Abstract
The fruitless gene (fru) encodes a set of transcription factors (Fru) that display sexually dimorphic gene expression in the brain of the fruit-fly; Drosophila melanogaster. Behavioural studies have demonstrated that fru is essential for courtship behaviour in the male fly and is thought to act by directing the development of sex-specific neural circuitry that encodes this innate behavioural response. This study reports the identification of direct regulatory targets of the sexually dimorphic isoforms of the Fru protein using an in vitro model system. Genome wide binding sites were identified for each of the isoforms using Chromatin Immunoprecipitation coupled to deep sequencing (ChIP-Seq). Putative target genes were found to be involved in processes such as neurotransmission, ion-channel signalling and neuron development. All isoforms showed a significant bias towards genes located on the X-chromosome, which may reflect a specific role for Fru in regulating x-linked genes. Taken together with expression analysis carried out in Fru positive neurons specifically isolated from the male fly brain, it appears that the Fru protein acts as a transcriptional activator. Understanding the regulatory cascades induced by Fru will help to shed light on the molecular mechanisms that are important for specification of neural circuitry underlying complex behaviour.
Background
Sex specific differences in reproductive behaviour between males and females in Drosophila are encoded via the sex determination hierarchy1,2, a genetic cascade that is initially specified by the differential expression of proteins that regulate mRNA splicing (Sxl, Tra) to produce sex-specific expression of the transcription factors fruitless (fru) and doublesex (dsx)2. The forms of these transcription factors produced in the male fly (DsxM and FruM) are thought to produce male specific behavioural phenotypes, such as courtship behaviour, via the regulation of networks of downstream target genes in the relevant neuronal circuitry2,3,4.
Drosophila male courtship is a robust, innate behaviour that requires the integration of multiple sensory inputs to elicit a stereotyped motor output5. The male courtship ritual is readily quantified, performed without instruction, and progresses through a series of well-defined steps. Mutations in the fruitless (fru) gene can lead to disruption at each of these steps; males display reduced courtship success with females, court males and females equally vigorously and the most severe fru alleles completely disrupt courtship6,7.
Expression of the fru gene is sexually dimorphic, with alternative splicing occurring in the male and female5,8. Forcing female specific splicing in the male prevents courtship behaviour, whilst driving ectopic male specific splicing in a female induces male-like courtship behaviour5,8. Thus, although a number of genes contribute to aspects of courtship, fru was shown to be able to direct this complex innate behaviour5,8. It is thought that fru acts by specifying sex-specific neural circuits within the CNS, which encode this stereotyped behavioural response5,8,9,10, however it is unclear at a molecular level how this occurs.
A great deal is known about the formation of sex-specific fru positive neural circuits10 and the sexual dimorphisms controlled by fru at a neuronal level. One of the best characterised examples is given by a cluster of neurons known as the mAL. Fru directs three types of phenotypic differences in the male mAL; cell numbers, projection laterality and neurite branching11,12. In the male brain, the mAL is composed of 30 neurons, whereas cell death induced in the female brain results in an mAL cluster containing only 5 neurons. Secondly, this neuronal cluster projects neurites ipsilaterally and contralaterally in the male, but in the female, only contralateral projections are present. Finally, the neurite branching of the contralateral projection is affected by Fru status, in the Fru positive male a simple ‘horsetail like shape’ forms, whereas in the Fru negative female, ‘Y’ shaped terminal projections form11,12. It is also well established that fru positive circuitry is essential for the formation of a male specific muscle in the abdomen known as the muscle of Lawrence (MOL)13,14. The MOL only develops when synaptic connections from a Fru positive (masculinised) motoneuron in the ventral ganglia (the MOL-inducing Mind motoneuron) innervate the muscle14,15.
Fruitless is a BTB-zinc finger (BTB-ZnF) protein that encodes a set of isoforms encoding putative transcription factors expressed from four possible promoters (P1–P4)5,6,16. Sexually dimorphic expression of fru occurs via expression from its P1 promoter which drives production of the FruM transcript in the male fly and FruF in the female17,18,19. The FruF transcript is alternatively spliced and is not translated, whereas the FruM transcript produces a protein product in approximately 3% of CNS neurons17,18,19. For simplicity, future references to fruitless herein indicate the male spliced form (FruM) unless explicitly stated. All alleles of fru that disrupt male courtship behaviour affect the P1 transcript6,7,20. Expression from other known promoters (P2–4) at the fruitless locus is not sexually dimorphic and these forms of fru are not thought to be involved in courtship behaviour but play developmental roles6,21.
Fru also undergoes alternative splicing at the 3′ terminus, such that five different forms of the protein (named FruA-E) can be produced, each carrying an alternative C-terminal zinc finger (ZnF) DNA binding5,15,22. Three of these isoforms, FruA-C (Figure 1A), represent the predominantly expressed forms in the fly nervous system that are responsible for the sexually dimophic functions of fruitless22. Other BTB-ZnF transcription factors with homology to fru, such as ttk and BR-C, also display alternatively spliced C-terminal zinc finger domains that allow distinct DNA binding specificities for each of the isoforms.
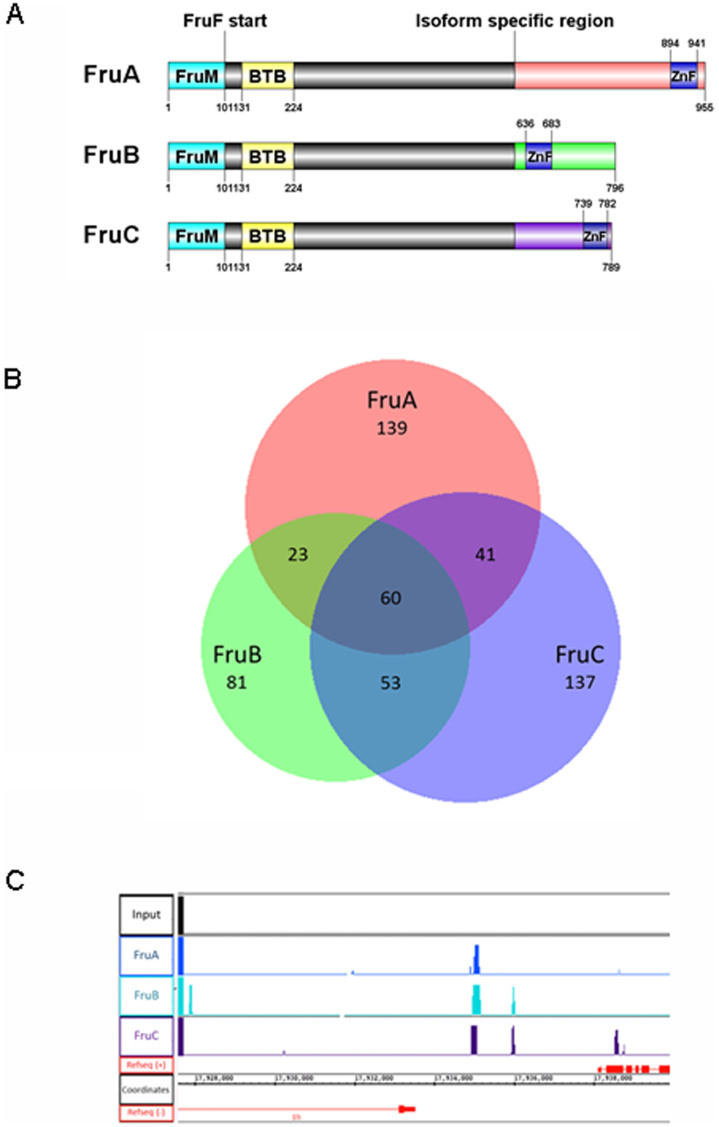
Figure 1. Genome wide identification of fru binding sites.
