Ihh expression at the early stage of skeletal development. These findings indicate that Runx2 and Runx3 are not essential for, but are partly involved in early chondrocyte differentiation and that they begin to induce chondrocyte differentiation as soon as mesenchymal condensation is completed.
Figure 3.
Early skeletal development in wild-type, Runx2–/–, and Runx2–/–3–/– embryos and in situ hybridization for Sox9 and Ihh expression. We used the forelimbs (A–C,G–R) and hind limbs (D–F) of wild-type (A,D,G,J,M,P), Runx2–/– (B,E,H,K,N,Q), and Runx2–/–3–/– (C,F,I,L,O,R) embryos for the analyses. (A–C) H&E staining at E12.5. (D–F) Whole-mount Alcian blue staining at E13.5. (G–L) In situ hybridization using Sox9 antisense probe at E13.5. (J–L) Higher magnification of boxed regions in G–I, respectively. (M–R) In situ hybridization using Ihh antisense probe at E12.5 (M–O) and E13.5 (P–R). We detected no signal using the sense probe of Sox9 and Ihh (data not shown). Cartilaginous anlagen was clearly seen in the wild-type (A) and Runx2–/– (B) embryos but not in the Runx2–/–3–/– embryos (C) at E12.5. Alcian blue staining of the hind limbs of E13.5 Runx2–/–3–/– embryos (F) was weak, compared with that of the wild-type (D) and Runx2–/– (E) embryos. (G,H,J,K) In the forelimb skeletons of wild-type and Runx2–/– embryos, the peripheral regions of the epiphyses of the humeri, radii, and ulnae (arrows in G,H), and most regions of the carpal and metacarpal bones (arrowheads in G) were composed of immature chondrocytes that strongly expressed Sox9, and the remaining regions of the humeri, radii, and ulnae, which were composed of differentiating chondrocytes, weakly expressed Sox9. (I,L) In the Runx2–/–3–/– forelimb skeletons, all of the chondrocytes strongly expressed Sox9. Ihh was strongly detected in immature chondrocytes. especially in digits of wild-type, Runx2–/–, and Runx2–/–3–/– mice. (h) humerus; (r) radius; (u) ulna; (d) digit. Bars: A–C,G–I,M–R, 100 μm; D–F, 500 μm; J–L, 10 μm.
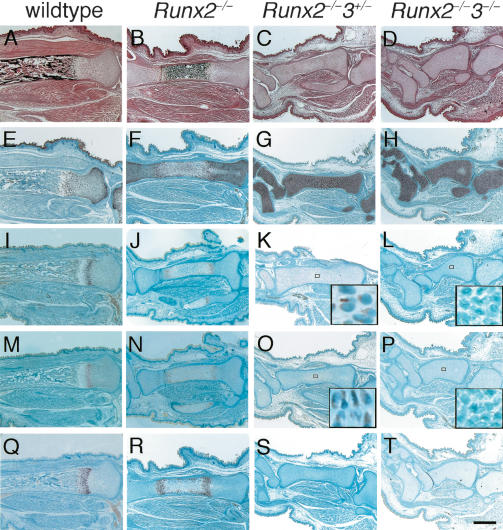
Complete absence of chondrocyte maturation in Runx2–/–3–/– mice
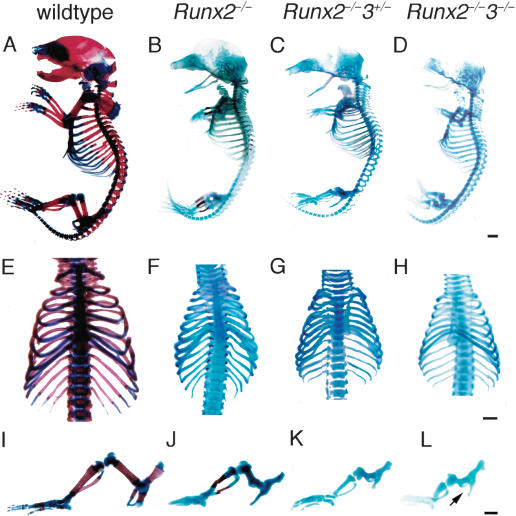
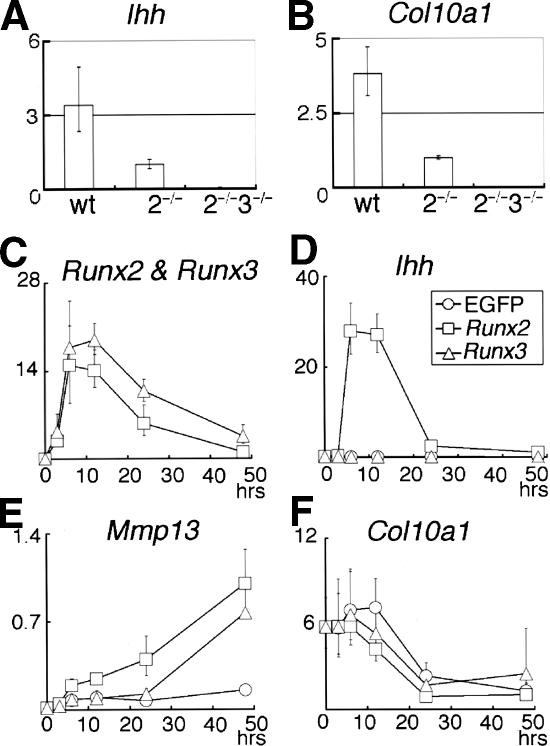
Mineralization, which is observed in restricted regions of the limbs, vertebrae, and ribs of Runx2–/– newborns, was completely absent throughout the entire skeletons of both Runx2–/–3+/– and Runx2–/–3–/– newborns (Fig. 4). Histological analyses of Runx2–/– tibiae at E18.5 showed that the diaphyses were composed of calcified cartilage, and the chondrocytes in the metaphyses expressed Pthr1, Ihh, or Col10a1 on in situ hybridization (Fig. 5). The level of Ihh expression in Runx2–/– limbs at E18.5 was less than one-third of that in wild-type limbs on real-time RT–PCR analysis (see Fig. 7A, below). In the tibiae of Runx2–/–3+/– and Runx2–/–3–/– embryos at E18.5, however, there was no mineralization, and all of the chondrocytes expressed Col2a1 but not Col10a1 (Fig. 5). Pthr1 and Ihh expression were very weakly detected in the diaphyses of Runx2–/–3+/– tibiae, but were not detected in Runx2–/–3–/– tibiae (Fig. 5K,L,O,P). The absence of Ihh and Col10a1 expression in Runx2–/–3–/– limbs was confirmed by real-time RT–PCR (see Fig. 7A,B, below). The chondrocytes in the diaphyses of Runx2–/–3+/– tibiae at E18.5 were slightly enlarged, whereas the chondrocytes throughout the Runx2–/–3–/– tibiae were homogeneously small (Fig. 6C,D). We observed that there was complete maturational blockage of chondrocytes throughout the entire skeletons of Runx2–/–3–/– embryos at E18.5 (data not shown). These findings indicate that the degree of maturational blockage of chondrocytes is dependent on the dosages of Runx2 and Runx3, and demonstrate that Runx2 and/or Runx3 must be present for chondrocyte maturation to occur.
