Abstract
Most organs scale proportionally with body size through regulation of individual cell size and/or cell number. Here we addressed how postmitotic and morphologically complex cells such as neurons scale with the body size by using the dendritic arbor of one Drosophila sensory neuron as an assay system. In small adults eclosed under a limited-nutrition condition, the wild-type neuron preserved the branching complexity of the arbor, but scaled down the entire arbor, making a “miniature”. In contrast, mutant neurons for the Insulin/IGF signaling (IIS) or TORC1 pathway exhibited “undergrowth”, which was characterized by decreases in both the branching complexity and the arbor size, despite a normal diet. These contrasting phenotypes hinted that a novel regulatory mechanism contributes to the dendritic scaling in wild-type neurons. Indeed, we isolated a mutation in the gene CHORD/morgana that uncoupled the neuron size and the body size: CHORD mutant neurons generated miniature dendritic arbors regardless of the body size. CHORD encodes an evolutionarily conserved co-chaperone of HSP90. Our results support the notion that dendritic growth and branching are controlled by partly separate mechanisms. The IIS/TORC1 pathways control both growth and branching to avert underdevelopment, whereas CHORD together with TORC2 realizes proportional scaling of the entire arbor.
The organ size is controlled through changes in rates of cell growth and division1,2. The underlying mechanisms of organ size control, such as Insulin/IGF signaling (IIS) and Target of Rapamycin complex 1 (TORC1) signaling pathways, have been intensively investigated by using proliferating and morphologically simple cells, such as those in epithelial monolayers of the Drosophila imaginal disc3,4. However, it is still largely unknown how the size of postmitotic and morphologically complex cells such as neurons scale with body size and which molecules control such scaling. We therefore explored these scaling mechanisms in Drosophila neurons.
For our investigation, we used the dendritic arbor of one class of sensory neurons in Drosophila adults as a readout (Figure 1A and 1B)5,6,7,8,9. Dendrites are the antennae of neurons that receive and integrate sensory and/or synaptic inputs10,11. Our model neuron in this study, previously designated as v'ada, is one of the dendritic arborization (da) neurons, whose arbor for adult life is regenerated during pupal stages and entirely covers the lateral plate (pleura) in the abdomen (Figure 1A–1C)7,8.
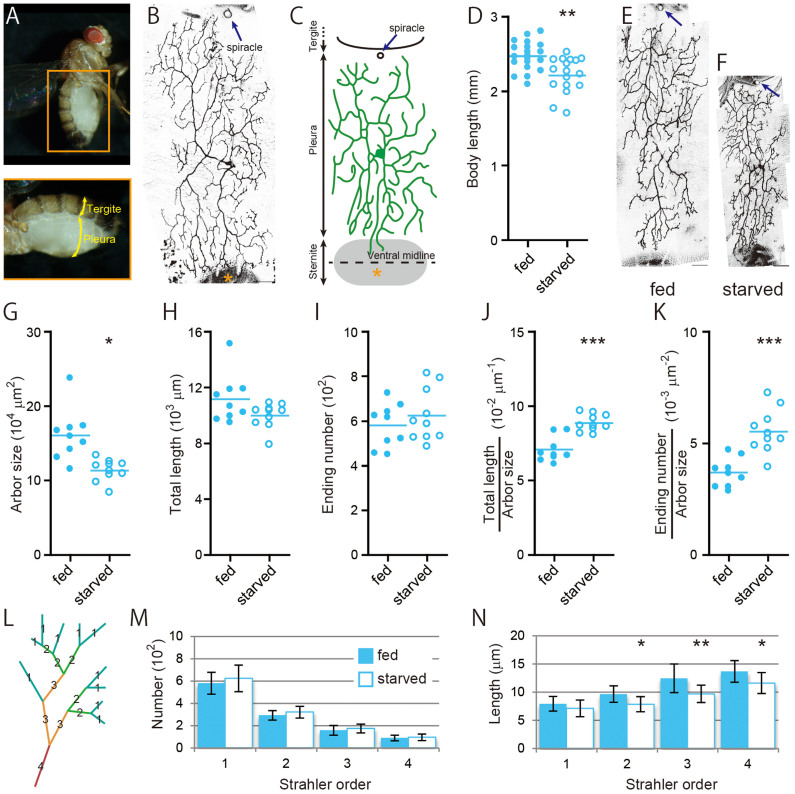
Figure 1. Scaling of dendritic arbors of the da neuron in the wild-type adult.
(A) A lateral view of an adult female (top). Its abdomen is highlighted at the bottom. Courtesy of Naoyuki Fuse. (B and C) A representative image of a MARCM clone of da neuron v'ada (B) and a diagram that illustrates the spatial arrangement of a dendritic arbor in an adult abdomen (C). A spiracle and sternite are indicated by an arrow and an asterisk, respectively. (D) Anterior-posterior body length of adult flies under the fed or starved condition. (E and F) Representative images of MARCM clones of the wild-type neurons under fed (E) and starved (F) conditions. In these and subsequent images of the neurons, posterior is to the right and dorsal is at the top, and dark blue arrows indicate spiracles. Scale bars, 50 μm. (G–N) Quantitative analysis of individual dendritic-arbor patterns. (G) Dendritic arbor size. (H) Total length of dendritic branches. (I) The number of endings of dendritic branches. (J and K) The branch density: total length/arbor size (J) and ending number/arbor size (K). (L) A diagram showing Strahler order. (M and N) The number (M) and segment length (N) of branches of each order. All data are presented as means ± standard deviation (SD). *p < 0.05, **p < 0.01, and ***p < 0.001.
Results
Scaling of dendritic arbors of the wild-type da neuron with body size
To examine how body-size changes affect the size and the branching pattern of the dendritic arbor of the wild-type neuron, we starved larvae beyond 91–96 hr after egg laying (AEL). This is because it has been shown that larvae exposed to such starvation after the “critical weight” stop body growth, but develop to become fertile adults that are smaller than normal, without any developmental delay12,13,14,15. Under this mild starvation condition, the size of the dendritic arbor was significantly decreased in proportion to the decreased body size (Figure 1D–1G). Importantly, the number of branch endings was not changed and the branch density (total length/arbor size and ending number/arbor size) was significantly increased (Figure 1H–1K). We also quantified the branching complexity, by assigning Strahler orders to individual segments (intervals between branching points; see Figure 1L)16,17,18. The numbers of individual order segments were similar under the fed and starved conditions (Figure 1M), while 2nd to 4th order segments became shorter under the starved condition (Figure 1N). These results are consistent with the hypothesis that the wild-type neuron can respond commensurately to the decrease in body size and/or the starvation and can form a “miniature” dendritic arbor by tuning its dendritic segment length.