(A) There are three sexually dimorphic, neuronally expressed fru isoforms; FruA, FruB and FruC (also known as FruMA, FruMB and FruMC to indicate the male form). FruM denotes the male specific N-terminal peptide, spanning amino acids 1–101 which is followed by a BTB domain (amino acids 131–224). Fru protein that is expressed in the female lacks the first 101 amino acids and thus the FruM epitope. The three C-terminal isoforms share the same sequence until amino acid 617, at which point alternative splicing produces isform specific domains containing the zinc finger (ZnF) DNA binding domain that are thought to confer different DNA binding specificity for each isoform. DOG (Domain Graph, version 1.0) was used to visualise protein structures42 (B) ChIP-Seq performed in S2 cells identified putative target genes for each of the Fru isoforms. 263, 217 and 291 genes were identified for FruA, FruB and FruC, respectively. Although each isoform list represented a unique set of genes, a high degree of overlap was seen between the respective lists, with 60 genes forming a ‘common list’ of genes identified for all three isoforms. Overlap was visualised using BioVenn43 (C) An example of raw ChIP-Seq reads demonstrating a peak region observed close to the Shaker (Sh) gene. Reads were visualised using the Integrated Genome Browser (IGB) for a FruA, FruB, FruC and input sample.
Recently, Fru was shown to form a complex with the transcriptional co-factor Bonus (bon)23. This Fru-Bon complex recruits the chromatin modifying factors HDAC1 and HP1a in order to associate with chromatin and it is though that this association may lead to modification of chromatin structure23,24, which in turn may be an important mechanism for Fru mediated gene regulation. Fru expression has been shown to result in downstream gene expression changes of genes such as defective proboscis extension response (dpr), hunchback (hb), yellow (y) and takeout (to)2,4,25,26,27, however thus far it is unclear if these genes are directly or indirectly regulated by Fru and a genome wide identification of genes directly targeted by Fru has yet to be reported.
This study aims to identify direct regulatory targets of Fru in order to begin to elucidate the molecular networks required for setting up the neural circuitry underlying sex-specific courtship behaviour. Chromatin immunoprecipitation coupled to high throughput sequencing (ChIP-Seq) was used to identify the genome wide binding sites of the sexually dimorphic isoforms of the Fru protein (FruA-C). From this dataset, a list of putative target genes were identified that are involved in cellular processes such as ion channel signalling, neuromuscular junction development and neurotransmission. The Fru isoforms displayed distinct binding patterns and target genes, but also demonstrated a high degree of overlap, suggesting a core set of genes that may be regulated by all isoforms. For all three isoforms of Fru, the target gene lists contained a significant over-representation of genes located on the X-chromosome, pointing to a specific role for Fru in regulating X-linked genes. Finally, the putative target gene lists were compared to a recent studies examining Fru target genes and Fru dependent gene expression changes in the fly brain25,28. A high degree of overlap was observed and for all isoforms more than 90% of the overlapping genes were found to be upregulated. This data, taken together with expression analysis carried out herein from specifically isolated Fru positive neurons, suggests that direct interaction of Fru with target DNA results in transcriptional activation of genes important for neural circuit formation.
Results & Discussion
Expression of tagged isoforms of Fru
In order to identify the direct regulatory targets of Fru, each of the sexually dimorphic Fru protein encoding isoforms (FruA, FruB and FruC) were cloned to carry a tag that would allow their specific isolation during biochemical studies. This tag contains a biotin-ligase recognition peptide (BLRP) that undergoes biotinylation when co-expressed with the bacterial biotin ligase protein; BirA29. Once biotinylated, these tagged proteins (and associated complexes) can be efficiently isolated using streptavidin coupled beads, due to the extremely high affinity streptavidin has for biotin. Chromatin immunoprecipitation of these tagged fru protein-DNA complexes coupled to high throughput sequencing allowed the identification of fruitless binding sites throughout the fly genome.
Fru protein isoforms interact with specific regions of the fly genome
Drosophila S2 cells were co-transfected with BirA and one of the BLRP tagged versions of Fru (FruA, FruB or FruC; Figure 1A). Streptavidin coupled to magnetic beads was used to immunoprecipitate the tagged, biotinylated Fru protein isoforms. To confirm both the expression, the tagging of the protein and to validate the pull down technique, the immunoprecipitated samples were detected via western blotting using an antibody specific for the male specific epitope (FruM)9 (Supp Figure 1). Chromatin Immunoprecipitation coupled to high throughput sequencing (ChIP-Seq) was performed as described, in order to identify the direct regulatory targets of each of the Fru isoforms in this model system. Peaks of enrichment, indicative of Fru binding, were identified via the Model-based Analysis of ChIP-Seq (MACS) program30. Using a p-value cutoff of p < 10−10 there were 791, 449 and 662 peak regions identified that were enriched over input DNA for FruA, FruB and FruC, respectively (Supp Tables 1–3).
Fru isoforms target ion channels important for neural circuit function
Peaks identified via ChIP-Seq were screened to discover proximal target genes, defined as having transcriptional start sites that lay within 2 kb of the peak region. This generated putative target gene lists for the isoforms of 263, 217 and 291 genes, respectively (Supp tables 4–6). These lists were first assessed for the presence of any genes that were already thought to be regulated, directly or indirectly by Fru. Previously, dpr and a number of its family members had been shown to reduce their expression when fru is mutated4, suggesting that these genes are normally upregulated by the Fru protein. Futhermore dpr mutations have been shown to have a phenotypic effect on wing extension initiation, an early aspect of courtship behaviour4. The ChIP-Seq datasets contained 8 of the dpr family genes, many of which were represented in more than one isoform gene list - including dpr itself, suggesting that Fru directly regulates the transcription of dpr and some dpr family members.