Figure 4.
Examination of the skeletal system of wild-type, Runx2–/–, Runx2–/–3+/–, and Runx2–/–3–/– mice at the newborn stage. Skeletons of wild-type (A,E,I), Runx2–/– (B,F,J), Runx2–/–3+/– (C,G,K), and Runx2–/–3–/– (D,H,L) newborns. (A–D) Whole skeletons. (E–H) Chest walls. (I–L) Hind limbs. Although mineralization was observed in restricted skeletal parts including the limbs, vertebrae, and ribs of Runx2–/– mice (B,F,J), it was completely absent in Runx2–/–3+/– (C,G,K) and Runx2–/–3–/– mice (D,H,L). Note that the limb length was reduced depending on the dosages of Runx2 and Runx3, and that Runx2–/–3–/– mice have severely shortened but relatively thick limbs (I–L). Runx2–/–3–/– mice completely lack a pubic bone structure (arrow in L). Bars: A–L, 1 mm.
Figure 5.
Histological examination and in situ hybridization for Col2a1, Pthr1, Ihh, and Col10a1 in the tibiae of wild-type, Runx2–/–, Runx2–/–3+/–, and Runx2–/–3–/– embryos at E18.5. Sections of tibiae from wild-type (A,E,I,M,Q), Runx2–/– (B,F,J,N,R), Runx2–/–3+/– (C,G,K,O,S), and Runx2–/–3–/– (D,H,L,P,T) embryos were examined by H&E and Kossa staining (A–D) and in situ hybridization using the antisense probes of Col2a1 (E–H), Pthr1 (I–L), Ihh (M–P), or Col10a1 (Q–T). The boxed regions in K, L, O, and P are magnified in the respective insets. On in situ hybridization of Pthr1 and Ihh, signal detection using the sections from Runx2–/– and Runx2–/–3+/– tibiae always took a longer period of time than that using sections from wild-type tibiae. Further, we failed to detect Pthr1 and Ihh expression in Runx2–/–3–/– tibiae even upon prolonged incubation. We detected no signal using sense probes of Col2a1, Pthr1, Ihh, and Col10a1 (data not shown). In the tibiae of wild-type embryos, the cartilage was largely replaced by bone, whereas all of the Runx2–/–, Runx2–/–3+/–, and Runx2–/–3–/– tibiae were cartilaginous. In the Runx2–/– tibiae, the diaphyses were composed of calcified cartilage and chondrocytes in the metaphyses expressed Pthr1, Ihh, or Col10a1. In contrast, Runx2–/– 3+/– and Runx2–/–3–/– tibiae showed no mineralization and the chondrocytes in the entire tibiae expressed Col2a1 but not Col10a1. Pthr1 and Ihh expression were very weakly detected in the diaphyses of Runx2–/–3+/– tibiae, but they were not detected in Runx2–/–3–/– tibiae (see insets in K,L,O,P). Bar, 500 μm.
Figure 7.
Regulation of Ihh expression by Runx2 and Runx3. (A,B) Real-time RT–PCR analysis of Ihh (A) and Col10a1 (B) expression using RNA extracted from wild-type (wt), Runx2–/– (2–/–), and Runx2–/–3–/– (2–/–3–/–) limbs at E18.5. The mean of two to three embryos is shown. The values in Runx2–/– limbs were defined as 1, and relative values are shown. (C–F) Real-time RT–PCR analysis of Runx2–/– chondrocyte cultures. Runx2–/– chondrocytes were infected with EGFP-expressing (○), Runx2-and-EGFP-expressing (□), or Runx3-and-EGFP-expressing (▵) adenovirus. Infected cells were harvested at the indicated times after the onset of viral infection, and Runx2 (C), Runx3 (C), Ihh (D), Mmp13 (E), and Col10a1 (F) expression were examined by real-time RT–PCR. The value of Runx2 mRNA expression at 48 h was defined as 1, and the relative values of Runx2 and Runx3 mRNA expression are shown in the same graph (Runx2 and Runx3). In the cells infected with EGFP-expressing adenovirus, the values of Runx2 mRNA expression were nearly zero and the values of Runx3 mRNA expression were very low; therefore, they were not included in the graph. The value of Ihh, Mmp13, or Col10a1 mRNA expression in Runx2-and-EGFP-expressing adenovirus infection at 48 h was defined as 1, and relative values are shown. Adenoviral introduction of Runx2 strongly up-regulated Ihh expression, whereas adenoviral introduction of Runx3 did not up-regulate Ihh expression. In the cells infected with any of the three adenoviruses, the level of Col10a1 expression was at the background level during the culture period examined. Data represent the mean of three to six wells, and representative data of three independent experiments are shown.
Figure 6.