Neurons with defective IIS/TORC1 signaling pathways show dendritic “undergrowth”
The above result suggested that the wild-type neuron is able to scale with the body size and/or the nutrition condition, while keeping the branching complexity intact. We wondered if the IIS/TORC1 pathways (Supplementary Figure S1A) participate in this scaling, and whether defects in the pathways would affect the size and the branching pattern of the dendritic arbor in adults, when larvae were raised under the normal food condition. Neurons with disruptions of Drosophila insulin receptor (dinr) or Akt downsized and simplified the dendritic arbors, as evidenced by decreases in the arbor size, the ending number, and the total length (Supplementary Figure S1B–S1D and S1H–S1J). In contrast, the branch density (total length/arbor size and terminal number/arbor size) was not significantly altered from that of the wild-type neuron (Supplementary Figure S1K and S1L). A loss of function mutation of tor, overexpression of a dominant negative form of Phosphoinositide 3-kinase (PI3K), or a knockdown of raptor encoding an essential component of TORC1 not only decreased the arbor size, but also the branching complexity (Supplementary Figure S1E–S1G). Thus this “undergrowth” phenotype is distinct from the “miniature” dendrite of the wild-type neuron under the starved condition (Figure 2L), suggesting that a regulatory mechanism other than the IIS/TORC1 pathways may contribute to the dendritic scaling in the normal da neuron.
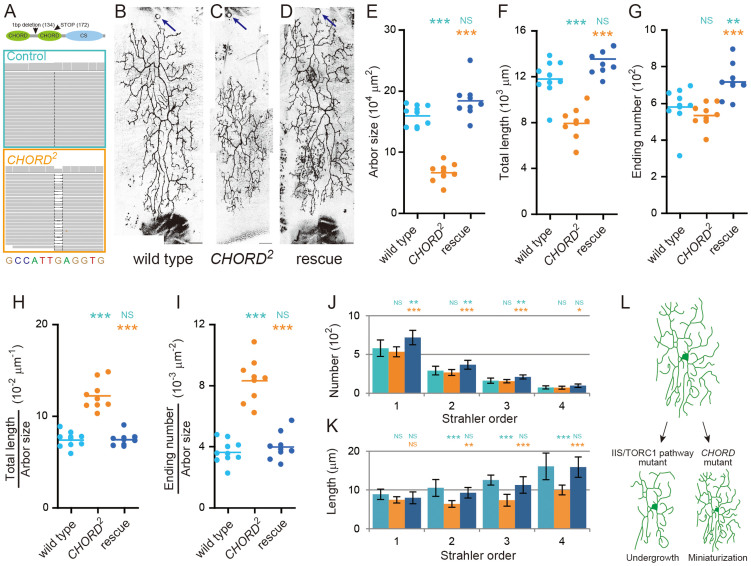
Figure 2. CHORD mutant neurons form “miniature” dendrites.
(A) Whole-genome sequencing identified a 1 bp deletion in the CHORD gene unique to the mutant strain that was isolated from our forward genetic screening (bottom). We denoted this allele as CHORD2 hereafter. Note that we determined the sequences of flies heterozygous for the mutation. (Top) The domain structure of CHORD protein20. The 1 bp deletion caused a frame shift that resulted in a premature stop codon (M172 → STOP) in the 2nd CHORD domain. (B–D) Representative images of MARCM clones of the wild-type neuron (B), the CHORD2 mutant neuron (C) and the CHORD2 mutant neuron to which a genomic fragment including the wild-type CHORD gene was introduced (D). The CHORD2 mutation is early larval lethal, and note that all of the data throughout this study are from single neurons homozygous for individual mutations in otherwise heterozygous animals, using the MARCM system (hence, a mosaic analysis), except for the data from RNAi. Scale bars, 50 μm. (E–K) Quantification of branching patterns of dendritic arbors. (E) Dendritic arbor size. (F) Total length of dendritic branches. (G) The number of endings of dendritic branches. (H and I) The branch density: total length/arbor size (H) and ending number/arbor size (I). (J and K) The number (J) and length (K) of branches of each order. All data are presented as means ± standard deviation (SD). *p < 0.05, **p < 0.01, and ***p < 0.001. Blue asterisks indicate statistically significant differences of the cohort from the wild-type neuron; and orange asterisks indicate statistically significant differences of the cohort from the CHORD mutant neuron. NS: Statistically not significant (P > 0.05). (L) Schematic representation of two partly distinct ways of downsizing dendritic arbors. In neurons with defective IIS/TORC1 signaling pathways showed the final arbor pattern similar to the proximal arbor of the normal neuron (left). In contrast CHORD mutant neurons proportionally downsize the original structure, making a miniature form (right).
CHORD mutant neurons form miniature dendrites
To explore the hypothetical mechanism of the dendritic scaling, we conducted a forward genetic screen by employing the mosaic analysis with a repressible cell marker (MARCM) system19. To facilitate the generation of mosaic clones, we made “SOP-FLP” lines that express FLP recombinase in sensory organ precursors (SOPs; see details in Methods). In our screening under the fed condition, we found a mutant chromosome that generated miniature dendritic patterns in homozygous neurons (Figure 2B and 2C). Indeed a number of quantitative features indicated that the arbor of this mutant neuron was a proportionally scaled down miniature or a microcopy of the wild-type neuron (Figure 2E–2I): decreases in the arbor size and the total dendritic length, an unaltered ending number, an increase in branch density, profiles of the Strahler-order analysis similar to those of the wild-type neuron under the starvation condition (compare Figure 1M–1N with Figure 2J–2K), and an unaltered distribution of angles at individual branching points (data not shown). In a word, the mutant neuron reduced the dendritic segment length (Figure 2K), without simplifying the branching pattern, which is reminiscent of how the wild-type neuron constructed its arbor under the starved condition (Figure 1F).
Whole-genomic sequencing and complementation mapping using Drosophila deficiency stocks identified a 1 bp deletion that leads to a frameshift in the CHORD/morgana gene (denoted CHORD2 hereafter; Figure 2A). CHORD is an evolutionarily conserved co-chaperone of HSP90, and it negatively regulates Rho-kinase (Rok) activity to suppress overduplication of centrosomes20,21. Introduction of the 4757-base-pair (bp) genomic fragment including the CHORD gene almost fully rescued the above phenotypes to normal, demonstrating that CHORD is the causative gene for the miniature dendrite phenotype (Figure 2D and 2E–2K).
