Abstract
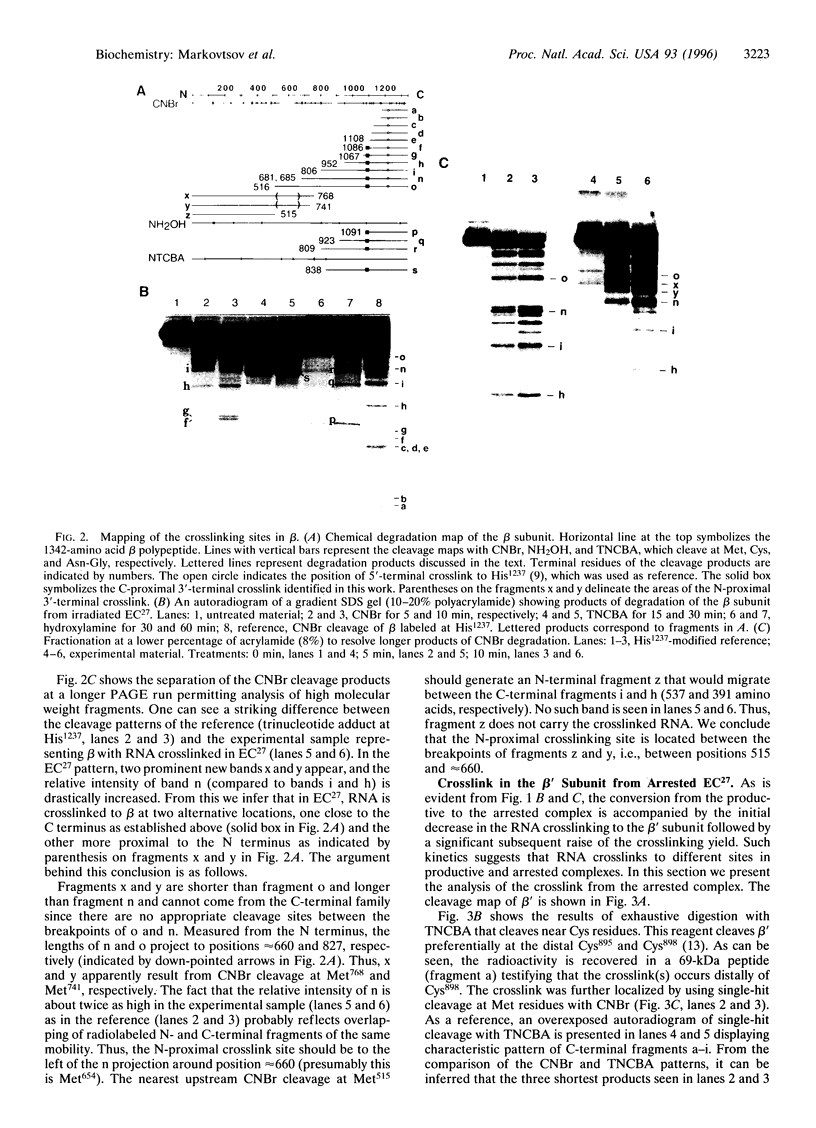
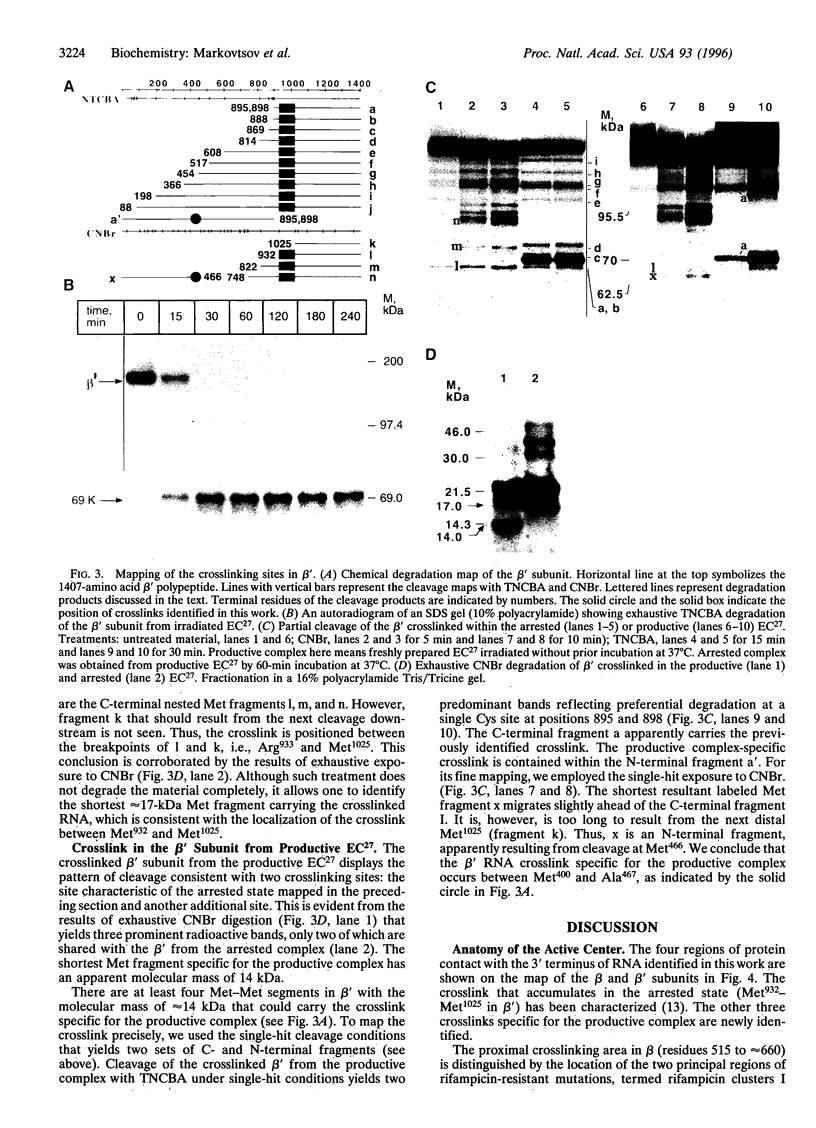
By using a crosslinkable probe incorporated into the 3' terminus of nascent transcript, three sites were mapped in Escherichia coli RNA polymerase that are contacted by the RNA in the productive elongation complex. Two of these sites are in the beta subunit and one is in the beta' subunit. During elongation, the transcription complex occasionally undergoes an arrest whereby it can neither extend nor release the RNA transcript. It is demonstrated that in an arrested complex, the three contacts of RNA 3' terminus are lost, while a new beta' subunit contact becomes prominent. Thus, elongation arrest appears to involve the disengagement of the bulk of the active center from the 3' terminus of RNA and the transfer of the terminus into a new protein environment.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berghöfer B., Kröckel L., Körtner C., Truss M., Schallenberg J., Klein A. Relatedness of archaebacterial RNA polymerase core subunits to their eubacterial and eukaryotic equivalents. Nucleic Acids Res. 1988 Aug 25;16(16):8113–8128. doi: 10.1093/nar/16.16.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S., Lee J., Goldfarb A. Mapping of a contact for the RNA 3' terminus in the largest subunit of RNA polymerase. J Biol Chem. 1991 Dec 15;266(35):23932–23935. [PubMed] [Google Scholar]
- Borukhov S., Sagitov V., Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993 Feb 12;72(3):459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Cassani G., Burgess R. R., Goodman H. M., Gold L. Inhibition of RNA polymerase by streptolydigin. Nat New Biol. 1971 Apr 14;230(15):197–200. doi: 10.1038/newbio230197a0. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J. New models for the mechanism of transcription elongation and its regulation. Harvey Lect. 1992 1993;88:1–21. [PubMed] [Google Scholar]
- Darst S. A., Edwards A. M., Kubalek E. W., Kornberg R. D. Three-dimensional structure of yeast RNA polymerase II at 16 A resolution. Cell. 1991 Jul 12;66(1):121–128. doi: 10.1016/0092-8674(91)90144-n. [DOI] [PubMed] [Google Scholar]
- Darst S. A., Kubalek E. W., Kornberg R. D. Three-dimensional structure of Escherichia coli RNA polymerase holoenzyme determined by electron crystallography. Nature. 1989 Aug 31;340(6236):730–732. doi: 10.1038/340730a0. [DOI] [PubMed] [Google Scholar]