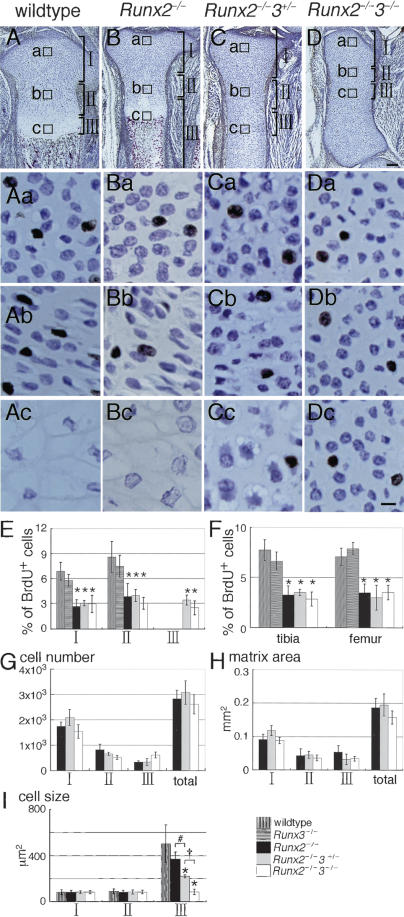
Analysis of chondrocyte proliferation in the limbs by BrdU labeling. (A–D) Immunohistochemistry of tibiae from BrdU-labeled wild-type (A), Runx2–/– (B), Runx2–/–3+/– (C), and Runx2–/– 3–/– (D) embryos at E18.5 using anti-BrdU antibody. The sections were counterstained with toluidine blue. Magnified views of the boxed regions (a,b,c) are shown in the same columns. (A,B) In wild-type and Runx2–/– tibiae, the boxed regions a, b, and c represent resting, proliferating, and hypertrophic chondrocytes, respectively. (C,D) In the Runx2–/–3+/– and Runx2–/–3–/– tibiae, the boxed regions a, b, and c represent chondrocytes in the epiphyses, metaphyses, and diaphyses, respectively. The growth plates were well formed in wild-type tibiae (A) and in Runx2–/– tibiae (B) but not in Runx2–/– 3+/– tibiae (C) or Runx2–/–3–/– tibiae (D). (Ab–Db) The columnar alignment of chondrocytes, which is seen in the layer of proliferating chondrocytes, is well formed in the wild-type and Runx2–/– tibiae, deformed in the Runx2–/–3+/– tibiae, and completely absent in the Runx2–/–3–/– tibiae. (C,D) The diaphyses of Runx2–/–3+/– tibiae were composed of slightly enlarged chondrocytes, whereas the entire tibiae of Runx2–/–3–/– embryos were composed of homogeneously small chondrocytes. (E–I) Measurement of the frequency of BrdU-positive cells (E,F), cell number (G), matrix area (H), and cell size (I) using the tibiae of six wild-type embryos, two Runx3–/– embryos, six Runx2–/– embryos, one Runx2–/–3+/– embryo, and two Runx2–/–3–/– embryos. We measured these parameters in each region shown in A–D. (A,B) In wild-type and Runx2–/– tibiae, the regions I, II, and III represent resting, proliferating, and hypertrophic and terminal hypertrophic chondrocytes, respectively. (C,D) In the Runx2–/–3+/– and Runx2–/–3–/– tibiae, the regions I, II, and III were arbitrarily determined in the proximal half of the tibiae and represent chondrocytes in the epiphysial, metaphysical, and diaphysial parts of the tibiae, respectively. (F) We also counted the number of BrdU-positive cells in femurs, because chondrocyte maturation in Runx2–/– femurs is more severely inhibited than that in Runx2–/– tibiae (Inada et al. 1999). (F) In the measurement of BrdU-positive cells in whole tibiae and femurs, we counted the number of BrdU-positive cells and the total number of cells in regions I and II in wild-type and Runx2–/– mice and in regions I, II, and III in Runx2–/– 3+/– and Runx2–/–3–/– mice, and the mean of the percentage of BrdU-positive cells ± standard deviation (std. dev.) is shown. We measured all of the parameters in three sections of each bone, and the mean ± std. dev. is shown. (*) p < 0.0001 compared with wild-type mice; (#) p < 0.0001; (†) p < 0.0001 as determined by one-way ANOVA. Bars: A–D, 100 μm; Aa–Dc, 10 μm.
Reduced and disorganized chondrocyte proliferation in Runx2–/–3–/– limb skeletons
The limbs of Runx2–/–3–/– embryos at E13.5 were slightly shorter than those of wild-type and Runx2–/– embryos (Fig. 3D–F), and the limb length was apparently reduced in the Runx2–/– and Runx2–/–3–/– newborns according to the dosages of Runx2 and Runx3 (Fig. 4I–L). This suggested that the reduction in limb length was coupled with inhibition of chondrocyte maturation. We labeled proliferating chondrocytes with BrdU and examined the relationship between chondrocyte proliferation and maturation of limb skeletons (Fig. 6). The chondrocytes in Runx2–/– tibiae differentiated to the terminal stage, and all stages of chondrocyte maturation were observed (Fig. 6B; Inada et al. 1999; Kim et al. 1999). In the Runx2–/– tibiae as well as the wild-type tibiae at E18.5, BrdU-positive cells were restricted to the layers of resting and proliferating chondrocytes (Fig. 6A,B), whereas BrdU-positive cells were detected throughout the entire tibiae of Runx2–/–3+/– and Runx2–/–3–/– embryos (Fig. 6C,D). Further, in the Runx2–/–3–/– tibiae, the BrdU-positive cells were distributed homogeneously and the columnar alignment of chondrocytes, which is formed by proliferation along a proximal–distal axis, was completely absent (Fig. 6D), indicating that the maturational blockage resulted in disorganized chondrocyte proliferation. In regions I and II, which represent the layers of resting and proliferating chondrocytes, respectively, in wild-type mice, Runx2 deficiency, but not Runx3 deficiency, significantly reduced chondrocyte proliferation (Fig. 6E). In region III, which represents the layers of hypertrophic and terminal hypertrophic chondrocytes in wild-type mice, there were no BrdU-positive cells in wild-type, Runx3–/–, or Runx2–/– mice, whereas numbers of BrdU-positive cells similar to those in regions I and II were observed in Runx2–/–3+/– and Runx2–/–3–/– mice in the diaphyses (region III; Fig. 6E). In both tibiae and femurs, the decrease in proliferation was related to the Runx2 deficiency but not to the Runx3 deficiency in combined regions, which contained BrdU-positive cells (Fig. 6F). These findings indicate that the reduction in limb length of Runx2–/– mice was due to the reduced proliferation, demonstrating that Runx2 is involved in enhancing chondrocyte proliferation.
As the gene dosage of Runx3 further affected the limb length in Runx2–/– mice, we examined the cell number, matrix area, and cell size in tibiae of Runx2–/–, Runx2–/–3+/–, and Runx2–/–3–/– mice. The cell number in each region was not significantly different among these mice, resulting in similar numbers of total cells in tibiae in these mice (Fig. 6G). Although the matrix area was similar among these mice (Fig. 6H), the cell size was reduced in region III in dependence on the gene dosage of Runx3 (Fig. 6I). Further, the growth of the limbs to a longitudinal direction but not to a radial direction was disturbed in Runx2–/–3–/– mice, leading to the further reduction in the limb length of Runx2–/–3–/– mice. These findings indicate that the reduction in cell size in the diaphyses and the disorganized chondrocyte proliferation led to the severe reduction in the limb length of Runx2–/–3–/– mice, and demonstrate that Runx2 and Runx3 play an important role in organizing chondrocyte proliferation in the limb skeletons to a longitudinal direction. However, apoptosis was not a major cause of the reduced limb length in Runx2–/–3+/– and Runx2–/–3–/– mice, because we detected TUNEL-positive cells in the terminal hypertrophic chondrocytes of Runx2–/– mice but not in the chondrocytes of Runx2–/–3+/– or Runx2–/–3–/– mice (data not shown).