To understand the molecular functions of the genes regulated by fruitless, gene ontology analysis was carried out on each individual target gene list. Significant over representation of a number of related gene categories was observed, as summarised in Table 1. All three isoforms demonstrated enrichment for genes involved in ‘ion gated channel activity’ [GO:0022839] and many related ontology categories were enriched in one or more of the isoform gene lists, including ‘voltage-gated cation channel activity’ [GO:0022843] and ‘extracellular-glutamate-gated ion channel activity’ [GO:0005234]. These categories included putative Fru target genes such as an NMDA receptor (Nmdar2), multiple nicotinic Acetylcholine Receptor subunits (nAcRalpha-96Aa & nAcRalpha-7E) and an ionotropic glutamate receptor (GluRIB). FruA and FruC lists were enriched for genes reported to have ‘receptor activity’ [GO:0004872] and the FruA list was further enriched for ‘transmembrane signalling receptor activity’ [GO:0004888] including genes such as sevenless (sev) and white (w). The FruC gene list was also enriched for ‘neurotransmitter receptor activity’ [GO:0030594] due to the inclusion of genes such as Dopamine 2-like receptor (Dop2R) and sex peptide receptor (SPR). All three gene lists also showed a highly statistically significant enrichment for genes that carried an Immunoglobulin-like domain [IPR007110] and Immunoglobulin-like fold [IPR013783] (Supp table 7), consistent with findings from previously published data that recently reported Fru dependent gene expression changes in the fly brain and saw similar protein domain enrichment25. Full lists of gene ontology and protein domain enrichment for each isoform can be found in supplementary tables 7–10.
Table 1. Molecular function gene ontology categories for the Fru isoform target gene lists.
| FruA | FruB | FruC | ||||
|---|---|---|---|---|---|---|
| GO Term | p-value | # genes | p-value | # genes | p-value | # genes |
| channel activity [GO:0015267] | 3.09e-5 | 20 | 0.0017 | 15 | - | - |
| ion channel activity [GO:0005216] | 6.16e-5 | 19 | 0.0244 | 13 | - | - |
| cation channel activity [GO:0005261] | 0.0145 | 11 | 6.99e-4 | 11 | - | - |
| gated channel activity [GO:0022836] | 2.60e-6 | 18 | 0.0052 | 12 | 0.0489 | 13 |
| ion gated channel activity [GO:0022839] | 2.60e-6 | 18 | 0.0052 | 12 | 0.0489 | 13 |
| voltage-gated channel activity [GO:0022832] | - | - | 0.0406 | 6 | - | - |
| voltage-gated ion channel activity [GO:0005244] | - | - | 0.0406 | 6 | - | - |
| voltage-gated cation channel activity [GO:0022843] | - | - | 0.0127 | 6 | - | - |
| substrate-specific channel activity [GO:0022838] | 8.89e-5 | 19 | 0.0311 | 13 | - | - |
| ligand-gated channel activity [GO:0022834] | 9.54e-5 | 14 | - | - | - | - |
| ligand-gated ion channel activity [GO:0015276] | 9.546e- | 14 | - | - | - | - |
| excitatory extracellular ligand-gated ion channel activity [GO:0005231] | 1.23e-4 | 10 | - | - | - | - |
| extracellular-glutamate-gated ion channel activity [GO:0005234] | 0.0096 | 7 | - | - | - | - |
| extracellular ligand-gated ion channel activity [GO:0005230] | 3.45e-5 | 11 | - | - | - | - |
| receptor activity [GO:0004872] | 2.11e-7 | 32 | - | - | 0.0377 | 24 |
| transmembrane signaling receptor activity [GO:0004888] | 2.53e-5 | 25 | - | - | - | - |
| passive transmembrane transporter activity [GO:0022803] | 3.09e-5 | 20 | 0.0017 | 15 | - | - |
| signaling receptor activity [GO:0038023] | 4.27e-5 | 26 | - | - | - | - |
| signal transducer activity [GO:0004871] | 5.59e-5 | 29 | - | - | - | - |
| molecular transducer activity [GO:0060089] | 5.59e-5 | 29 | - | - | - | - |
| neurotransmitter receptor activity [GO:0030594] | - | - | - | - | 0.0228 | 8 |
| dipeptidyl-peptidase activity [GO:0008239] | - | - | - | - | 0.0279 | 5 |
The highly significant ontology categories that were identified across these datasets suggests that Fru mediated transcriptional regulation is important for cellular communication mediated by ion channels. The appropriate expression of combinations of ion channels in neuronal subtypes is essential for the correct formation of and signalling through neuronal circuits. For example, NMDA receptors have been shown to be important for synapse refinement, an essential process required during circuit development to produce the appropriate connectivity31. In total, four acetylcholine receptor subunits were identified across the datasets (nAcRalpha-7E, nAcRalpha-30D, nAcRalpha-80B & nAcRalpha-96Aa). Signalling mediated via acetylcholine is central to insect nervous system function and nicotinic acetylcholine receptors have been implicated in instinctive behaviours such as the escape reflex in Drosophila32 as well as the integration of information in the visual system33. Thus the control of expression of specific ion channels such as Nmdar2, GluRIB and nAc receptor subunits by fruitless may represent an important mechanism by which sex-specific circuitry develops downstream of Fru.
Despite the differences in the DNA binding domains of the three isoforms, a high degree of overlap was observed between the three gene lists. Many genes were identified as putative targets for more than one isoform and 60 genes were shared across all three datasets (Figure 1B; Supp table 11), which equates to more than 20% of each individual gene list being common to all isoforms tested. This common list contained a number of interesting genes including genes implicated in courtship behaviour such as the Sex peptide receptor (SPR) and Shaker (Sh) (see Figure 1C), as well as genes implicated in synaptic transmission (Snap-25), and axon outgrowth (Dscam3). The common gene list showed significant enrichment of a number of the same gene ontology categories as the individual isoforms including ‘ion gated channel activity’ [GO:0022839] but also ‘passive transmembrane transporter activity’ [GO:0022803] (Table 2 & Supp table 12). The target genes in the common list were also significantly enriched for protein domains including ‘immunoglobulin-like domain’ (IPR007110) and ‘p53/RUNT-type transcription factor, DNA-binding domain’ [IPR012346] (Supp Table 13). Thus, despite very different DNA binding domains, a core set of genes involved in common pathways seem to be targeted by all three Fru isoforms.
Table 2. Molecular function gene ontology categories for the ‘common’ target gene list.
| ‘Common’ gene list | ||
|---|---|---|
| GO Term | p-value | # genes |
| channel activity [GO:0015267] | 3.26e-3 | 9 |
| ion channel activity [GO:0005216] | 2.07e-2 | 8 |
| cation channel activity [GO:0005261] | 1.93e-3 | 7 |
| gated channel activity [GO:0022836] | 2.62e-2 | 7 |
| ion gated channel activity [GO:0022839] | 2.62e-2 | 7 |
| passive transmembrane transporter activity [GO:0022803] | 3.26e-3 | 9 |
| substrate-specific channel activity [GO:0022838] | 2.44e-2 | 8 |
During review of this manuscript, an in vivo study identifying targets of FruM isoforms (FruA, FruB, FruC) at different timepoints (larvae, pupae and adult) in neurons was published by Neville and colleagues28. To estimate the biological relevance of the dataset described herein, the S2 Fru-ChIP targets were compared with the neuronal Fru targets28. A high degree of overlap was observed, despite the differences in model systems used. Since S2 cells do not represent any particular developmental timepoint, the complete list of targets for each isoform (larvae, pupae and adult) identified by Neville et al28 were overlapped with the S2 Fru-ChIP targets for the corresponding isoform. 29% percent of the S2 FruMA-ChIP targets were represented in the in vivo dataset, while 45% and 49% of the S2 ChIP targets overlapped for FruMB and FruMC, respectively (Table 3 and Supp table 14). Interestingly the overlapping genes included Dpr family members (dpr and dpr6, 8, 10, 11, 13 & 16), nicotinic Acetylcholine Receptor subunits (nAcRalpha-7E, -30D, 80B & -96Aa), sevenless (sev), white (w), sex peptide receptor (SPR), shaker (sh) and Dscam3. This high degree of overlap in targets independently identified using an in vivo system, supports the biological relevance of the genes identified in this study.
