We recently reported that the combination of Gata4, Hand2, Tbx5, and the fusion gene MM3 between Mef2c and the transactivation domain of MyoD (MM3-GHT) produces 18 times as many clusters of beating cells (induced cardiomyocyte-like cells or iCMs) as the wild-type combination (M-GHT).1 In the current study, we added the chemicals GSK126 and UNC0638 to MM3-GHT to examine whether depletion of suppressive histone markers further increases the efficiency of making iCMs. GSK126 inhibits Enhancer of Zeste Homolog 2 (Ezh2), which induces di- and trimethylation of Lys27 on histone H3 (H3K27me2 and H3K27me3, respectively) as a catalytic subunit of the Polycomb Repressive Complex 2 (PRC2).2 UNC0638 inhibits two closely related enzymes, G9a and GLP, which form a heterodimer in vivo and mediate mono- and dimethylation of Lys9 on histone H3 (H3K9me and H3K9me2) and methylation of non-histone substrates such as p53.3 H3K27me3 and H3K9me2 are typically associated with suppressed genes although the underlying mechanisms have not been fully characterized.
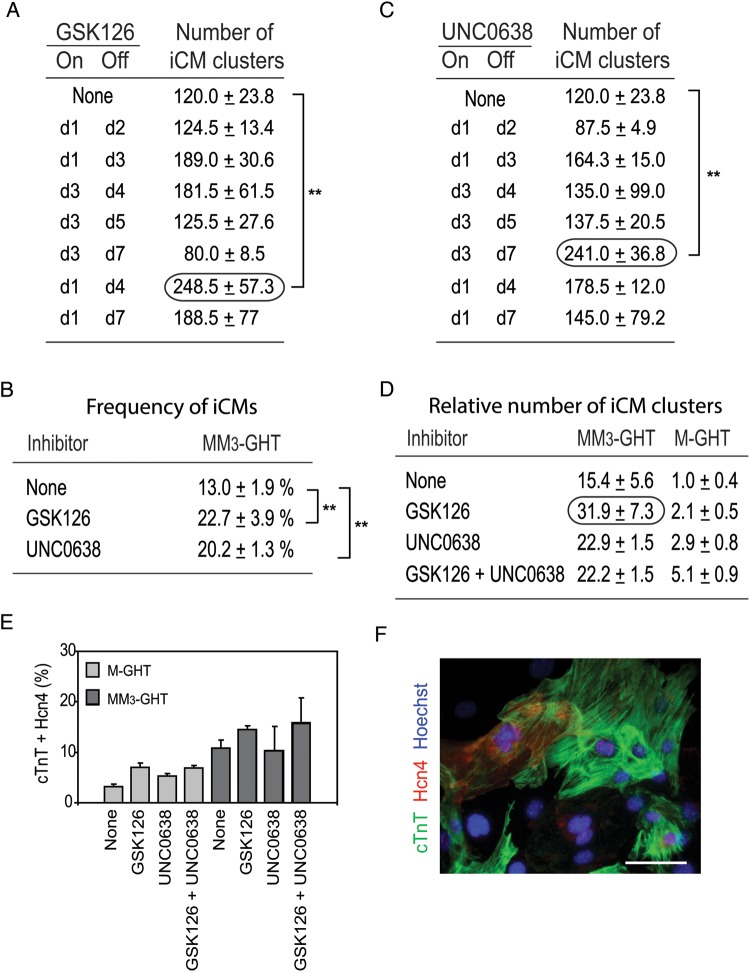
We prepared fibroblasts from the heads of Day 13.5 mouse embryos and incubated them with 1 μM GSK126 for 2 days to verify a decrease of H3K27me3 (see Supplementary material online, Figure S1A). To test the effect of GSK126 on reprogramming, fibroblasts were transduced with MM3-GHT on Day −1 and 0, and GSK126 was added to the culture medium at different time points (Figure 1A). The most effective schedule was to add GSK126 from Day 1 to 4, resulting in 2.1 times as many iCM clusters as negative control without GSK126 (Figure 1A, encircled and Supplementary material online, Figure S1B). Ca2+ oscillations in the cells were monitored using a transgene encoding the Ca2+ indicator protein GCaMP5, which emits stronger green fluorescence in the presence of a higher intracellular Ca2+ concentration.4 When GSK126 was used from Day 1 to 4, numerous cells exhibited asynchronous oscillation of the fluorescence intensity, and thus the intracellular Ca2+ concentration, on Day 14 (see Supplementary material online, Figure S1C and Videos 1A and 2A). Some cells were spontaneously beating (Phase and Hoechst panels in Supplementary material online, Figure S1C and Videos 1B and 2B). The prevalence of cells that showed beating or oscillation of fluorescence was calculated by dividing the number of these cells by the number of nuclei detected by the Hoechst dye. Although mature cardiomyocytes are generally binucleated,5 we did not take this into consideration because distinguishing which two nuclei belong to a single cell was difficult. With this method, 55% of the cells in a well displayed Ca2+ oscillation (see Supplementary material online, Figure S1D) and 23% of the cells beat spontaneously (Figure 1B). Although the effect of GSK126 on the number of cells with Ca2+ oscillation was not statistically significant, it generated 1.8 times as many iCMs (cell number, not cluster number) as control. No cell demonstrated fluorescence oscillation or spontaneous beating without MM3-GHT.