The miniature arbor of the CHORD mutant neuron may mimic a default state of the wild-type neuron
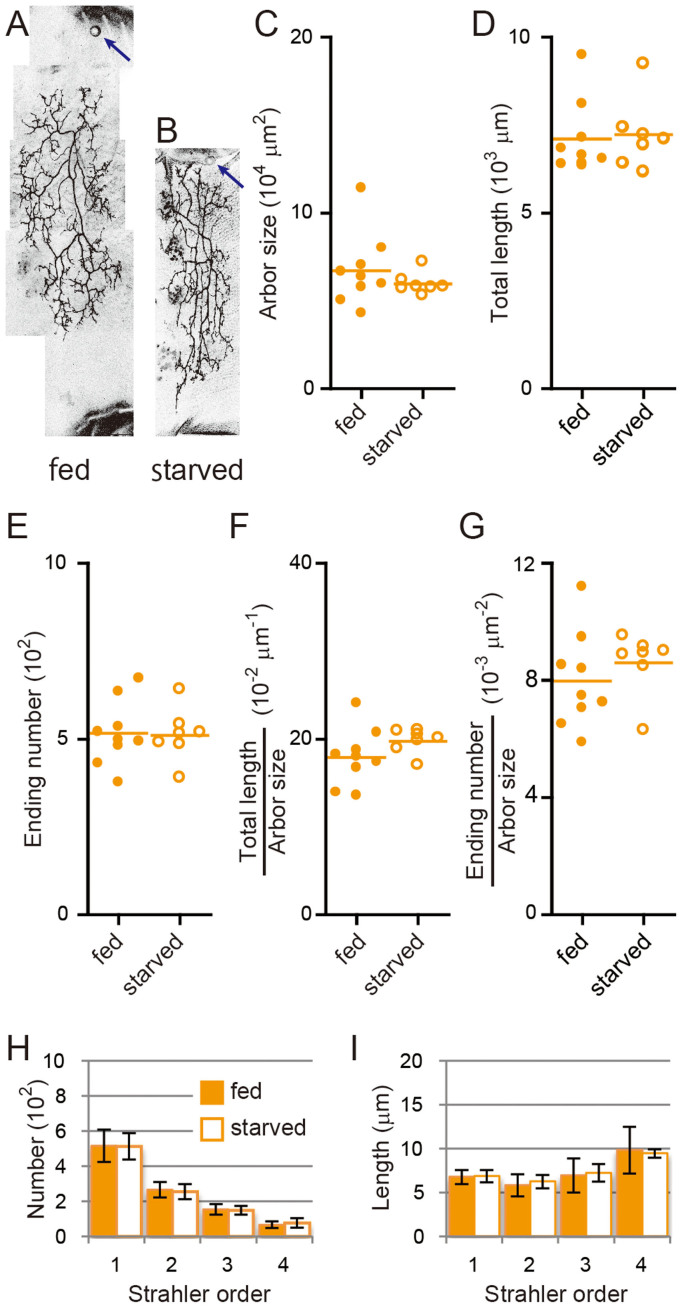
One possible explanation of the CHORD phenotype is that the mutant neuron no longer interprets favorable or unfavorable extracellular conditions (either the nutrition and/or body size) and generates the miniature arbor as an invariable default. Alternatively, the CHORD mutant neuron misreads the size of the abdominal lateral plate; consequently, the arbor occupies only a portion (e.g., 50% instead of 100%) of the body surface. To distinguish these possibilities, we examined dendritic arbors of CHORD mutant neurons under the starved condition (Figure 3A and 3B). Neither the size nor the branching complexity significantly changed between the two nutrition conditions (Figure 3C–3I), showing that the CHORD mutant neuron formed the miniature arbor irrespective of the extracellular condition. This result suggested that the latter possibility was less likely. It appears that the wild-type neuron possesses a CHORD-dependent mechanism that extends the branch segment length beyond a preset value in response to a favorable environment.
Figure 3. CHORD mutant neurons cannot respond to body size changes.
(A and B) Representative images of MARCM clones of CHORD2 mutant neurons under the fed (A) or starved condition (B). Scale bars, 50 μm. (C–I) Quantification of dendritic arbors showed no statistical significance between the two conditions. (C) Dendritic arbor size. (D) Total length of dendritic branches. (E) The number of endings of dendritic branches. (F and G) The branch density: total length/arbor size (F) and ending number/arbor size (G). (H and I) The number (H) and length (I) of branches of each order. All data are presented as means ± standard deviation (SD). *p < 0.05, **p < 0.01, and ***p < 0.001.
Elongation/retraction rate of terminal branches is critical for reproducing the miniature phenotype of the CHORD mutant neuron
We next investigated the dynamics of dendrite formation whereby the CHORD mutant neuron produced the miniature arbor during pupal stages. First we found that the mutant arbor underwent a persistent increase in complexity and size, but the overall appearance had already become miniature-like before eclosion (Supplementary Figure S2A–S2H). Then we performed time-lapse recordings of the wild-type and mutant neurons at 70–75 hr after puparium formation (APF), when dendritic arbors were under active construction, and dissected the dendrite dynamics of elongation/retraction and branching.
We quantified frequencies of elongations and retractions of terminal branches (branches of Strahler order 1, see Figure 1L) and found that these parameters were not altered in CHORD mutant neurons (Supplementary Figure S2I and S2J). We also used a Fano Factor as a quantitative descriptor of branch tip dynamics (see details in Methods)22 and observed that there was no significant difference between the wild-type and CHORD mutant neurons (Supplementary Figure S2K). In contrast, the rates of branch–length changes (μm/min) were significantly decreased in the mutant neurons (Supplementary Figure S2L). In this sensory neuron, most of the branches are collaterals that sprout from stalks, rather than bifurcations of branch tips9. Therefore, we quantified the frequency of lateral branching and found that wild-type and CHORD mutant neurons showed similar values (Supplementary Figure S2M). Altogether, dendritic dynamics of CHORD mutant neurons are very similar to that of wild-type neurons, with the exception that the rates of change of branch lengths decrease in the mutant neurons.
We then asked whether these changes can explain the miniature phenotype of the mutant neurons or not. We addressed this question with a computer-assisted simulation of dendrite growth using experimental values. The elongation and retraction of the terminals was represented as the addition and removal, respectively, of one unit segment per 1 minute ( = 1 step in simulation) at the terminal of the dendrites, and lateral branching was reproduced by the addition of unit segments to the existing branches23,24,25. The probabilities of elongation, retraction, and lateral branching were determined by experimental data (see details in Methods and Supplementary Table S3). We defined the unit length of a segment as 0.9 μm or 0.75 μm, based on the experimental data for the wild-type or CHORD mutant neurons, respectively (Supplementary Figure S2L). Representative images of the wild-type and mutant model neurons closely resembled those of in vivo neurons (Supplementary Figure S2N). In fact, our quantification showed decreases in the arbor size and the total length, an unaltered ending number, and an increase in the branch density of the mutant model arbors, compared to the wild-type model arbors (Supplementary Figure S2O–S2S). Thus, these results support the hypothesis that subtle but significant differences in the rates of branch-length changes play a critical role in scaling the dendritic arbor, and that the miniature arbor of the CHORD mutant neuron can be attributed to the scaled-down elongation/retraction dynamics of dendritic branches.