- Das A. Control of transcription termination by RNA-binding proteins. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- Dieci G., Hermann-Le Denmat S., Lukhtanov E., Thuriaux P., Werner M., Sentenac A. A universally conserved region of the largest subunit participates in the active site of RNA polymerase III. EMBO J. 1995 Aug 1;14(15):3766–3776. doi: 10.1002/j.1460-2075.1995.tb00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburg D., Dworniczak B., Faust D. M., Bautz E. K. RNA polymerase II of Drosophila. Relation of its 140,000 Mr subunit to the beta subunit of Escherichia coli RNA polymerase. J Mol Biol. 1987 Jun 20;195(4):929–937. doi: 10.1016/0022-2836(87)90496-7. [DOI] [PubMed] [Google Scholar]
- Grachev M. A., Kolocheva T. I., Lukhtanov E. A., Mustaev A. A. Studies on the functional topography of Escherichia coli RNA polymerase. Highly selective affinity labelling by analogues of initiating substrates. Eur J Biochem. 1987 Feb 16;163(1):113–121. doi: 10.1111/j.1432-1033.1987.tb10743.x. [DOI] [PubMed] [Google Scholar]
- Grachev M. A., Lukhtanov E. A., Mustaev A. A., Zaychikov E. F., Abdukayumov M. N., Rabinov I. V., Richter V. I., Skoblov Y. S., Chistyakov P. G. Studies of the functional topography of Escherichia coli RNA polymerase. A method for localization of the sites of affinity labelling. Eur J Biochem. 1989 Apr 1;180(3):577–585. doi: 10.1111/j.1432-1033.1989.tb14684.x. [DOI] [PubMed] [Google Scholar]
- Gu W., Reines D. Identification of a decay in transcription potential that results in elongation factor dependence of RNA polymerase II. J Biol Chem. 1995 May 12;270(19):11238–11244. doi: 10.1074/jbc.270.19.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOARD D. E., OTT D. G. CONVERSION OF MONO- AND OLIGODEOXYRIBONUCLEOTIDES TO 5-TRIPHOSPHATES. J Am Chem Soc. 1965 Apr 20;87:1785–1788. doi: 10.1021/ja01086a031. [DOI] [PubMed] [Google Scholar]
- Heisler L. M., Suzuki H., Landick R., Gross C. A. Four contiguous amino acids define the target for streptolydigin resistance in the beta subunit of Escherichia coli RNA polymerase. J Biol Chem. 1993 Dec 5;268(34):25369–25375. [PubMed] [Google Scholar]