Table 3. Overlap of direct Fru targets identified via S2 ChIP-Seq with direct Fru targets identified via fly brain ChIP-Seq (Neville et al, 2014)28.
| Fly brain RNA-Seq genes | ||||||
|---|---|---|---|---|---|---|
| Isoform | S2 ChIP-Seq genes | Larvae | Pupae | Adult | Overlap | % S2 ChIP-Seq overlap |
| FruA | 263 | 385 | 1722 | 2572 | 77 | 29.2% |
| FruB | 217 | 2074 | 2076 | 809 | 97 | 45% |
| FruC | 291 | 1387 | 2771 | 3583 | 143 | 49% |
A specific role for Fru in regulating X-linked genes
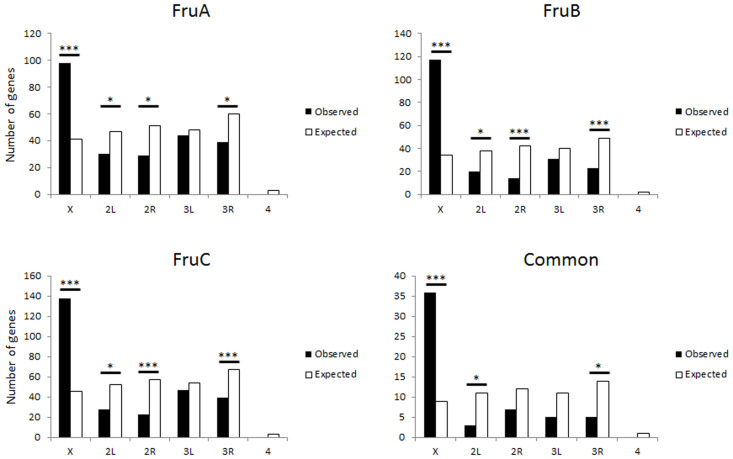
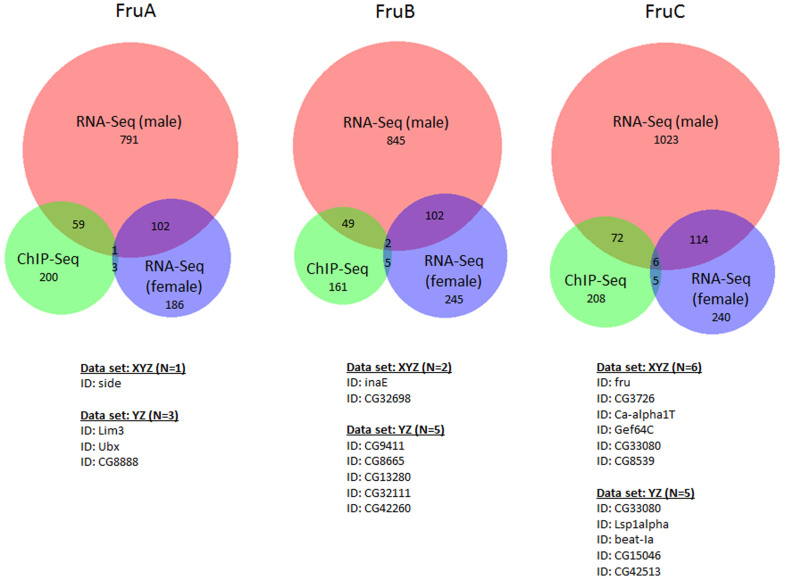
Of particular interest, when the Fru binding sites were assessed for genomic distribution, a highly significant enrichment of binding on the X chromosome was observed (Figure 2). ChIP-Seq is an unbiased screen for transcription factor binding, and although accessibility of the epitope/tag or local chromatin structure may affect the ability to pull down protein-DNA complexes, it is not expected that this would result in a chromosome specific bias. Indeed ChIP-Seq studies using other transcription factors have not observed this sort of X-chromosome specific bias. Rather, this over-representation of target sites may represent a specific role for X-linked genes in fruitless directed gene networks. Indeed this finding is consistent with results from another paper that was published while this manuscript was in preparation25. Dalton et al25 showed that fru overexpression resulted in changes in expression for hundreds of genes in neurons of the male fly. A significant over-representation of genes encoded on the X chromosome was observed for those genes that increased due to Fru overexpression, but not for down-regulated genes25.
Figure 2. Chromosomal distribution of putative target genes.
For each of the isoform lists and the ‘common’ list, the chromosomal distribution of the genes identified as putative targets was mapped. In all gene sets, there was a significant over-representation of loci on the X-chromosome. Signficance was calculated using χ-squared test for observed vs expected values based on genomic distribution. * = p < 0.01, ** = p < 0.0001.