Finally, we examined whether the dendrite dynamics in starved wild-type animals is comparable to that of CHORD mutant neurons. We found that the wild-type neurons under the starved condition showed significantly decreased rates of branch–length changes compared to those under the fed condition, while other parameters were not significantly changed (Supplementary Figure S2I–S2M), very much resembling CHORD mutant neurons under the fed condition. These results strengthened our hypothesis that coordinated regulation of the elongation/retraction dynamics of branches contributes to scaling of the dendritic arbor.
Genetic interaction between CHORD and the TORC2 component Rictor
We next asked how body size/nutrition state is conveyed to CHORD. As a candidate of such an upstream molecule, we examined the involvement of TORC2, another TOR complex, for the following reasons: (1) TORC2 is activated by association with ribosomes in a growth factor dependent manner, thus it may mediate between the extracellular environment and growth capacity of individual cells26,27,28. (2) TORC2 regulates the actin cytoskeleton and controls dendrite pattern formation27,28,29,30.
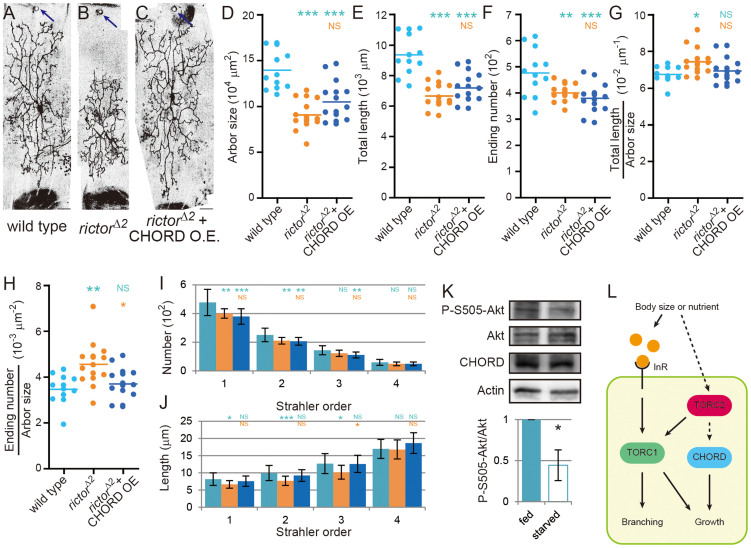
We assessed the phenotype of mutant neurons of rictor, which encodes an essential and specific component of TORC2 (Figure 4A and 4B). rictor mutant neurons showed complicated but intriguing phenotypes; that is, combined features of undergrowth and miniaturization. They showed a decrease in the arbor size, the total length, and the ending number (Figure 4D–F), which is characteristic of the undergrowth phenotype. On the other hand, they also exhibited features of the miniature phenotype: increases in the branch density (both the total length/arbor size and the ending number/arbor size, as shown in Figure 4G and 4H) and a decrease in segment length (Figure 4J). Importantly, overexpression of the CHORD transgene in rictor mutant neurons partially restored the features of the miniature phenotype (Figure 4G–4J), but not those of the undergrowth phenotype (Figure 4D–4F). In contrast to this genetic interaction between CHORD and TORC2 (Rictor), CHORD overexpression did not rescue simplified phenotype of the dinr knockdown (Supplementary Figure S1M and S1N). All these results could be explained by the hypothesis that CHORD functions downstream of TORC2, being at least partly separate from TORC1. The partially separate nature was further supported by our KD experiment using S2 cells (Supplementary Figure S1O). Even when CHORD was knocked down, phosphorylation of S6K (a readout of TORC1 activity) was up-regulated in an insulin-dependent manner, as in the control knockdown cells.
Figure 4. Phenotype of neurons defective in TORC2, and a genetic interaction between CHORD and TORC2.
(A–C) Representative images of MARCM clones of the wild-type neuron (A), the rictor2 mutant neuron (B), and the rictor2 mutant neuron in which a CHORD transgene was overexpressed (C). Scale bars, 50 μm. (D–J) Quantification of branching patterns of dendritic arbors. (D) Dendritic arbor size. (E) Total length of dendritic branches. (F) The number of endings of dendritic branches. (G and H) The branch density: total length/arbor size (G) and ending number/arbor size (H). (I and J) The number (I) and length (J) of branches of each order. (K) Western blot analysis of extracts of larvae under the fed or starved condition with the indicated antibodies (top). Larvae of 91–97 hr AEL were collected, placed under either the fed or starved condition for 8 hrs, and then homogenized. Levels of phospho-S505 Akt were normalized to the total Akt levels in three independent preparations and the ratios were plotted (bottom). Uncropped images of the blots are shown in Supplementary Figure S4. All data are presented as means ± standard deviation (SD). *p < 0.05, **p < 0.01, and ***p < 0.001. Blue asterisks indicate statistically significant differences of the cohort from the wild-type neuron; and orange asterisks indicate statistically significant differences of the cohort from the rictor mutant neuron. NS: Statistically not significant (P > 0.05). (L) Hypothetical molecular complexes that regulate dendrite growth and branching in response to the extracellular environment (the body size and/or the nutrition condition). The environmental cue(s) controls CHORD downstream of TORC2, which tunes the segment length (growth); on the other hand, the cue(s) also regulates the IIS/TORC1 pathway and affects both growth and branching.
We then asked whether the TORC2 activity is altered under the nutrient-limited condition that was employed to decrease body size. We starved larvae for 8 hrs beyond 91–96 hr AEL and analyzed the phosphorylation of Akt at Ser505 as an established readout of the TORC2 activity31. The level of S505 phosphorylation was significantly and reproducibly reduced under this condition compared to that in well-fed flies (Figure 4K). All these results suggest the possibility that TORC2 may communicate the extracellular conditions (body size and/or nutrients) to CHORD proteins (Figure 4L).
Discussion
This study provides novel mechanistic insights into size control of neuronal dendritic arbors. To sample sensory input precisely, certain types of neurons should adjust their dendritic arbor size to cover the receptive field completely. We showed that there are two distinct ways of downsizing dendritic arbors when the field size is reduced: one way is to arrest both growth and branching during maturation of dendrite morphologies, causing the “undergrowth” phenotype. The other way is to regulate the elongation of dendrites (more specifically speaking, the length of branch segments) selectively, thereby making a “miniature” form (Figure 2L). Our results support the notion that dendritic growth and branching are controlled by at least partly separate mechanisms, which is also seen in other instances such as the development of postembryonic dendritic architecture of motorneurons32. At the molecular level, the IIS/TORC1 pathways control both growth and branching to avert underdevelopment, whereas CHORD and TORC2 tunes the segment length to realize proportional scaling of the entire arbor (Figure 4L). Ablation of the TORC2 component Rictor in mouse Purkinje cells causes multiple structural changes of dendritic arbors, including a decrease in total dendrite length and an increase in the number of primary branches30. It will require further investigation to address how the overall branching complexity and the segment length is affected in the absence of Rictor, and to fully characterize the loss of function phenotype of CHORD in this subtype of neurons.