Figure 1.
GSK126 and UNC0638 facilitate iCM formation from mouse embryonic fibroblasts with MM3-GHT and M-GHT. (A) The number of iCM clusters obtained with the addition of GSK126 to the culture medium of fibroblasts during iCM formation with MM3-GHT. The day of the addition (On) and removal (Off) of GSK126 is listed on the left; the maximum number of iCM clusters obtained between Day 1 and 14 is listed on the right. A total of 10 000 fibroblasts were seeded in each well of 48-well plates on Day –2 and the total number of iCM clusters was counted in each well. Mean ± SD of three independent experiments is shown. **P < 0.01 (Student's t-test). The highest number of iCM clusters is encircled. (B) Frequency of iCMs on Day 14 after the addion of GSK126 or UNC0638 from Day 1 to 4. The percentage of spontaneously beating cells (iCMs) among more than 500 Hoechst-positive cells is shown. **P < 0.01 (Student's t-test). (C) The number of iCM clusters obtained with the addition of UNC0638 to the culture medium of fibroblasts during iCM formation with MM3-GHT. The highest number of iCM clusters is encircled. **P < 0.01 (Student's t-test). (D) Maximum number of iCM clusters obtained with 1 μM GSK126 and/or 0.25 μM UNC0638 added from Day 1 to 4 relative to the number of iCM clusters obtained with M-GHT without inhibitors (defined as 1.0). The values shown in (A) and (C), and Supplementary material online, Figure S2C (for MM3-GHT), and Supplementary material online, Figure S3A (for M-GHT) were used to calculate these ratios. (E) Frequency of cells double-positive for cardiac troponin T (cTnT) and hyperpolarization-activated cation channel 4 (Hcn4) on Day 14. 1 μM GSK126 and/or 0.25 μM UNC0638 were used from Day 1 to 4. (F) Immunofluorescence staining of cTnT and Hcn4 on Day 14. Cells were transduced with MM3-GHT, and 1 μM GSK126 and 0.25 μM UNC0638 were added from Day 1 to 4. DNA was counterstained with Hoechst 33342. Bars, 50 µm.
We next tested the effects of 0.25 μM UNC0638 after verifying a decrease of H3K9me2 by western blotting (see Supplementary material online, Figure S2A). In contrast to GSK126, UNC0638 was most effective when used from Day 3 to 7, when it produced twice as many iCM clusters as control (Figure 1C, encircled and Supplementary material online, Figure S2B), with 20% of individual cells beating (Figure 1B). We further tested a cumulative effect of the two inhibitors by using them simultaneously from Day 1 to 4 (see Supplementary material online, Figure S2C) or from Day 1 to 7 (see Supplementary material online, Figure S2D) or sequentially during Day 1 to 7 (see Supplementary material online, Figure S2E). However, none of these combinations was more effective than GSK126 alone.
Because M-GHT was much less efficient in inducing iCMs than MM3-GHT,1 the two inhibitors might display more profound effects when used with M-GHT. Although GSK126 increased iCM cluster formation two-fold with M-GHT (see Supplementary material online, Figure S3A) like MM3-GHT, UNC0638 increased the number of iCM clusters 2.9-fold with M-GHT when used between Day 1 and 4 (see Supplementary material online, Figure S3A), compared with a 1.5-fold increase with MM3-GHT (Figure 1C). The combination of the two inhibitors increased the number of iCM clusters 5.1-fold when used with M-GHT (see Supplementary material online, Figure S3A). Raising the concentration of UNC0638 from 0.25 to 1.0 μM further increased the number of iCM clusters (see Supplementary material online, Figure S3B, black) when the inhibitors were used from Day 1 to 4. Longer use from Day 1 to 7 (see Supplementary material online, Figure S3C), sequential addition (see Supplementary material online, Figure S3D), and a single late addition of UNC0638 (see Supplementary material online, Figure S3E) were all less effective than simultaneous use. Overall, the most effective combination was to transduce MM3-GHT and add GSK126 from Day 1 to 4 (Figure 1D, encircled), resulting in a 32-fold increase in the number of iCM clusters in comparison to M-GHT without inhibitors.