Transcripts of genes targeted by Fru are enriched in fru positive neurons
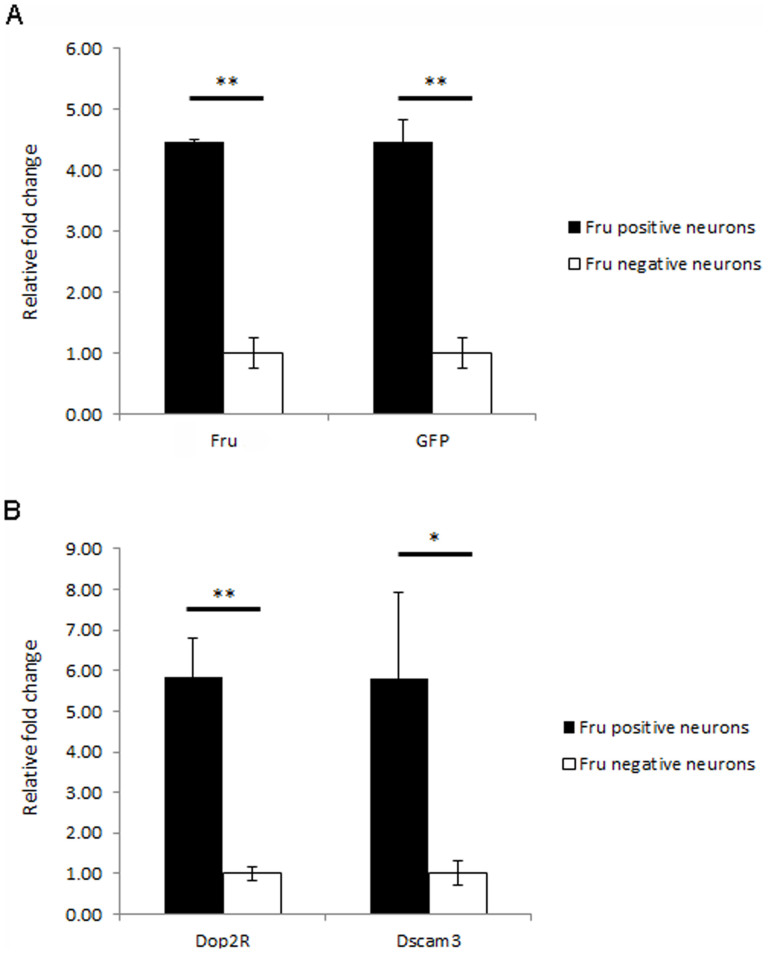
To demonstrate that the binding sites identified in S2 cells could translate to real gene expression changes in the fly brain, a method was employed to specifically isolate RNA from Fru positive neurons. This system utilised the GAL4/UAS system to drive expression of a membrane bound GFP signal (CD8-GFP) in subsets of neurons (as described by Iyer et al34). Here, the GFP signal was expressed in all Fru positive neurons by coupling the Fru-GAL4 driver line with the UAS-CD8-GFP line19, however it would be possible to drive expression in subsets of Fru positive neurons by using combinations of driver lines in an intersectional approach10. The expression of a cell surface CD8-GFP protein, allowed dissociated neurons to be isolated via antibody coupled magnetic cell sorting34. Techniques such as FACS (Fluorescence activated cell sorting) have also been used to isolate tagged populations of cells for analysis, however FACS is a more harsh technique that can cause stress and/or damage to the cells which could affect the results of molecular assays such as transcript analysis. Following magnetic cell sorting, RNA was extracted from the two populations of neurons harvested from the fly; Fru positive neurons (that express CD8-GFP) and all other neurons in the brain. If Fru acts to upregulate a target gene, it would be expected to have enriched levels of transcript in Fru positive neuron sample compared to the baseline (the sample containing all other neurons in the brain). First, the enrichment of fru and GFP transcripts in the cell sorted samples compared to baseline was confirmed via qPCR (Figure 3A). Next a small number of target genes were chosen for validation. Two genes Dop2R (Dopamine 2 receptor) and Dscam3 (Down syndrome cell adhesion molecule 3) showed significant enrichment in the Fru positive neurons compared to baseline, suggesting that Fru binding results in their upregulation (Figure 3B). Two further genes were also tested (Shaker and Nmdar2) however the transcripts levels were too low to be reliably detected and were thus excluded. In addition to validating targets identified in the S2 Fru-ChIP experiments, these results demonstrate the utility of this method, particularly when coupled to an intersectional genetic approach10, to specifically isolate intact populations of neurons for biochemical study eg. RNA-Seq or ChIP-Seq. In this way, the regulatory cascades that are necessary to specify different aspects of the sexually dimorphic circuitry underlying courtship behaviour could be defined.
Figure 3. Transcript enrichment in Fru positive neurons of the male fly brain.
(A) Using magnetic cell sorting, Fru/CD8-GFP positive neurons were isolated via the presence of a CD8-GFP cell surface tag for transcript analysis and compared to samples containing all Fru/CD8-GFP negative neurons. qPCR demonstrated that a more than 4 fold enrichment of both Fru and GFP mRNAs were observed in the Fru positive samples, suggesting that the cell sorting had been successful. (B) Putative target genes identified via ChIP-Seq in S2 cells were tested for transcript enrichment in the cell sorted samples. Significant enrichment of both Dop2R and Dscam3 transcripts was observed in Fru positive neurons compared to baseline (Fru negative neurons), suggesting that Fru acts to upregulate the expression of these genes. Results are representative of two independent biological replicates. Significance was calculated using students t-test where * = p < 0.05 and ** = p < 0.01.
Fru isoform target genes display Fru dependent expression differences in neurons of the male fly brain
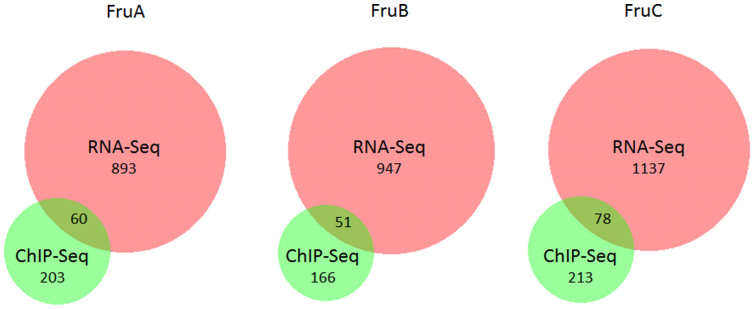
A recent study by Dalton et al interrogated mRNA changes in the fly brain resulting from Fru isoform overexpression via RNA-Seq25. The gene lists identified therein are expected to include genes that are downstream of Fru, but that may represent either direct transcriptional targets or indirectly regulated genes. By contrast, the work detailed herein reports exclusively those genes putatively targeted by Fru via direct interaction with DNA. By comparing these two datasets, we can determine which of the RNA-Seq genes are directly regulated by Fru isoforms, and in turn, further validate our ChIP-Seq targets in an in vivo system. Table 4 demonstrates the overlap for each isoform between the ChIP-Seq target gene identification detailed herein and the RNA-Seq expression analysis performed in male flies25. A very high degree of overlap was observed between these independent datasets, much more than would be expected based on chance alone (all p-values < 1.5e-19). Between ~23–27% of the direct ChIP-Seq targets were shown to change their expression in Fru P1-expressing neurons of the male fly brain in response to Fru isoform overexpression (Figure 4, Table 4 and Supp table 15). Of particular note, the vast majority of S2 Fru-ChIP targets that are also represented in the RNA-Seq data (more than 90%) were upregulated in the fly brain (Table 5). Only a handful of direct targets in each list were downregulated. This suggests that when Fru binds to a gene promoter it acts as a transcriptional activator, inducing expression of the target gene unlike some other BTB-ZnF transcription factors such as ttk that mediate gene repression35. This is further supported by finding that in the in vivo RNA-Seq experiments both Fru binding motif enrichment and X-chromosome enrichment were only observed for those genes that were upregulated in the fly brain25. The finding that putative Fru target genes identified via ChIP-Seq were upregulated in the fly brain both in this study as well as in independent studies of gene expression4,12,25 together with the enrichment of Fru binding motifs in upregulated target genes25 supports the hypothesis that direct Fru binding induces the expression of target genes in the fly brain.