CHORD was originally discovered in plants as a key player in the innate immune response33,34; in animals, CHORD negatively regulates Rho kinase activity, thereby suppressing overduplication of centrosomes20,21. CHORD is also expressed in tissues that are populated by postmitotic cells, such as brains35, and has been proposed to function beyond regulating cell division. Here, we revealed that CHORD regulates the size of dendritic arbors: cells defective in CHORD showed a decreased elongation/retraction rate of terminal branches; thus, by extrapolation, the normal function of CHORD must be to accelerate the elongation and retraction of terminal branches. Rho-kinase (Rok) is reported to inhibit neurite outgrowth, dendritic branching, and spine formation36. Therefore, we pursued the possibility that Rok acts downstream of CHORD. However, we couldn't find any genetic and biochemical evidence for the interaction between Rok and CHORD (Supplementary Figure S3; see details in the legend). Instead, our results suggested that CHORD is functionally related to the TORC2 signaling pathway, which regulates the actin cytoskeleton28. It has been reported in mammalian fibroblasts that phosphorylation by TORC2 facilitates folding of some AGC kinases, such as Akt and conventional PKCs, and stabilizes them, and that newly synthesized, unphosphorylated Akt is protected by HSP90 from degradation37,38. Therefore, CHORD may contribute as a co-chaperone to the stability of the AGC kinases by recruiting HSP90 to those clients, or it may employ its own chaperone activity39 to perform this task. This hypothetical role of CHORD might underlie the partial rescue of the rictor mutant neuron by CHORD overexpression.
Comparative anatomical studies have reported that dendrites of some types of neurons, such as sympathetic neurons, become larger and increase branching complexity as the brain or the body size increases across species40,41, which we designate overgrowth as opposed to undergrowth. In contrast, other types of neurons such as somatosensory thalamocortical projection neurons increase their dendritic arbor size with larger brains, while preserving key features of the dendritic branching pattern42,43, which is proportional magnification. These two distinct ways of scaling, overgrowth/undergrowth and miniaturization/magnification, might be regulated by IIS/TORC1 and TORC2/CHORD, respectively, and may contribute to neuronal cell-type specific information processing.
It should be noted that “scaling growth” of dendritic arbors does take place during larval development, but mechanistically it is distinct from the arbor scaling at postlarval stages, which we reported in this study. Larval da neurons regulate their growth in coordination with the expanding body whose mass increases by approximately 200-fold during the complete larval development18,44. This is accomplished by increasing both branching numbers and the total length, where IIS/TORC1 plays a critical role29,45, but CHORD appears to be dispensable (Supplementary Figure S2), and TORC2 is required for dendritic tiling for larval da neurons29. On the other hand, the da neuron during pupal development first prunes its dendrites and then starts constructing arbors for adult life; thus this neuron completes this task in a body whose volume has been predetermined by the nutritional status during larval development. It is this task that requires the role of CHORD, which fits the dendritic arbor to the final adult body size.
We speculate that this hypothetical CHORD-dependent mechanism somehow senses body size by receiving as-yet undetermined signals, either local or systemic, from other tissues. Candidate tissues for sources of such signals would be either those adjacent to the neuron, such as lateral tergosternal muscles, abdominal histoblasts (epitherial cells), and glial cells, and/or tissues that secrete growth factors, such as fat body and insulin producing cells8,45,46,47,48. Therefore the dendrite scaling of the adult da neuron might provide a useful model system to study interactions between neurons and other organs.
Methods
Molecular cloning
Six tandem repeats of a 20-base-pair (bp) sequence that includes a proneural binding site ([scE1]6)49 were used to build a SOP-FLP transgene. The hsp70 minimal promoter and the entire coding sequence of flp from the UAS-FLP vector (DGRC) were inserted between [scE1]6 and a SV40-polyA sequence. This SOP-FLP transgene was cloned into the pHStinger vector, or a pUAST vector from which the UAS sequence and hsp70 minimal promoter had been removed. To generate a rescue construct, the 4754-bp sequence (3R: 20009616-20013777 in version FB2013_05) that includes CHORD was amplified from yw genomic DNA and cloned into the vector pCasper. To generate UAS shRNA for each gene, we followed a protocol previously described50. The target sequences of the shRNAs are as follows: 5′-CACCGAGTTCCTCAACATCAA-3′ and 5′-TTCGACCTGGATGACATTAAA-3′. These shRNAs were cloned into the vector pUAST-attB. All constructs were injected, in accordance with standard protocols, to generate transgenic fly lines.
Drosophila strains
Detailed genotypes of the animals and clones are described and summarized in Supplementary Table S1. To visualize dendrites and /or express transgenes, we used the following Gal4 drivers: Gr28b.c51,52 and Gal45-4053. To express fluorescent proteins, we used UAS-mCD8:GFP (#5137 of the Bloomington Stock Center) or UAS-Venus-pm44,54,55. Other strains used were dinr33956, Aktq57, TorΔP58, rok259, UAS-dicer2 (#60009 of Vienna Drosophila RNAi Center), UAS-raptorRNAi (HMS00124/#34814 of the Bloomington Stock Center), UAS-mCherryRNAi (#35785 of the Bloomington Stock Center), UAS-CHORDRNAi (this study), UAS-Dp110[D954A] (#25918 of the Bloomington Stock Center), UAS-rokRNAi (GD1522/#3793 and KK107802/#104675 of the Vienna Drosophila RNAi Center; JF03225/#28797, HMS01311/#34324, and GL00209/#35305 of the Bloomington Stock Center), UAS-hsp90RNAi (HMS00899/#33947 of the Bloomington Stock Center) and UAS-Rok.CAT48.2 60.