Regulation of Ihh expression by Runx2 and Runx3
In the present study, Ihh expression was reduced depending on the degree of maturational blockage of chondrocytes, and it was absent in Runx2–/–3–/– skeletons (Figs. 5M–P; 7A). Thus, we next examined whether Runx2 and Runx3 directly regulate Ihh expression using Runx2–/– chondrocyte cultures (Fig. 7C–F). Adenoviral introduction of Runx2 into Runx2–/– chondrocytes strongly induced Ihh expression at 6 h after the onset of infection, whereas the introduction of Runx3 into Runx2–/– chondrocytes failed to induce Ihh expression (Fig. 7D). Introduction of Runx2 or Runx3 into Runx2–/– chondrocytes induced matrix metalloproteinase 13 (Mmp13) expression (Fig. 7E), which is a downstream gene of Runx2 (Inada et al. 1999; Jimenez et al. 1999), but did not up-regulate Col10a1 expression (Fig. 7F). As Runx2 and Ihh expression are both up-regulated in prehypertrophic chondrocytes (Bitgood and McMahon 1995; Vortkamp et al. 1996; Iwasaki et al. 1997; Zou et al. 1997; Inada et al. 1999; Kim et al. 1999), these findings suggest that Runx2 directly induces Ihh expression. As there was weak Ihh expression in Runx2–/– tibiae (Figs. 5N, 7A), this finding indicates that either Runx3 promotes Ihh expression indirectly or additional factors are required for Ihh induction by Runx3.
Regulation of the promoter of Ihh by Runx2
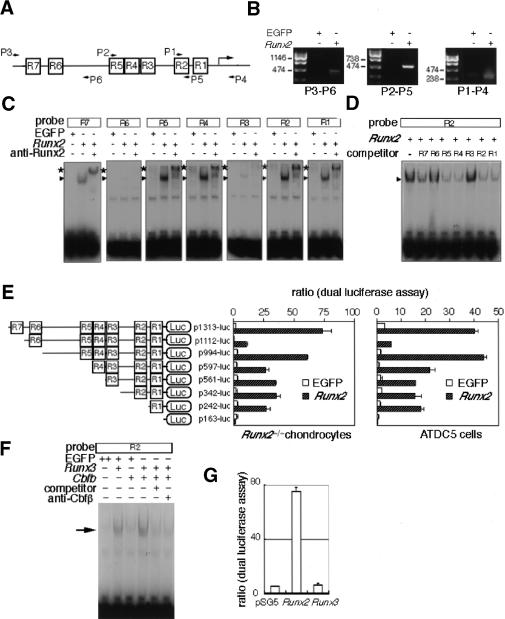
A database search revealed that the 1.2-kb promoter region of the Ihh gene was highly conserved between mouse and human (Mus musculus chromosome 1 genomic contig, NT_039170; Homo sapiens chromosome 2 genomic contig, NT_005403). The 1.2-kb promoter region of the mouse Ihh gene contained seven putative Runx binding sites (R1–R7), in which R1, R2, R4, R5, and R7 were conserved between mouse and human (Fig. 8A). The binding of Runx2 to the 1.2 kb Ihh promoter was confirmed by chromatin immunoprecipitation (ChIP; Fig. 8B). Four putative Runx binding sites (R1, R2, R4, and R5) had perfectly corresponding sequences with the binding sequences of Runx, and Runx2 strongly bound to each of the four oligonucleotides in the electrophoresis mobility shift assay (EMSA; Fig. 8C). The other putative Runx binding sites (R3, R6, and R7) included one less common base, and Runx2 bound relatively strongly to the R7 oligonucleotides and weakly to the R3 and R6 oligonucleotides. Competition assays also showed that Runx2 bound strongly to the R1, R2, R4, and R5 oligonucleotides, to the R7 oligonucleotides less strongly, but to the R3 and R6 oligonucleotides barely (Fig. 8D). Further, reporter assays of various deletion constructs of the 1.2-kb promoter region of the Ihh gene using Runx2–/– chondrocytes and chondrogenic ATDC5 cells showed that Runx2 strongly activated the Ihh promoter, and that R1, R5, and R7 are important Runx2 binding sites for transcriptional activation (Fig. 8E). As the reporter activity was significantly reduced in the p1112-luc construct, inhibitory factors seem to bind around R6 in the Ihh promoter region. In contrast to Runx2, Runx3 failed to activate the 1.3-kb Ihh promoter, although Runx3 bound to the R2 oligonucleotides (Fig. 8F,G).
Figure 8.
Regulation of the Ihh promoter by Runx2. (A) Schematic presentation of seven putative Runx-binding sites (R1–R7) and primers used for ChIP in the promoter region of the mouse Ihh gene. (B) ChIP. Runx2–/– chondrocytes were infected with EGFP- or Runx2-and-EGFP-expressing adenovirus. After sonication, the cell lysates were immunoprecipitated by anti-Runx2 antibody, and the binding of Runx2 to the Ihh promoter region was examined by PCR using the primers shown in A. Bands of correct sizes were detected in Runx2-expressing cells, and the amplified DNA contained each Ihh promoter fragment, which was confirmed by sequencing (data not shown). (C) Binding of Runx2 to the putative Runx binding sequences. Runx2–/– chondrocytes were infected with EGFP- or Runx2-and-EGFP-expressing adenovirus, and EMSA was performed using each of the R1–R7 probes. Runx2 strongly bound to R1, R2, R4, and R5, less strongly to R7, and weakly to R3 and R6. All of the specific bands (arrowheads) were super-shifted with anti-Runx2 antibody (*). All of the specific bands were competed with the respective unlabeled oligonucleotide (data not shown). (D) The affinity of Runx2 for each putative Runx binding sequence. Runx2–/– chondrocytes were infected with Runx2-and-EGFP-expressing adenovirus, and EMSA was performed using labeled R2 probe in the presence of each unlabeled oligonucleotide (R1–R7/×200). The formation of the Runx2–R2 complex was strongly inhibited in the presence of unlabeled R1, R2, R4, or R5 oligonucleotides, less strongly inhibited in the presence of unlabeled R7 oligonucleotides, but barely inhibited in the presence of unlabeled R3 or R6 oligonucleotides. (E) Reporter assays of the Ihh promoter. The reporter plasmid containing each Ihh promoter construct and pRL-CMV vector were cotransfected into Runx2–/– chondrocytes (left) or ATDC5 cells (right). After culture for 24 h, Runx2 was adenovirally introduced into the cells at an MOI of 10. The data show the relative level of luciferase activity against the level of Renilla luciferase activity. The values are mean ± S.E. of four wells, and representative data from four independent experiments are shown. (F) Binding of Runx3 to the R2 oligonucleotides. Runx2–/– chondrocytes were infected with retrovirus expressing EGFP alone or Cbfb and EGFP. After 1 d, the cells were infected with EGFP- or Runx3-and-EGFP-expressing adenovirus, and EMSA was performed using the R2 probe. The introduction of Runx3 alone showed a specific band (arrow), and the intensity of the specific band was increased by the introduction of Cbfb. The specific band was competed with unlabeled R2 oligonucleotides (×200). Addition of antibody against Cbfβ prevented formation of the specific Runx3–Cbfβ–DNA complexes. (G) Reporter assay using the 1.3-kb Ihh promoter. To compare the capacities of Runx2 and Runx3 in the transcriptional activation of Ihh promoter, Runx2–/– chondrocytes were transiently transfected with DNA mixture containing p1313-luc, Runx2- or Runx3-expressing vector or empty pSG5, and pRL-CMV. The luciferase activity was normalized to Renilla luciferase activity.