Table 4. Overlap of direct Fru targets identified via ChIP-Seq with downstream expression changes induced by Fru isoforms in the male fly brain (Dalton et al, 2013)25.
| Isoform | ChIP-Seq genes | RNA-Seq genes | Overlap | % ChIP-Seq gene overlap | Significance of overlap |
|---|---|---|---|---|---|
| FruA | 263 | 953 | 60 | 22.8% | p < 5.51e-30 |
| FruB | 217 | 998 | 51 | 23.5% | p < 1.49e-19 |
| FruC | 291 | 1215 | 78 | 26.8% | p < 2.92e-28 |
Figure 4. ChIP-Seq identified genes demonstrate expression changes in the male fly brain.
The genes identified in this study via ChIP-Seq were compared to a recent RNA-Seq study25 that assessed gene expression changes in the male fly brain following overexpression of each of the male specific isoforms FruA, FruB and FruC. A highly significant degree of overlap was observed between these datasets. Approximately one quarter of the genes identified via ChIP-Seq also showed expression changes in the male fly brain for the corresponding isoform, a level of overlap that was much higher than would be expected by chance alone. Overlap was visualised using BioVenn43.
Table 5. Overlap of direct Fru targets identified via ChIP-Seq with genes upregulated by Fru isoforms in the male fly brain (Dalton et al, 2013)25.
| Isoform | ChIP-Seq genes | Overlap with RNA-Seq data | Overlap with induced genes in RNA-Seq data | % overlap due to induced genes |
|---|---|---|---|---|
| FruA | 263 | 60 | 57 | 95% |
| FruB | 217 | 51 | 48 | 94% |
| FruC | 291 | 78 | 72 | 92% |
The ChIP target genes that overlap with the Dalton et al RNA-Seq experiments25 represent a subset of high confidence target genes, in that these genes have a ChIP-Seq signal indicative of Fru protein binding and also change their expression in neurons in response to the presence of one or more of the Fru isoforms. Indeed the two target genes (Dscam3 and Dop2R) that showed enrichment in Fru positive neurons herein (Figure 3B), also showed upregulation by all three isoforms tested in the Dalton et al study25. Thus, to better understand the pathways regulated by Fru, the genes that overlapped between the ChIP-Seq and RNA-Seq experiments were explored via gene ontology and protein domain enrichment analaysis (Supp Tables 16–18). The FruA overlap list was significantly enriched for categories relating to neuron development [GO:0048666] and differentiation [GO:0030182], and more specifically axonogenesis [GO:0007409] and axon guidance [GO:0007411]. Significant over-representation was also observed for genes involved in cell projection organisation [GO:0030030] (FruC overlap), as well as cell communication [GO:0007154] (FruC overlap), synapse assembly [GO:0007416] (FruB overlap) and synaptic target attraction [GO:0016200] (FruB & FruC overlap).
The FruB overlap list was significantly enriched for genes involved in neuromuscular junction development [GO:0007528]. This enrichment was observed for both the FruB overlap list (Supp Table 17), as well as the FruB ChIP-Seq list (Supp Table 9), but not for other isoform lists. Genes shared between the two datasets include Nlg1 (Neuroligin 1), futch and cac (cacophony), which have previously been implicated in synapse development at neuromuscular junctions and were also identified as Fru targets in vivo28. In addition to courtship behaviour, the male specific functions of fru include directing the formation of the MOL36. The MOL is a large abdominal muscle found exclusively in male flies and its development is dependent upon direct innervation by masculinised, fru positive, glutamatergic motor neurons14,15,19. Thus, the putative target genes identified herein that are involved in neuromuscular junction development, such as Nlg1, futch and cac may contribute to the molecular mechanism by which fru is able to affect MOL innervation.
The overlap between the in vitro lists identified herein and the recent in vivo studies is particularly striking when considering the vastly different model systems used. In this study an in vitro model (S2 cells) was used to investigate the binding of the Fru protein throughout the genome. By contrast the Fru-neuron target identification was performed in CNS tissue28 and Fru RNA-Seq transcript analysis was prepared from fly heads25 and thus both reflect the changes occurring in Fru positive neurons in vivo. S2 cells are derived from late stage D.melanogaster embryos and grow as a monolayer of cells with epithelial-like morphology and as such do not reflect a neuronal identity37. An advantage of using a cell line such as this is that protein constructs can be tagged and overexpressed to allow high occupancy rates throughout the genome (even at low affinity sites) and efficient isolation of protein-DNA complexes, which reduces background. Hence despite these cells not being neuronal in origin, we hypothesised that it would be possible to identify some biologically relevant target sites using this methodology. For each isoform, between 30–50% of the targets identified in S2 cells were identified as Fru targets in the CNS28 and around one quarter showed FruM dependent neuronal gene expression changes25 suggesting that a subset of the targets identified herein might be important for the sexual dimorphism induced in the developing fly nervous system and thus warrant further, in vivo investigation. Some of the genes identified herein that did not show overlap with the in vivo ChIP or expression data may represent non-neuronal or developmental, non-sexually dimorphic targets, or reflect the technical limitations of the model system. Although some may be true neuronal targets of Fru that could not be detected by the in vivo assays. Given that Fru is expressed in a range of different neurons, such as mAL, median bundle and descending neurons19 it is likely that the genes regulated by Fru in these neuronal subsets will differ. Thus, targets that are regulated only in small subsets of Fru positive neurons are unlikely to show expression changes dramatic enough to be detected as significant when considering whole head RNA samples, as was done by Dalton and colleagues25. In order to discover these changes, it may be necessary to directly assay the transcripts from only these specific subsets of neurons, using a technique such as the cell sorting method described herein.
Fru isoform target genes do not correlate with gene expression differences observed in the female brain
Dalton et al. also assayed gene expression changes induced when FruM isoforms were introduced into the female fly25. In contrast to the male fly, here they observed more genes being repressed, rather than upregulated, and only a small proportion of the genes that were regulated in the male were replicated in the female brain (14% overlap between male/female), suggesting that the regulatory activity of FruM is in part determined by its environment25. To determine if the S2 Fru-ChIP targets identified herein reflected the genes that were regulated in the male, female or overlapping male/female dataset, the respective datasets were compared. In stark contrast to the high degree of overlap observed with the male RNA-Seq gene lists (Table 4), very little overlap was observed with the female derived RNA-Seq data. Only 1.5%, 3.2% and 3.8% of genes were shared for the FruA, FruB and FruC datasets, respectively (Table 6 & Figure 5). This was unexpected given that the ChIP-Seq data was generated in a cell model system rather than a sexually dimorphic brain. Chromosome analysis of S2 cells have shown that they are male in origin and have an X/A ration of 0.5, as is found in male Drosophila, demonstrating similar dosage compensation as is seen for X-linked genes in the male fly38 This might, in part, explain the bias towards target genes that are specifically regulated in the male brain. In any case, we demonstrate here that the in vitro S2 model system represents a powerful starting point providing ease of manipulation for investigating the activity of transcription factors such as Fru, that are involved in complex behavioural programs.