MARCM-based forward genetic screen
The piggyBac insertion collection with FRT insertion61 was used for genetic screening. We crossed individual insertion stocks to “SOP-FLP based MARCM-ready” fly stocks (for 2nd-chromosome left arm screen, Gal45-40 UAS-Venus:pm SOP-FLP#42; tubP-Gal80 FRT40A; for 3rd-chromosome left arm screen, SOP-FLP#42; Gal4109(2)80 UAS-mCD8:GFP SOP-FLP#73/CyO; tubP-Gal80 FRT2A; and for 3rd-chromosome right arm screen, hsFLP UAS-mCD8:GFP; Gal4109(2)80 UAS-mCD8:GFP SOP-FLP#73/CyO; FRT82B tubP-Gal80). We then mounted adult abdomens, and dendrites were imaged under a laser scanning microscope. We screened 1537 piggyBac stocks and isolated 3 stocks (LL04611, LL04133, and LL03277) that showed the “miniature” phenotype, and 19 that showed the “undergrowth” phenotype (Supplementary Table S2). Both LL04611 and LL04133 were homozygous lethal.
Mapping and identification of the mutation that was responsible for the “miniature” phenotype
Starting from the above LL04611 and LL04133 stocks of the piggyBac collection, we generated precise excision strains. These precise excision strains were still homozygous lethal, and MARCM clones of those strains showed the miniature dendritic phenotypes. We performed a complementation test for lethality between LL04611 and LL04133, and found that they belong to the same complementation group. These results suggested that the lethality and the “miniature” dendrite phenotype were caused by a background mutation(s) that was not linked to the piggyBac insertion. We backcrossed the excision strains to the control FRT82B stock three times and used the progeny for the phenotypic analysis below.
To identify the mutated gene that gave rise to the phenotype, we determined whole-genomic sequences of the two mutant strains with an Illumina next-generation sequencer and directly compared them with each other. For the sequencing, we prepared genomic DNA from larvae that were heterozygous for LL04611 or LL04133 and employed paired-end Illumina sequencing technology. We also determined the sequences of LL02779 and LL00232, whose MARCM clones did not show a miniature phenotype, as negative controls. Then we mapped the sequence data to the wild-type reference genome and identified variants relative to the wild-type reference genome by using Burrows-Wheeler Aligner (BWA) and SAMtools. In addition to the whole genome sequencing, we performed conventional deficiency mapping of the lethality by using the Bloomington Deficiency Kit, and mapped the mutation responsible for the lethality (and possibly the clone phenotype) within a ~45 kb region in the right arm of the 3rd chromosome. Within this ~45 kbp mapped region, we found only one mutation unique to LL04611 and LL04133: a 1 bp deletion in the CHORD/morgana gene that caused a frame shift (denoted as CHORD2), which was confirmed by the Sanger method.
Image collection and image analysis
Imaging da neurons in whole-mount adults was done as described earlier7. Briefly, we collected adult females within 12-hour after eclosion unless described otherwise, washed them in 0.7% NaCl and 0.3% Triton X-100, removed the heads and legs of adult flies, and mounted the abdomens in 50% glycerol on slides, between spacers made of vinyl tape. All of the images were acquired using a Nikon C1 laser-scanning confocal microscope or a Zeiss LSM 510 META laser-scanning confocal microscope.
For quantification of dendritic patterns of da neurons, live imaged dendrites were first traced using Simple Neurite Tracer plugin in Fiji and tracing data was exported as csv files. Quantification of total length, terminal number, total length/arbor size, and terminal number/arbor size and analysis of Strahler order were performed using Excel. Arbor size was quantified by analyzing skeletonized tracing images by using the convex-hull selection in Fiji. The data of different genotypes were compared by one-way ANOVA with Tukey's HSD post hoc analysis, or by Student's t-test using KaleidaGraph (version 4.0; Synergy Software).
Time-lapse recording
Time-lapse recordings of branch formation (Supplementary Figure S2I–S2M and Supplementary Video S1–S3) were performed as described previously9. Briefly, each pupa between 70–74 hour after puparium formation (APF) was taken out of its puparium and mounted on a 35-mm glass-bottomed dish. In mounting, we folded legs and put abdomens on the dish and tilted them to retain an appropriate angle. All images were collected at 2-minute intervals for 1-hour with a 2-μm Z-step using a Nikon C1 laser-scanning confocal microscope. After image acquisition, each pupa was kept at 25°C, and their survival was confirmed to at least the pharate stage or the adult stage. Acquired movies were traced manually using Simple Neurite Tracer plugin in Fiji and the frequency and rate of elongation/retraction were quantified using Excel. As an unbiased indicator of branch dynamics, a Fano factor (FF) value was calculated based on branch tip length measurements. The FF is defined as the variance in the measured length of an individual branch across all imaging sessions, divided by the mean length of that branch22. We quantified neurons whose FF was above 0.35, which is defined as dynamic neurons22.
Reconstruction of dendrite dynamics in a computer-aided simulation
Dendrite growth dynamics was modeled in two-dimensional space following previous mathematical models23,24,25. The ‘elongation’ and ‘retraction’ were represented by the addition and removal of 1 unit segment, respectively. As a length of 1 unit segment, we employed the 0.9 μm and 0.75 μm to recapitulate the wild-type and CHORD mutant neurons, respectively. Lateral branching was reproduced by the addition of unit segments to the existed branches. The angle of elongation was assumed to follow a homogenous distribution from −7.5° to +7.5°, while that of branching was assumed to follow a normal distribution with average 97° and standard deviation 38°. The initial condition was as follows: 2 branches with 20-segment lengths sprout from the origin in anti-parallel directions along the y axis and bifurcate with angle 30°, and each of these 4 branches elongate 10-segments in length. Other parameters used in the simulation are summarized in Supplementary Table S3.
Production of antibodies, RNAi, and Western blotting
Guinea pigs were immunized with GST proteins that had been fused to the N-terminal 200 amino acids of CHORD to generate anti-CHORD antibody. For RNAi experiments, S2 cells were cultured with 20 μg/ml dsRNA of the full-length CHORD or GFP-coding sequence for 3 days. Lysates were separated on a 8% polyacrylamide gel and transferred to PVDF (Millipore). Antibodies were used at the following concentrations: guinea pig anti-CHORD, 1:1000 (this study); rabbit anti-phospho-Myosin light chain 2 (Ser-19), 1:1000 (Cell Signaling #3671); rabbit anti-phospho-Akt (Ser-505), 1:1000 (Cell Signaling #4054); rabbit anti-Akt, 1:1000 (Cell Signaling #9272); mouse anti-actin, 1:1000 (Millipore MAB1501). Signals were detected with ECL plus (Amersham). To obtain stronger signals, we employed Can Get Signal Immunoreaction Enhancer solution (TOYOBO).
Author Contributions
K.S. carried out most of the experiments; K.F., T.N., M.O., T. Usui and M.K. helped some experiments. K.S. and T. Uemura wrote the paper.