Discussion
We demonstrated here that chondrocyte maturation is dependent on the dosages of Runx2 and Runx3, and that the absence of both Runx2 and Runx3 causes a complete lack of chondrocyte maturation. We further showed that Runx2 enhances chondrocyte proliferation through the direct induction of Ihh expression. Thus, Runx2 and Runx3 play fundamental roles in skeletal development by regulating and coordinating chondrocyte maturation and proliferation to form endochondral bones of appropriate size and shape.
The degree of the disturbance of chondrocyte maturation in Runx2–/– mice depended on the skeletal part (Inada et al. 1999; Kim et al. 1999), whereas the chondrocytes in all skeletal parts of Runx2–/–3–/– mice failed to mature (Figs. 5, 6; data not shown). This indicates that the level of Runx3 expression or Runx3 activation differs in different parts of the skeleton, because the deficiency of Runx2 has no significant effect on the expression level of Runx3 (Stricker et al. 2002). Indeed, Runx2 plays a larger role in promoting chondrocyte maturation than Runx3, because chondrocyte maturation was more delayed in Runx2–/– mice than in Runx3–/– mice in all skeletal parts (Figs. 2, 5; data not shown). Although Sox9, which is an inhibitor of chondrocyte maturation (Akiyama et al. 2002), was strongly expressed in E13.5 Runx2–/–3–/– cartilage (Fig. 3I,L), its expression decreased during development, indicating that the chondrocytes in Runx2–/–3–/– cartilage were released from the inhibitory effect of Sox9 on maturation. However, the chondrocytes in Runx2–/–3–/– cartilage failed to mature, as shown by the homogeneously small chondrocytes and the absence of expression of all of the chondrocyte maturation marker genes, including Pthr1, Ihh, and Col10a1, at E18.5 (Figs. 5, 6A–D, 7A,B), demonstrating that Runx2 and Runx3 are essential transcription factors for chondrocyte maturation.
The Runx family consists of three genes, Runx1, Runx2, and Runx3 (Komori 2002). Runx1–/– mice die at approximately E12.5 due to the absence of fetal liver hematopoiesis, demonstrating that Runx1 is essential for definitive hematopoiesis (Okuda et al. 1996; Wang et al. 1996; Okada et al. 1998). As Runx1 was also expressed in cartilage (Levanon et al. 2001), we generated Runx1+/–2–/– mice and examined their skeletal development. The degree of maturational delay of the chondrocytes in Runx1+/–2–/– mice was the same as that in Runx2–/– mice (C. Yoshida and T. Komori, unpubl.), whereas the maturational delay of the chondrocytes in Runx2–/–3+/– mice was more severe than that in Runx2–/– mice (Fig. 5), indicating that the contribution of Runx1 in chondrocyte maturation, if any, seems to be limited.
Expression of a dominant negative form of Runx2 blocked maturation but increased the proliferation of cultured chick primary chondrocytes (Enomoto-Iwamoto et al. 2001). However, chondrocyte proliferation was reduced in Runx2–/– mice (Fig. 6), suggesting that regulatory factors that are induced during maturation promote chondrocyte proliferation in vivo. Ihh is the most plausible factor based on the following results: chondrocyte proliferation was reduced in Ihh–/– mice (St-Jacques et al. 1999); addition of Sonic hedgehog (Shh) and bone morphogenetic protein (BMP)-2 synergistically promoted the proliferation of cultured primary chondrocytes (Takamoto et al. 2003); and Ihh expression is up-regulated during chondrocyte maturation (Bitgood and McMahon 1995; Vortkamp et al. 1996; Iwasaki et al. 1997; Zou et al. 1997). Although Ihh expression was detected in immature chondrocytes in Runx2–/– and Runx2–/–3–/– mice as well as wild-type mice at E12.5 and E13.5 (Fig. 3M–R), Ihh expression in chondrocytes was reduced in Runx2–/– mice and was undetectable in Runx2–/–3–/– mice at E18.5 (Figs. 5N,P, 7A). Therefore, it must be clarified whether the reduction in Ihh expression was a result of maturational blockage of chondrocytes or whether Runx2 and Runx3 are directly involved in Ihh expression. In the present study, Runx2 strongly induced Ihh expression shortly after adenoviral introduction of Runx2 in Runx2–/– chondrocyte cultures (Fig. 7D). Further, the 1.2-kb promoter region of the mouse Ihh gene contained seven putative Runx binding sites, five of which were conserved in human, and Runx2 bound to the five Runx binding sites, thereby activating the promoter (Fig. 8A–E). These findings demonstrate that Runx2 directly regulates Ihh expression during chondrocyte maturation, but that neither Runx2 nor Runx3 is essential for the Ihh expression in the immature chondrocytes at the early stage of skeletal development. Thus, our findings indicate that Runx2 enhances chondrocyte proliferation through the induction of Ihh expression during chondrocyte maturation. Interestingly, Shh induced Runx2 expression and activated the Runx2 promoter in primary chondrocyte cultures (Takamoto et al. 2003). Therefore, there may be a positive feedback loop between Ihh and Runx2. In contrast, Runx3 failed to induce Ihh expression in Runx2–/– chondrocyte cultures (Fig. 7D), and it failed to activate the Ihh promoter (Fig. 8G). Therefore, either the induction of Ihh by Runx3 is indirect or other factors are required for the induction of Ihh expression. We examined the requirement of soluble factors in Runx3-dependent Ihh expression. However, the addition of BMP-7, transforming growth factor-β (TGF-β), retinoic acid, insulin-like growth factor 1 (IGF-1), basic fibroblast growth factor (bFGF), platelet-derived growth factor (PDGF), parathyroid hormone (PTH), or T3 to Runx3-overexpressing Runx2–/– chondrocyte cultures failed to induce Ihh expression, although BMP-2 had a mild effect on Ihh expression irrespective of the Runx3 overexpression (data not shown).
Ihh was weakly expressed in Runx2–/– cartilage but not at all in Runx2–/–3–/– cartilage (Figs. 5N,P, 7A). However, the absence of Ihh expression in Runx2–/–3–/– cartilage did not result in further reduction in the chondrocyte proliferation, indicating that there was no complete correlation between the degree of inhibition of proliferation and the level of Ihh expression (Fig. 6E,F). As immature chondrocytes have more capacity for proliferation than mature chondrocytes in vitro (Enomoto et al. 2000; Enomoto-Iwamoto et al. 2001), the severe inhibition of chondrocyte maturation in Runx2–/–3+/– and Runx2–/–3–/– mice may have enhanced the chondrocyte proliferation in an Ihh-independent manner and compensated for the reduction in proliferation due to the reduced Ihh expression. However, Ihh, even at a low dose mediated by Runx3, may play an important role in organizing chondrocyte proliferation to a longitudinal direction, because the loss of columnar alignment of chondrocytes was well correlated with the disappearance of Ihh expression (Figs. 5N–P, 6B–D).