- Izban M. G., Luse D. S. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3'----5' direction in the presence of elongation factor SII. Genes Dev. 1992 Jul;6(7):1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- Jin D. J., Gross C. A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988 Jul 5;202(1):45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- Joyce C. M., Steitz T. A. Function and structure relationships in DNA polymerases. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. RNA polymerase: regulation of transcript elongation and termination. FASEB J. 1991 Oct;5(13):2833–2842. doi: 10.1096/fasebj.5.13.1916107. [DOI] [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Individual complexes halted along different transcription units have distinct and unexpected biochemical properties. J Mol Biol. 1992 May 20;225(2):221–237. doi: 10.1016/0022-2836(92)90917-9. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mustaev A., Kashlev M., Lee J. Y., Polyakov A., Lebedev A., Zalenskaya K., Grachev M., Goldfarb A., Nikiforov V. Mapping of the priming substrate contacts in the active center of Escherichia coli RNA polymerase. J Biol Chem. 1991 Dec 15;266(35):23927–23931. [PubMed] [Google Scholar]
- Mustaev A., Kashlev M., Zaychikov E., Grachev M., Goldfarb A. Active center rearrangement in RNA polymerase initiation complex. J Biol Chem. 1993 Sep 15;268(26):19185–19187. [PubMed] [Google Scholar]
- Mustaev A., Zaychikov E., Severinov K., Kashlev M., Polyakov A., Nikiforov V., Goldfarb A. Topology of the RNA polymerase active center probed by chimeric rifampicin-nucleotide compounds. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):12036–12040. doi: 10.1073/pnas.91.25.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustaev A., Zaychikov E., Severinov K., Kashlev M., Polyakov A., Nikiforov V., Goldfarb A. Topology of the RNA polymerase active center probed by chimeric rifampicin-nucleotide compounds. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):12036–12040. doi: 10.1073/pnas.91.25.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E., Goldfarb A., Kashlev M. Discontinuous mechanism of transcription elongation. Science. 1994 Aug 5;265(5173):793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- Nudler E., Kashlev M., Nikiforov V., Goldfarb A. Coupling between transcription termination and RNA polymerase inchworming. Cell. 1995 May 5;81(3):351–357. doi: 10.1016/0092-8674(95)90388-7. [DOI] [PubMed] [Google Scholar]