Supplementary Material
Recording of dendritic dynamics of the wild-type neuron under the fed condition
Recording of dendritic dynamics of the CHORD mutant neuron
Recording of dendritic dynamics of the wild-type neuron under the starved condition
Supplementary Figures and Tables
Acknowledgments
The reagents were provided by the Drosophila Genetic Resource Center at Kyoto Institute of Technology, the Bloomington Stock Center, Y.N. Jan, B. Yi, T. Chihara, K. Emoto, L.M. Powell, B. Lemaitre, S. Cohen, R. Niwa, T. Nishimura, R. Karess, N. Fuse, S. Hayashi, and J. Zallen. We thank O. Nishimura, N. Fuse, and R. Ueda for help with next-generation sequencing analysis, S. Yonehara for use of the DNA sequencer, N. Yamamoto for discussion about the neuron size, J.A. Hejna for his efforts to polish the manuscript, and M. Futamata, Y. Miyake, K. Shimizu, J. Mizukoshi, and K. Oki for their technical assistance. We also thank the members of Comparative Genomics Laboratory in National Institute of Genetics for technical and computational assistance. This study was supported by a Grant-in-Aid for Scientific Research (A) and for Scientific Research on Innovative Areas “Mesoscopic neurocircuitry” (22115006) to T. Uemura, by the Funding Program for Next Generation World-Leading Researchers (NEXT Program) to M. K., by Research and Education Platform on Dynamic Living Systems, by NIG Collaborative Research Program (2011-A48), and by a Grant-in-aid for Scientific Research on Innovative Areas ‘Genome Science’ and ‘Comprehensive Brain Science Network’ from the Ministry of Education, Science, Sports and Culture of Japan. K.S. was a recipient of a Fellowship of the Japan Society for the Promotion of Science for Young Scientists.
References
- Gomer R. H. Not being the wrong size. Nat Rev Mol Cell Biol 2, 48–54, 10.1038/35048058 (2001). [DOI] [PubMed] [Google Scholar]
- Neufeld T. P. Body building: regulation of shape and size by PI3K/TOR signaling during development. Mech Dev 120, 1283–1296 (2003). [DOI] [PubMed] [Google Scholar]
- Stocker H. & Hafen E. Genetic control of cell size. Curr Opin Genet Dev 10, 529–535 (2000). [DOI] [PubMed] [Google Scholar]
- Hietakangas V. & Cohen S. M. Regulation of tissue growth through nutrient sensing. Annu Rev Genet 43, 389–410, 10.1146/annurev-genet-102108-134815 (2009). [DOI] [PubMed] [Google Scholar]
- Williams D. W. & Truman J. W. Remodeling dendrites during insect metamorphosis. J Neurobiol 64, 24–33, 10.1002/neu.20151 (2005). [DOI] [PubMed] [Google Scholar]
- Kuo C. T., Jan L. Y. & Jan Y. N. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci U S A 102, 15230–15235, 10.1073/pnas.0507393102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono K. et al. Multidendritic sensory neurons in the adult Drosophila abdomen: origins, dendritic morphology, and segment- and age-dependent programmed cell death. Neural Dev 4, 37, 10.1186/1749-8104-4-37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga K., Kanamori T., Morikawa R., Suzuki E. & Emoto K. Dendrite reshaping of adult Drosophila sensory neurons requires matrix metalloproteinase-mediated modification of the basement membranes. Dev Cell 18, 621–632, 10.1016/j.devcel.2010.02.010 (2010). [DOI] [PubMed] [Google Scholar]
- Satoh D., Suyama R., Kimura K. & Uemura T. High-resolution in vivo imaging of regenerating dendrites of Drosophila sensory neurons during metamorphosis: local filopodial degeneration and heterotypic dendrite-dendrite contacts. Genes Cells 17, 939–951, 10.1111/gtc.12008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan Y. N. & Jan L. Y. Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci 11, 316–328, 10.1038/nrn2836 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- London M. & Häusser M. Dendritic computation. Annu Rev Neurosci 28, 503–532, 10.1146/annurev.neuro.28.061604.135703 (2005). [DOI] [PubMed] [Google Scholar]
- Beadle G. W., Tatum E. L. & Clancy C. W. Food level in relation to rate of development and eye pigmentation in Drosophila melanogaster. Biological Bulletin 75, 447–462 (1938). [Google Scholar]
- Edgar B. A. How flies get their size: genetics meets physiology. Nat Rev Genet 7, 907–916, 10.1038/nrg1989 (2006). [DOI] [PubMed] [Google Scholar]
- Mirth C. K. & Riddiford L. M. Size assessment and growth control: how adult size is determined in insects. Bioessays 29, 344–355, 10.1002/bies.20552 (2007). [DOI] [PubMed] [Google Scholar]
- Tennessen J. M. & Thummel C. S. Coordinating growth and maturation - insights from Drosophila. Curr Biol 21, R750–757, 10.1016/j.cub.2011.06.033 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahler A. N. Revisions of Horton's quantitative factors in erosional terrain. Transactions of the American Geophysical Union 34, 345 (1953). [Google Scholar]
- Uylings H. B., Smit G. J. & Veltman W. A. Ordering methods in quantitative analysis of branching structures of dendritic trees. Adv Neurol 12, 347–354 (1975). [PubMed] [Google Scholar]
- Grueber W. B., Jan L. Y. & Jan Y. N. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development 129, 2867–2878 (2002). [DOI] [PubMed] [Google Scholar]
- Lee T. & Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461 (1999). [DOI] [PubMed] [Google Scholar]
- Ferretti R. et al. Morgana/chp-1, a ROCK inhibitor involved in centrosome duplication and tumorigenesis. Dev Cell 18, 486–495, 10.1016/j.devcel.2009.12.020 (2010). [DOI] [PubMed] [Google Scholar]
- Ferretti R. et al. Morgana and melusin: two fairies chaperoning signal transduction. Cell Cycle 10, 3678–3683, 10.4161/cc.10.21.18202 (2011). [DOI] [PubMed] [Google Scholar]
- Lee W. C. et al. A dynamic zone defines interneuron remodeling in the adult neocortex. Proc Natl Acad Sci U S A 105, 19968–19973, 10.1073/pnas.0810149105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono K., Sugimura K., Kengaku M., Uemura T. & Mochizuki A. Computational modeling of dendritic tiling by diffusible extracellular suppressor. Genes Cells 15, 137–149, 10.1111/j.1365-2443.2009.01367.x (2010). [DOI] [PubMed] [Google Scholar]
- Fujishima K., Horie R., Mochizuki A. & Kengaku M. Principles of branch dynamics governing shape characteristics of cerebellar Purkinje cell dendrites. Development 139, 3442–3455, 10.1242/dev.081315 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pelt J., Graham B. & Uylings H. in Modeling Neural Development (ed van Ooyen, A.) Ch. 4, 75–94 (The MIT Press, 2003). [Google Scholar]
- Zinzalla V., Stracka D., Oppliger W. & Hall M. N. Activation of mTORC2 by association with the ribosome. Cell 144, 757–768, 10.1016/j.cell.2011.02.014 (2011). [DOI] [PubMed] [Google Scholar]