As the addition of Pthlh inhibited Runx2-induced maturation of cultured chick primary chondrocytes (Iwamoto et al. 2003), up-regulation of Ihh expression by Runx2 is also important as a negative feedback pathway through Pthlh to acquire an appropriate rate of chondrocyte maturation. Thus, in addition to the essential role of Runx2 and Runx3 in chondrocyte maturation, Runx2 is involved in coordinating the maturation and proliferation of chondrocytes, which is required for the formation of skeletal parts of appropriate size and shape, through the regulation of Ihh expression.
Materials and methods
Generation of Runx2–/–3–/– mice
We generated Runx2+/– mice and Runx3+/– mice as described (Komori et al. 1997; Li et al. 2002). We mated Runx2+/– mice with Runx3+/– mice to generate Runx2+/–3+/– mice. We further mated Runx2+/–3+/– males with Runx2+/–3+/– females to generate Runx2–/–3–/– mice. We determined each genotype of Runx2 and Runx3 by PCR using tail DNA using the following primers: Runx2, 5′-AGCTTTAGCGTCGTCAGACC-3′ and 5′-CAGGT TCAGCAGTCCATCTC-3′, 5′-AGCTTTAGCGTCGTCAGA CC-3′ and 5′-CAAGCGAAACATCGCATCGAGC-3′; Runx3, 5′-GACTGTGCATGCACCTTTCACCAA-3′ and 5′-TAGGGC TCAGTAGCACTTACGTCG-3′, 5′-GACTGTGCATGCACC TTTCACCAA-3′ and 5′-ATGAAACGCCGAGTTAACGCCA TCA-3′. Prior to the study, all experiments were reviewed and approved by the Osaka University Medical School Animal Care and Use Committee.
Detection of β-galactosidase activity, skeletal preparation, and analyses of histology, BrdU incorporation, and matrix and cell parameters
To study the localization of Runx3 expression, we examined the activity of β-galactosidase, whose gene was integrated in exon 3 of Runx3 in Runx3+/– mice (Li et al. 2002) as described (Ueta et al. 2001). We embedded stained embryos in paraffin and generated 7-μm sections. Newborn skeletons were stained with Alcian blue and Alizarin red as described (Komori et al. 1997). For histological analysis, we fixed embryos in 4% paraformaldehyde, embedded them in paraffin, and prepared 7-μm sections to perform hematoxylin and eosin (H&E) staining, double staining with H&E and von Kossa, or Alcian blue staining as described (Ueta et al. 2001). For analysis of BrdU incorporation, we intraperitoneally injected pregnant mice with 50 μg BrdU/g body weight 1 h before sacrifice. We processed the embryos for histological analysis and detected BrdU incorporation by immunohistochemistry using anti-BrdU antibody (Dako). We counter-stained the sections with toluidine blue. For the analyses of matrix and cell parameters, we prepared 4-μm sections and stained the sections with safranin O. The cell number, area of matrix, and cell size were analyzed using Mac Scope 2.6 software (Mitani).
In situ hybridization
For in situ hybridization, we prepared digoxigenin-11-UTP-labeled single-stranded RNA probes using a DIG RNA labeling kit (Roche Biochemica) according to the manufacturer's instructions. We used a 0.4-kb fragment of Col2a1 cDNA (Inada et al. 1999), a 0.65-kb fragment of mouse Col10a1 cDNA (Inada et al. 1999), a 1.2-kb fragment of osteopontin cDNA (Komori et al. 1997), a 0.4-kb fragment of mouse Sox9 cDNA (nt 723–1263, NM-011448), a 0.8-kb fragment of mouse Pthr1 cDNA (Inada et al. 1999), and a 0.6-kb fragment of mouse Ihh cDNA (Inada et al. 1999) to generate antisense and sense probes. We carried out hybridization as described previously and counterstained the sections with methyl green.
Cell culture and adenoviral transfer
We isolated chondrocytes from the skeletons of E18.5 Runx2–/– embryos, plated them at a density of 1 × 105 cells/well in 24-well plates (Corning), and cultured them in DMEM/Ham's F12 (1:1) hybrid medium (GIBCO-BRL) containing 10% fetal bovine serum (FBS; GIBCO-BRL) as described (Enomoto et al. 2004). When the cells reached confluence, we infected the cells with enhanced green fluorescent protein (EGFP)-expressing adenovirus, Runx2-and-EGFP-expressing adenovirus, or Runx3-and-EGFP-expressing adenovirus for 2 h at a multiplicity of infection (MOI) of 10 in 0.2 mL medium. After the medium was added up to 1 mL, the cells were cultured for 46 h. The infected cells were harvested for real-time RT–PCR at the indicated times after the onset of viral infection. We generated bicistronic adenovirus vectors expressing Runx2 and EGFP or EGFP alone as described (Enomoto et al. 2003). We generated a bicistronic adenovirus vector expressing Runx3 and EGFP using human Runx3 cDNA (Bae et al. 1995).
Real-time RT–PCR
Total RNA that had been extracted from wild-type, Runx2–/–, and Runx2–/–3–/– limb skeletons at E18.5 and Runx2–/– chondrocytes was reverse transcribed using M-MLV reverse transcriptase. We mixed the cDNA (7.5 ng total RNA equivalent) with SYBR Green PCR Master Mix (Applied Biosystems) and analyzed it by real-time PCR using the ABI7700 (Applied Biosystems). Primers used for amplification were as follows: Ihh, 5′-TTCAAGGACGAGGAGAACACG-3′ and 5′-TTCAGACG GTCCTTGCAGC-3′; Col10a1, 5′-AGAACGGCACGCCTAC GAT-3′ and 5′-AGGTAGCCTTTGCTGTACTCATCAT-3′; Runx2, 5′-CCGCACGACAACCGCACCAT-3′ and 5′-CGCTC CGGCCCACAAATCTC-3′; Runx3, 5′-GGTTCAACGACCTT CGCTTC-3′ and 5′-CACGGTCACCTTGATGGCT-3′; Mmp13, 5′-TCACCTGATTCTTGCGTGCT-3′ and 5′-CTGTGGGTT ATTATCAATCTTGTTTCTT-3′. We normalized the obtained CT (cycle number at which amplification threshold of detection was reached) values for Ihh, Col10a1, Runx2, Runx3, and Mmp13 to that of rodent Gapdh (Applied Biosystems) expression by the ΔΔCT method. The mean ΔΔCT was converted to relative expression value by the equation 2–ΔΔCt, and the range was calculated by the equation 2–(ΔΔCt + StdevΔΔCt).