Immunofluorescence staining on Day 14 after transduction of MM3-GHT without inhibitors demonstrated that 19% of the cells were positive for the cardiac marker cardiac troponin T (cTnT) (see Supplementary material online, Figure S4A, dark grey, None). In addition, 11% of the cells were double-positive for cTnT and the ion channel protein Hcn4 (hyperpolarization-activated cation channel 4) (Figure 1E, dark grey, None). Although the addition of the inhibitors increased the number of cells positive for these proteins, the frequency was always less than twice of that seen with control which did not include the inhibitors (Figure 1E and Supplementary material online, Figure S4A). An example of the double staining is shown in Figure 1F and Supplementary material online, Figure S4B after transduction with MM3-GHT and the addition of the two inhibitors. Regularly distributed sarcomeres were evident with the cTnT staining (see Supplementary material online, Figure S4B, magnified panel). Thus, an increase in iCM clusters does not directly translate to an increase in the number of cardiac protein-positive cells. This is consistent with results obtained in our previous study with immunostaining of myosin light chain 2v and cardiac myosin heavy chain.1
This work has two implications for the use of chemical inhibitors of epigenetic enzymes to promote direct reprogramming. First, the effective time window could be narrow (Figure 1A and C). Although this is not surprising, it underscores that effective candidates could be overlooked during large-scale screening of small molecules if they are used at the wrong time. Details of the epigenetic reprogramming during iCM formation remain largely elusive; however, the reprogramming during induced pluripotent stem cell (iPSC) formation is known to be a stepwise process of suppression of the cell-specific genes expressed in the source cell, followed by activation of pluripotency-specific genes.6 Global removal of suppressive histone markers does not necessarily benefit reprogramming because it potentially prevents suppression of parent cell-specific genes. Whether GSK126 and UNC0638 facilitate reprogramming or not will be determined by the cumulative effects of activating both cardiac genes and inhibitory genes for reprogramming. This complexity is highlighted by a recent report that inhibition of Ezh2 and other PRC2 components by short hairpin RNAs decreases the efficiency of making iPSCs from fibroblasts.7
The second implication is the importance of the combination between an inhibitor and a transgene. UNC0638 was more effective with M-GHT than with MM3-GHT when used between Day 1 and 4 [2.9-fold increase (see Supplementary material online, Figure S3A) vs. 1.5-fold increase (Figure 1C)]. In addition, the combination of the two inhibitors was more effective than a single inhibitor only for M-GHT. One potential explanation is that MM3 recruits H3K9me2 demethylases, and additional inhibition of H3K9me2 is unnecessary. Another possibility is that UNC0638 demethylates the MyoD transactivation domain in MM3, inhibiting the promotion of reprogramming by this fusion protein. However, although G9a methylates Lys104 on MyoD, inhibiting its transcriptional activity,8 our MyoD domain contains only amino acids 1–62. Methylation of Mef2c has not been reported as far as we know.
We found that 23% of the cells treated with MM3-GHT and GSK126 beat spontaneously. Although some studies counted the number of beating cell clusters,1,9 the number of beating cells has not been reported, partly because the number was generally very low. Because mature cardiomyocytes (other than the cells in the conduction system) generally do not beat spontaneously,10 beating cannot be used as the sole indicator for the formation of cardiomyocytes. However, it is a better indicator of the magnitude of cardiomyocyte formation than the expression of a few cardomyocyte-specific proteins, the measure used in most previous studies,9 because beating requires multiple cardiac features, including a reasonably assembled set of ion channels, a calcium signalling system, and a contractile cytoskeletal apparatus, however incomplete.
Fifty-five per cent of the cells treated with MM3-GHT and GSK126 exhibited Ca2+ oscillation (see Supplementary material online, Figure S1D), but many of them showed a wave-like spread of high Ca2+ concentration from one side of a cell to the other (see Supplementary material online, Videos 1A and 2A). This wave-like pattern is typical of cardiomyocytes created by direct reprogramming, whereas cardiomyocytes prepared from embryonic heart show a simultaneous increase of the fluorescence intensity in the entire cytoplasm.4 The wave-like pattern appears to represent partially reprogrammed cardiomyocyte-like cells. Indeed, only 30% of the cells expressed cTnT, a marker for terminally differentiated cardiomyocytes (see Supplementary material online, Figure S4A) on the same day that the wave-like pattern was observed. Thus, further improvement of the protocol is required to increase the population of mature cardiomyocytes through direct reprogramming.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This study was supported by the Engdahl Family Foundation and the National Institutes of Health (R01 GM098294).
Supplementary Material
Acknowledgements
We thank Toshio Kitamura for pMXs-IP, Meri Firpo for mice, and Atsushi Asakura for the video recording equipment.
References
- 1.Hirai H, Katoku-Kikyo N, Keirstead SA, Kikyo N. Accelerated direct reprogramming of fibroblasts into cardiomyocyte-like cells with the MyoD transactivation domain. Cardiovasc Res. 2013;100:105–113. doi: 10.1093/cvr/cvt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. Ezh2 inhibition as a therapeutic strategy for lymphoma with Ezh2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 3.Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, et al. A chemical probe selectively inhibits G9a and Glp methyltransferase activity in cells. Nat Chem Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Addis RC, Ifkovits JL, Pinto F, Kellam LD, Esteso P, Rentschler S, et al. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol. 2013;60:97–106. doi: 10.1016/j.yjmcc.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28:1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 6.Papp B, Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling BM, Bharathy N, Chung TK, Kok WK, Li S, Tan YH, et al. Lysine methyltransferase G9a methylates the transcription factor MyoD and regulates skeletal muscle differentiation. Proc Natl Acad Sci USA. 2012;109:841–846. doi: 10.1073/pnas.1111628109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mummery CL, Zhang J, Ng ES, Elliott DA, Elefanty AG, Kamp TJ. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circ Res. 2012;111:344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.