- Laplante M. & Sabatini D. M. mTOR signaling in growth control and disease. Cell 149, 274–293, 10.1016/j.cell.2012.03.017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh W. J. & Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle 10, 2305–2316 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Kumagai M., Yasunaga K., Morikawa R., Kanamori T. & Emoto K. The target of rapamycin complex 2 controls dendritic tiling of Drosophila sensory neurons through the Tricornered kinase signalling pathway. EMBO J 28, 3879–3892, 10.1038/emboj.2009.312 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomanetz V. et al. Ablation of the mTORC2 component rictor in brain or Purkinje cells affects size and neuron morphology. J Cell Biol 201, 293–308, 10.1083/jcb.201205030 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V. & Cohen S. M. Re-evaluating AKT regulation: role of TOR complex 2 in tissue growth. Genes Dev 21, 632–637, 10.1101/gad.416307 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duch C., Vonhoff F. & Ryglewski S. Dendrite elongation and dendritic branching are affected separately by different forms of intrinsic motoneuron excitability. J Neurophysiol 100, 2525–2536, 10.1152/jn.90758.2008 (2008). [DOI] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K. & Guerois R. NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem Sci 35, 199–207, 10.1016/j.tibs.2009.12.005 (2010). [DOI] [PubMed] [Google Scholar]
- Shirasu K. et al. A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99, 355–366 (1999). [DOI] [PubMed] [Google Scholar]
- Brancaccio M. et al. Chp-1 and melusin, two CHORD containing proteins in vertebrates. FEBS Lett 551, 47–52 (2003). [DOI] [PubMed] [Google Scholar]
- Amano M., Nakayama M. & Kaibuchi K. Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell polarity. Cytoskeleton (Hoboken) 67, 545–554, 10.1002/cm.20472 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchinetti V. et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J 27, 1932–1943, 10.1038/emboj.2008.120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenoue T., Inoki K., Yang Q., Zhou X. & Guan K. L. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J 27, 1919–1931, 10.1038/emboj.2008.119 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michowski W. et al. Morgana/CHP-1 is a novel chaperone able to protect cells from stress. Biochim Biophys Acta 1803, 1043–1049, 10.1016/j.bbamcr.2010.05.005 (2010). [DOI] [PubMed] [Google Scholar]
- Defelipe J. The evolution of the brain, the human nature of cortical circuits, and intellectual creativity. Front Neuroanat 5, 29, 10.3389/fnana.2011.00029 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg G. & Wang S. in Dendrites (eds Greg Stuart, Nelson Spruston & Michael Häusser) Ch. 2, 43–67 (Oxford University Press, 2007). [Google Scholar]
- Ohara P. T. & Havton L. A. Preserved features of thalamocortical projection neuron dendritic architecture in the somatosensory thalamus of the rat, cat and macaque. Brain Res 648, 259–264 (1994). [DOI] [PubMed] [Google Scholar]
- Teeter C. M. & Stevens C. F. A general principle of neural arbor branch density. Curr Biol 21, 2105–2108, 10.1016/j.cub.2011.11.013 (2011). [DOI] [PubMed] [Google Scholar]
- Sugimura K. et al. Distinct developmental modes and lesion-induced reactions of dendrites of two classes of Drosophila sensory neurons. J Neurosci 23, 3752–3760 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish J. Z., Xu P., Kim C. C., Jan L. Y. & Jan Y. N. The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in drosophila sensory neurons. Neuron 63, 788–802, 10.1016/j.neuron.2009.08.006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A. & Perrimon N. Drosophila as a model for interorgan communication: lessons from studies on energy homeostasis. Dev Cell 21, 29–31, 10.1016/j.devcel.2011.06.034 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth C. K. & Shingleton A. W. Integrating body and organ size in Drosophila: recent advances and outstanding problems. Front Endocrinol (Lausanne) 3, 49, 10.3389/fendo.2012.00049 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A. C. The regulation of cell size. Cell 154, 1194–1205, 10.1016/j.cell.2013.08.053 (2013). [DOI] [PubMed] [Google Scholar]
- Powell L. M., Zur Lage P. I., Prentice D. R., Senthinathan B. & Jarman A. P. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol Cell Biol 24, 9517–9526, 10.1128/MCB.24.21.9517-9526.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Q. et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8, 405–407, 10.1038/nmeth.1592 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne N. & Amrein H. Atypical expression of Drosophila gustatory receptor genes in sensory and central neurons. J Comp Neurol 506, 548–568, 10.1002/cne.21547 (2008). [DOI] [PubMed] [Google Scholar]
- Xiang Y. et al. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature 468, 921–926, 10.1038/nature09576 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Onishi M., Jan L. Y. & Jan Y. N. Peripheral multidendritic sensory neurons are necessary for rhythmic locomotion behavior in Drosophila larvae. Proc Natl Acad Sci U S A 104, 5199–5204, 10.1073/pnas.0700895104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato D., Sugimura K., Satoh D. & Uemura T. Crossveinless-c, the Drosophila homolog of tumor suppressor DLC1, regulates directional elongation of dendritic branches via down-regulating Rho1 activity. Genes Cells 15, 485–500, 10.1111/j.1365-2443.2010.01399.x (2010). [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Ueda R., Takahashi K., Saigo K. & Uemura T. Control of axonal sprouting and dendrite branching by the Nrg-Ank complex at the neuron-glia interface. Curr Biol 16, 1678–1683, 10.1016/j.cub.2006.06.061 (2006). [DOI] [PubMed] [Google Scholar]
- Brogiolo W. et al. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol 11, 213–221 (2001). [DOI] [PubMed] [Google Scholar]
- Staveley B. E. et al. Genetic analysis of protein kinase B (AKT) in Drosophila. Curr Biol 8, 599–602 (1998). [DOI] [PubMed] [Google Scholar]
- Zhang H., Stallock J. P., Ng J. C., Reinhard C. & Neufeld T. P. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev 14, 2712–2724 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C. G. et al. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell 105, 81–91 (2001). [DOI] [PubMed] [Google Scholar]
- Simões S. e. M. et al. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev Cell 19, 377–388, 10.1016/j.devcel.2010.08.011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O. et al. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell 14, 227–238, 10.1016/j.devcel.2007.11.001 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Recording of dendritic dynamics of the wild-type neuron under the fed condition
Recording of dendritic dynamics of the CHORD mutant neuron
Recording of dendritic dynamics of the wild-type neuron under the starved condition
Supplementary Figures and Tables