ChIP
Chondrocytes from Runx2–/– embryos at E18.5 were infected with EGFP- or Runx2-and-EGFP-expressing adenovirus at an MOI of 10. ChIP was performed as described (Takahashi et al. 2000). After immunoprecipitation using monoclonal anti-Runx2 antibody (Yoshida et al. 2002) and DNA extraction, PCR was performed using the following primers in the promoter region of the mouse Ihh gene: P1, 5′-ATGGATAGGGTGCG GTCTGC-3′; P2, 5′-CTGAGAAAGGGAATGTTGCC-3′; P3, 5′-GTGAGGGTGAGGGAGGAAGA-3′; P4, 5′-GCGTGCTGT CCCCCTCGGCG-3′; P5, 5′-AGAGGGGAAATGGAAGAGAT-3′; P6, 5′-TAGGGGATGCGGGTGGAGGC-3′.
EMSA
We prepared nuclear extracts from chondrocytes of Runx2–/– embryos at E18.5 as described (Yoshida et al. 2002). Briefly, Runx2–/– chondrocytes were infected with EGFP- or Runx2-and-EGFP-expressing adenovirus at an MOI of 10, and nuclear extracts were prepared 24 h later. The nuclear extracts were incubated with 10 fmol of each of 32P-labeled double-stranded R1–R7 oligonucleotides, and EMSA was performed as described (Yoshida et al. 2002). The oligonucleotide sequences of R1–R7 were as follows: R1, 5′-GGATCCGCTAACCGCGGGTGGAT CC-3′; R2, 5′-GGATCCGGGTGCGGTCTGCGGATCC-3′; R3, 5′-GGATCCAGGA GCCGCAAGGGGATCC-3′; R4, 5′-GGAT CCGGTTGCGGTCTCCGGATCC-3′; R5, 5′-GGATCCTTGT GCGGTCCGGGGATCC-3′; R6, 5′-GGATCCGTCCG TGGC TTCTGGATCC-3′; R7, 5′-GGATCCCATTGTGGTGGTGGA TCC-3′. For supershift experiments, the monoclonal antibody against Runx2 (Yoshida et al. 2002) was added to the entire mixture. We carried out a competition with 200-fold molar excess of each unlabeled oligonucleotide of R1–R7. In the analysis of DNA binding of Runx3, Runx2–/– chondrocytes were infected with retrovirus expressing Cbfb and EGFP or EGFP alone by the bicistronic retroviral expression vector (Yoshida et al. 2002). After 1 d, the cells were infected with EGFP- or Runx3-and-EGFP-expressing adenovirus at an MOI of 10, and nuclear extracts were prepared 24 h later. For supershift experiments, the polyclonal antibody against Cbfβ (Yoshida et al. 2002) was added to the entire mixture.
Reporter assay
The 1.3-kb promoter region (–1208/+105) of the mouse Ihh gene was amplified by PCR using liver DNA from C57BL/6 × C3H F1. Various lengths of Ihh promoter fragments were generated either by restriction digests of the 1.3-kb Ihh promoter region or by PCR, and they were inserted into the pGL3 vector. All of the constructs were confirmed by sequencing. Chondrocytes from Runx2–/– embryos at E18.5 or ATDC5 cells in 48-well multiplates were transiently transfected with a DNA mixture containing each Ihh promoter construct (0.2 μg) and pRL-CMV (0.002 μg) using FuGENE 6 (Roche Diagnostics). After culture for 24 h, the cells were infected with EGFP- or Runx2-and-EGFP-expressing adenovirus at an MOI of 10. After 24 h, we assayed the luciferase activity of the cell lysate using the Dual Luciferase Reporter Assay System (Promega) and a model TD20/20 luminometer (Promega). The level of luciferase activity was normalized to the level of Renilla luciferase activity. To compare the capacities of Runx2 and Runx3 in the transcriptional activation of Ihh promoter, the cDNA of Runx2 or Runx3 was inserted into pSG5, and Runx2–/– chondrocytes were transfected with DNA mixture containing p1313-luc (0.2 μg), Runx2- or Runx3-expressing vector or pSG5 (0.05 μg), and the control vector pRL-CMV (0.002 μg; Promega) using FuGENE 6 (Roche Diagnostics).
Acknowledgments
We thank M. Iwamoto for the Sox9 probe; Z. Maruyama for helpful discussion; R. Fukuyama, N. Kanatani, A. Ono, and H. Enomoto for technical assistance; R. Hiraiwa for maintaining mouse colonies; and M. Yanagita for secretarial assistance. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1174704.
References
- Akiyama, H., Chaboissier, M.C., Martin, J.F., Schedl, A., and de Crombrugghe, B. 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes & Dev. 16: 2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, S.C., Takahashi, E., Zhang, Y.W., Ogawa, E., Shigesada, K., Namba, Y., Satake, M., and Ito, Y. 1995. Cloning, mapping and expression of PEBP2 αC, a third gene encoding the mammalian Runt domain. Gene 159: 245–248. [DOI] [PubMed] [Google Scholar]
- Bi, W., Deng, J.M., Zhang, Z., Behringer, R.R., and de Crombrugghe, B. 1999. Sox9 is required for cartilage formation. Nat. Genet. 22: 85–89. [DOI] [PubMed] [Google Scholar]
- Bitgood, M.J. and McMahon, A.P. 1995. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell–cell interaction in the mouse embryo. Dev. Biol. 172: 126–138. [DOI] [PubMed] [Google Scholar]
- Enomoto, H., Enomoto-Iwamoto, M., Iwamoto, M., Nomura, S., Himeno, M., Kitamura, Y., Kishimoto, T., and Komori, T. 2000. Cbfa1 is a positive regulatory factor in chondrocyte maturation. J. Biol. Chem. 275: 8695–8702. [DOI] [PubMed] [Google Scholar]
- Enomoto, H., Shiojiri, S., Hoshi, K., Furuichi, T., Fukuyama, R., Yoshida, C., Kanatani, N., Nakamura, R., Mizuno, A., Zanma, A., et al. 2003. Induction of osteoclast differentiation by Runx2 through RANKL and OPG regulation and partial rescue of osteoclastogenesis in Runx2–/– mice by RANKL transgene. J. Biol. Chem. 278: 23971–23977. [DOI] [PubMed] [Google Scholar]
- Enomoto, H., Furuichi, T., Zanma, A., Yamana, K., Yoshida, C., Sumitani, S., Yamamoto, H., Enomoto-Iwamoto, M., Iwamoto, M., and Komori, T. 2004. Runx2 deficiency in chondrocytes causes adipogenic changes in vitro. J. Cell Sci. 117: 417–425. [DOI] [PubMed] [Google Scholar]
- Enomoto-Iwamoto, M., Enomoto, H., Komori, T., and Iwamoto, M. 2001. Participation of Cbfa1 in regulation of chondrocyte maturation. Osteoarthritis Cartilage 9: S76–S84. [DOI] [PubMed] [Google Scholar]
- Gilbert, S.F. 1997. Developmental Biology, 5th ed., pp. 351–357. Sinauer Associates, Sunderland, MA.