Table 6. Overlap of direct Fru targets identified via ChIP-Seq with downstream expression changes induced by Fru isoforms in female fly brains (Dalton et al, 2013)25.
| Isoform | ChIP-Seq genes | RNA-Seq genes (female) | Overlap | % ChIP-Seq gene overlap |
|---|---|---|---|---|
| FruA | 263 | 292 | 4 | 1.5% |
| FruB | 217 | 354 | 7 | 3.2% |
| FruC | 291 | 365 | 11 | 3.8% |
Figure 5. ChIP-Seq identified genes do not show expression changes in the female fly brain.
The ChIP-Seq gene lists were also compared to RNA-Seq performed in the female fly brain25 following overexpression of the male specific isoform. Although the male and female brain RNA-Seq experiments showed some overlap, very few of these genes were also identified in the ChIP-Seq experiments. In fact, less than 4% of genes identifed in any of the isoform specific ChIP-Seq gene lists demonstrated any expression differences in the female fly brain, compared to more than 20% of genes that were affected in the male brain. Overlap was visualised using BioVenn43.
Taken together, the in vitro and in vivo studies suggest that fruitless is able to initiate a cascade of expression changes of genes spread throughout the genome. As shown by Dalton et al25, the majority of these changes result in increased gene expression, however a small subset of genes are down-regulated. More than 90% of the genes that were shown to be both direct targets in this study and downstream of Fru in Dalton, et al. were upregulated, suggesting that the fruitless protein normally binds to promoter regions in order to upregulate target genes. This is further supported by our finding that there was a significant enrichment of genes encoded on the X-chromosome for all isoforms tested and the corresponding X-chromosome enrichment observed for upregulated genes in the RNA-Seq study25. Thus we can hypothesise that Fru may play a specific role in directly upregulating genes located on the X-chromosome. More work is needed to determine in which subsets of neurons and for which particular functions of Fru these targets play a role. By combining transcript profiling for specific neuronal subpopulations with neural circuit tracing and behavioural studies, true insight can be gained into the molecular mechanisms underlying Fru directed courtship behaviour.
Conclusions
This study has identified a set of direct regulatory targets for each of the sexually dimorphic isoforms of the fruitless gene (FruA, FruB & FruC). It has been hypothesised that Fru was able to directly control gene expression by binding to target DNA and the work herein directly demonstrates this capacity, suggesting that Fru upregulates a range of genes implicated in neural circuit formation. Understanding the regulatory cascades induced by Fru will help to shed light on the molecular mechanisms that are important for specification of neural circuitry underlying complex behaviours.
Methods
Immunoprecipitation and Western blotting
Drosophila S2 cells were co-transfected with BirA and a tagged fru isoform (FruA-BLRP, FruB-BLRP or FruC-BLRP). using FuGENE6 (Promega) according to the manufacturer's instructions. 48 hours post-transfection cells were washed twice in PBS and proteins were extracted via treatment with Lysis Buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 1 mM PMSF, protease inhibitor cocktail) at 4°C for 20 minutes. Cells were centrifuged at 10,000 g for 30 minutes at 4°C, allowing cell debris to be pelleted and discarded. 50 μl/ml of streptavidin coupled magnetic beads (M-280 Dynabeads; Life Technologies) were pre-blocked via incubation with 5% BSA, 0.5% Tween20, PBS, rotating at 4°C for 1 hour. Blocked streptavidin beads were combined with cell lysates and allowed to rotate at 4°C for 2 hours to capture the tagged protein complexes. After washing with lysis buffer, protein was eluted from the beads by boiling at 100°C for 5 minutes in the presence of sample buffer (100 mM Tris pH = 6.8, 10% SDS w/v, 20% glycerol, 0.1% bromophenol blue, 10% β-mercaptoethanol).
Proteins were resolved on 10% Polyacrylamide SDS gels and transferred to Polyvinylidene fluoride (PVDF) membrane (Invitrogen) at 25 volts for 90 minutes using a semi-dry transfer system (Invitrogen). Membranes were blocked in Western Blocking Buffer (5% Skim Milk Powder, 0.1% Tween 20 in PBS) to prevent non-specific antibody interactions. Proteins were detected using primary antibodies (FruM or BirA)9 at 4°C overnight. Secondary antibodies were applied for 1 hour at room temperature. Proteins were visualized using ‘ECL Plus’ Enhanced Chemiluminescence Reagents (Amersham Biosciences) and Kodak MXB Film.
Chromatin Immunoprecipitation
Drosophila S2 cells were co-transfected with BirA and a tagged fru isoform (FruA-BLRP, FruB-BLRP or FruC-BLRP) using FuGENE6 (Promega) according to the manufacturer's instructions. After 48 hours at 37°C and 5% CO2, cells from at least two biological replicates were cross-linked using 1% formaldehyde in cross-linking buffer (50 mM HEPES, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) at room temperature for 10 minutes. The cross linking reaction was halted via the addition of 125 mM glycine. Cells were washed in PBS and incubated for 10 minutes in ice-cold ChIP lysis buffer (10 mM Tris, 0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, protease inhibitors), and centrifuged at 10,000 g and 4°C for 5 minutes to pellet nuclei. Nuclei from approx 1 × 107 cells were resuspended in 1 ml Sonication Buffer (10 mM Tris, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, protease inhibitors) before undergoing 2 rounds of 30-second sonication pulses at 40% power, with 2 minutes on ice between each round. Cells were centrifuged at 10,000 g and 4°C for 5 minutes to remove cell debris. Cell lysates were pre-cleared via incubation with 20 μl protein-G dynabeads, rotating at 4°C for 1 hour.
50 μl of streptavidin coupled magnetic beads (M-280 Dynabeads; Life Technologies) that had been pre-blocked (via incubation with 5% BSA, 0.5% Tween20, PBS, rotating at 4°C for 1 hour) were incubated with the pre-cleared supernatants in IP buffer (0.1 M Tris, 10 mM EDTA 150 mM NaCl, 0.2% Triton X-100, 1% PMSF, protease inhibitors) rotating overnight at 4°C to capture the protein-DNA complexes. Protein was eluted from beads by incubation with elution buffer (10 mM Tris pH = 7.5, 1 mM EDTA, 1% SDS, 100 mM NaHCO3, 200 mM NaCl, 1.5 μg/ml RNaseA) at 37°C, shaking for 30 minutes. Cross links were reversed in the presence of 2.5 μg/ml proteinase K by incubating at 45°C for 1 hour, followed by 65°C overnight. DNA was isolated via Phenol-Chloroform extraction followed by ethanol precipitation. Concentration and purity of the DNA was evaluated by spectrophotometry and size was assessed via gel electrophoresis.
ChIP sequencing
ChIP isolated DNA was amplified according to the Illumina ChIP-Seq library preparation protocol to generate libraries with fragment size approx. 300–700 bp and quantified using the Agilent Bioanalyser. Cluster generation and sequencing was carried out via Illumina/Solexa Genome Analyser (GA) II according to the manufacturer's protocols.