- Orlova M., Newlands J., Das A., Goldfarb A., Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci U S A. 1995 May 9;92(10):4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pühler G., Leffers H., Gropp F., Palm P., Klenk H. P., Lottspeich F., Garrett R. A., Zillig W. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem. 1992 Feb 25;267(6):3795–3800. [PMC free article] [PubMed] [Google Scholar]
- Schultz P., Célia H., Riva M., Sentenac A., Oudet P. Three-dimensional model of yeast RNA polymerase I determined by electron microscopy of two-dimensional crystals. EMBO J. 1993 Jul;12(7):2601–2607. doi: 10.1002/j.1460-2075.1993.tb05920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Severinov K., Fenyö D., Severinova E., Mustaev A., Chait B. T., Goldfarb A., Darst S. A. The sigma subunit conserved region 3 is part of "5'-face" of active center of Escherichia coli RNA polymerase. J Biol Chem. 1994 Aug 19;269(33):20826–20828. [PubMed] [Google Scholar]
- Severinov K., Mustaev A., Severinova E., Kozlov M., Darst S. A., Goldfarb A. The beta subunit Rif-cluster I is only angstroms away from the active center of Escherichia coli RNA polymerase. J Biol Chem. 1995 Dec 8;270(49):29428–29432. doi: 10.1074/jbc.270.49.29428. [DOI] [PubMed] [Google Scholar]
- Severinov K., Soushko M., Goldfarb A., Nikiforov V. Rifampicin region revisited. New rifampicin-resistant and streptolydigin-resistant mutants in the beta subunit of Escherichia coli RNA polymerase. J Biol Chem. 1993 Jul 15;268(20):14820–14825. [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Meier T. I., Chan C. L., Feng G., Lee D. N., Landick R. Discontinuous movements of DNA and RNA in RNA polymerase accompany formation of a paused transcription complex. Cell. 1995 May 5;81(3):341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- Zaychikov E., Denissova L., Heumann H. Translocation of the Escherichia coli transcription complex observed in the registers 11 to 20: "jumping" of RNA polymerase and asymmetric expansion and contraction of the "transcription bubble". Proc Natl Acad Sci U S A. 1995 Feb 28;92(5):1739–1743. doi: 10.1073/pnas.92.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Helm K., Krakow J. S. Inhibition of RNA polymerase by streptolydigin. Nat New Biol. 1972 Jan 19;235(55):82–83. doi: 10.1038/newbio235082a0. [DOI] [PubMed] [Google Scholar]