- Inada, M., Yasui, T., Nomura, S., Miyake, S., Deguchi, K., Himeno, M., Sato, M., Yamagiwa, H., Kimura, T., Yasui, N., et al. 1999. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev. Dyn. 214: 279–290. [DOI] [PubMed] [Google Scholar]
- Inoue, K., Ozaki, S., Shiga, T., Ito, K., Masuda, T., Okado, N., Iseda, T., Kawaguchi, S., Ogawa, M., Bae, S.C., et al. 2002. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat. Neurosci. 5: 946–954. [DOI] [PubMed] [Google Scholar]
- Iwamoto, M., Kitagaki, J., Tamamura, Y., Gentili, C., Koyama, E., Enomoto, H., Komori, T., Pacifici, M., and Enomoto-Iwamoto. M. 2003. Runx2 expression and action in chondrocytes are regulated by retinoid signaling and parathyroid hormone-related peptide (PTHrP). Osteoarthritis Cartilage 11: 6–15. [DOI] [PubMed] [Google Scholar]
- Iwasaki, M., Le, A.X., and Helms, J.A. 1997. Expression of indian hedgehog, bone morphogenetic protein 6 and gli during skeletal morphogenesis. Mech. Dev. 69: 197–202. [DOI] [PubMed] [Google Scholar]
- Jiménez, M.J., Balbin, M., Lopez, J.M., Alvarez, J., Komori, T., and Lopez-Otin, C. 1999. Collagenase 3 is a target of Cbfa1, a transcription factor of the runt gene family involved in bone formation. Mol. Cell Biol. 19: 4431–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, I.S., Otto, F., Zabel, B., and Mundlos, S. 1999. Regulation of chondrocyte differentiation by Cbfa1. Mech. Dev. 80: 159–170. [DOI] [PubMed] [Google Scholar]
- Komori, T. 2002. Runx2, a multifunctional transcription factor in skeletal development. J. Cell. Biochem. 87: 1–8. [DOI] [PubMed] [Google Scholar]
- Komori, T., Yagi, H., Nomura, S., Yamaguchi, A., Sasaki, K., Deguchi, K., Shimizu, Y., Bronson, R.T., Gao, Y., Inada, M., et al. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89: 755–764. [DOI] [PubMed] [Google Scholar]
- Kundu, M., Javed, A., Jeon, J.P., Horner, A., Shum, L., Eckhaus, M., Muenk, M., Lian, J.B., Yang, Y., Nuckolls, G.H., et al. 2002. Cbfβ interacts with Runx2 and has a critical role in bone development. Nat. Genet. 32: 639–644. [DOI] [PubMed] [Google Scholar]
- Levanon, D., Brenner, O., Negreanu, V., Bettoun, D., Woolf, E., Eilam, R., Lotem, J., Gat, U., Otto, F., Speck N., et al. 2001. Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1 (Aml1) indicates non-redundant functions during mouse embryogenesis. Mech. Dev. 109: 413–417. [DOI] [PubMed] [Google Scholar]
- Levanon, D., Bettoun, D., Harris-Cerruti, C., Woolf, E., Negreanu, V., Eilam, R., Bernstein, Y., Goldenberg, D., Xiao, C., Fliegauf, M., et al. 2002. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 21: 3454–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q.L., Ito, K., Sakakura, C., Fukamachi, H., Inoue, K., Chi, X.Z., Lee, K.Y., Nomura, S., Lee, C.W., Han, S.B., et al. 2002. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109: 113–124. [DOI] [PubMed] [Google Scholar]
- Miller, J., Horner, A., Stacy, T., Lowrey, C., Lian, J.B, Stein G., Nuckolls, G.H., and Speck N. 2002. The core-binding factor b subunit is required for bone formation and hematopoietic maturation. Nat. Genet. 32: 645–649. [DOI] [PubMed] [Google Scholar]
- Okada, H., Watanabe, T., Niki, M., Takano, H., Chiba, N., Yanai, N., Tani, K., Hibino, H., Asano, S., Mucenski, M.L., et al. 1998. AML1(–/–) embryos do not express certain hematopoiesis-related gene transcripts including those of the PU.1 gene. Oncogene 17: 2287–2293. [DOI] [PubMed] [Google Scholar]
- Okuda, T., van Deursen, J., Hiebert, S.W., Grosveld, G., and Downing, J.R. 1996. AML1, the target of multiple chromosomal translocation in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84: 321–330. [DOI] [PubMed] [Google Scholar]
- Otto, F., Thornell, A.P., Crompton, T., Denzel, A., Gilmour, K.C., Rosewell, I.R., Stamp, G.W.H., Beddington, R.S.P., Mundlos, S., Olsen, B.R., et al. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89: 765–771. [DOI] [PubMed] [Google Scholar]
- St-Jacques, B., Hammerschmidt, M., and McMahon, A.P. 1999. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes & Dev. 13: 2072–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker, S., Fundele, R., Vortkamp, A., and Mundlos, S. 2002. Role of Runx genes in chondrocyte differentiation. Dev. Biol. 245: 95–108. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., Rayman, J.B., and Dynlacht BD. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: Distinct E2F proteins mediate activation and repression. Genes & Dev. 14: 804–16. [PMC free article] [PubMed] [Google Scholar]
- Takamoto, M., Tsuji, K., Yamashita, T., Sasaki, H., Yano, T., Taketani, Y., Komori, T., Nifuji, A., and Noda, M. 2003. Hedgehog signaling enhances core-binding factor a1 and receptor activator of nuclear factor-κB ligand (RANKL) gene expression in chondrocytes. J. Endocrinol. 177: 413–421. [DOI] [PubMed] [Google Scholar]
- Takeda, S., Bonnamy, J.P., Owen, M.J., Ducy, P., and Karsenty, G. 2001. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes & Dev. 15: 467–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta, C., Iwamoto, M., Kanatani, N., Yoshida, C., Liu, Y., Enomoto-Iwamoto, M., Ohmori, T., Enomoto, H., Nakata, K., Takada, K., et al. 2001. Skeletal malformations caused by overexpression of Cbfa1 or its dominant negative form in chondrocytes. J. Cell Biol. 153: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vortkamp, A., Lee, K., Lanske, B., Segre, G.V., Kronenberg, H.M., and Tabin, C.J. 1996. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 273: 613–622. [DOI] [PubMed] [Google Scholar]
- Wang, Q., Stancy, T., Binder, M., Marín-Padilla, M., Sharpe, A.H., and Speck, N.A. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. 93: 3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, C.A., Furuichi, T., Fujita, T., Fukuyama, R., Kanatani, N., Kobayashi, S., Satake, M., Takada, K., and Komori, T. 2002. Core-binding factor β interacts with Runx2 and is required for skeletal development. Nat. Genet. 32: 633–638. [DOI] [PubMed] [Google Scholar]
- Zou, H., Wieser, J., Massage, J., and Niswander, L. 1997. Distinct roles of types I bone morphogenetic protein receptors in the formation and differentiation of cartilage. Genes & Dev. 11: 2191–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]