ChIP-Seq data analysis
After quality filtering, sequence reads (between 20–22 million unique reads per sample) were mapped to the Drosophila melanogaster genome (2006 assembly; BDGP Release5/dm3) using Bowtie (version 0.12.2) and visualized using the integrated genome browser (IGB). Peak finding was performed using MACS version 1.3.6.130 using the default parameters and the settings: --tsize = 25, --bw = 300, --mfold = 32, --pvalue = 1e-10 for the ChIP-Seq replicates vs input. The proximity of peaks to genes was determined using the galaxy bioinformatics server (http://galaxyproject.org/)39.
Gene Ontology and protein domain enrichment
Gene Ontology (GO) and protein domain enrichment were carried out for each target gene list (FruA, FruB, FruC or ‘common’) within the Flymine portal40 using the Drosophila genome as background dataset and Holm-Bonferroni multiple testing correction.
Isolation of fruitless positive neurons
Freshly eclosed FruGAL4;UAS-CD8-GFP flies were collected and between 30–80 whole brains dissected from male flies. Brains were placed into ice cold PBS and gently spun down. Tissue was digested with 12 μl (30 μg) of liberase enzyme (Roche) in a total volume of 500 μl PBS (the liberase enzyme was prepared by making up a 2.5 mg/ml solution in water and agitating at 4°C for 15 minutes to disolve). Digestion took place at room temperature for 20 minutes, vortexing regularly. Tissue was washed twice in PBS before trituration with a P1000 pipette to produce single cells. Production of a single cell suspension was confirmed on a fluorescence microscope. The single cell solution was then combined with CD8 antibody (MHCD0800, Invitrogen) coupled protein-G dynabeads (antibody-bead coupling performed by combining 10 μg of antibody with 50 μl of beads and rotating at 4°C for 1 hour). Immunoprecipitation was allowed to take place by gently rotating the solution at 4°C for 30 minutes. The supernatant containing the Fru/CD8-GFP negative neurons were lysed in Trizol or QIAGEN RLT buffer to generate the baseline sample. The bead-neuron complexes were washed three times in PBS before being lysed in Trizol reagent or RLT buffer prior to RNA extraction (representing the Fru/CD8-GFP positive fraction). Enrichment for the GFP signal was also confirmed via fluorescence microscopy prior to lysis.
RNA extraction and expression analysis
RNA samples from two independent isolations of fruitless positive neurons were purified using the QIAGEN RNAeasy micro kit according to the manufacturers instructions. RNA samples were reverse transcribed into cDNA using the Invitrogen Superscript III first strand synthesis supermix for RT-PCR with random hexamer primers, as described previously41. qPCR was performed using SYBR green and gene specific primers for the house keeping gene RP49 (Fwd: CGAACAAGCGCACCCGC, Rev: CGCAGGCGACCGTTGGGG), as well as GFP (Fwd: AAAGGGCAGATTGTGTGGAC, Rev: TGGAAGCGTTCAACTAGCAG), Fru (Fwd: GGACTCTCAGGCCAACTTC, Rev: GAGCGGCGCTCGGCAAGTAATCTG), Dop2R (Fwd: GGACTTTCGCAGGGCCTTT, Rev: CGATCTGGTTCACCGAGTGG) and Dscam3 (Fwd: CCGGGCCTCAGGAAAATATCA, Rev: ATGGCGCACTTAATCAACGC). Normalisation was performed via the ΔΔ-Ct method, as described previously41. The RP49 housekeeping gene was used to normalise the amount of template cDNA present in each reaction. Fru and GFP were used to confirm the specific enrichment of fru positive (and therefore GFP positive) neuronal transcripts in the immunoprecipitated samples.
Availability of supporting data
The data sets supporting the results of this article are available in the additional files accompanying this manuscript.
Author Contributions
S.C.V. conceived the study design, carried out the experimental and analytical work and drafted the manuscript.
Supplementary Material
Supplementary information
Supplementary Tables 1-3
Supplementary Tables 4-6
Supplementary Table 8-10
Supplementary Table 11
Supplementary Tables 12
Supplementary Table 14
Supplementary Tables 15
Supplementary Tables 16-18
Acknowledgments
This research was carried out with in the laboratory of Barry Dickson at the IMP Vienna and with support from Boehringer Ingelheim. S.C.V. was supported by a Human Frontiers (HFSP) fellowship and Marie Curie IEF fellowship. Thank you to Andreas Sommer and the Campus Support Facilities (CSF Vienna) for Solexa sequencing and read mapping and Paolo Devanna for assistance with qPCR experiments.
References
- Yamamoto D. The neural and genetic substrates of sexual behavior in Drosophila. Adv Genet 59, 39–66, 10.1016/S0065-2660(07)59002-4 (2007). [DOI] [PubMed] [Google Scholar]
- Dauwalder B. The roles of fruitless and doublesex in the control of male courtship. Int Rev Neurobiol 99, 87–105, 10.1016/B978-0-12-387003-2.00004-5 (2011). [DOI] [PubMed] [Google Scholar]
- Villella A. & Hall J. C. Neurogenetics of courtship and mating in Drosophila. Adv Genet 62, 67–184, 10.1016/S0065-2660(08)00603-2 (2008). [DOI] [PubMed] [Google Scholar]
- Goldman T. D. & Arbeitman M. N. Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS genetics 3, e216, 10.1371/journal.pgen.0030216 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir E. & Dickson B. J. fruitless splicing specifies male courtship behavior in Drosophila. Cell 121, 785–794, 10.1016/j.cell.2005.04.027 (2005). [DOI] [PubMed] [Google Scholar]
- Anand A. et al. Molecular genetic dissection of the sex-specific and vital functions of the Drosophila melanogaster sex determination gene fruitless. Genetics 158, 1569–1595 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H. et al. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc Natl Acad Sci U S A 93, 9687–9692 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli D. S. et al. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature 436, 395–400, 10.1038/nature03859 (2005). [DOI] [PubMed] [Google Scholar]
- Stockinger P., Kvitsiani D., Rotkopf S., Tirian L. & Dickson B. J. Neural circuitry that governs Drosophila male courtship behavior. Cell 121, 795–807, 10.1016/j.cell.2005.04.026 (2005). [DOI] [PubMed] [Google Scholar]
- Yu J. Y., Kanai M. I., Demir E., Jefferis G. S. & Dickson B. J. Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr Biol 20, 1602–1614, 10.1016/j.cub.2010.08.025 (2010). [DOI] [PubMed] [Google Scholar]
- Kimura K., Ote M., Tazawa T. & Yamamoto D. Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature 438, 229–233, 10.1038/nature04229 (2005). [DOI] [PubMed] [Google Scholar]
- Goto J., Mikawa Y., Koganezawa M., Ito H. & Yamamoto D. Sexually dimorphic shaping of interneuron dendrites involves the hunchback transcription factor. J Neurosci 31, 5454–5459, 10.1523/JNEUROSCI.4861-10.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailey D. A., Taylor B. J. & Hall J. C. Elements of the fruitless locus regulate development of the muscle of Lawrence, a male-specific structure in the abdomen of Drosophila melanogaster adults. Development 113, 879–890 (1991). [DOI] [PubMed] [Google Scholar]
- Nojima T., Kimura K., Koganezawa M. & Yamamoto D. Neuronal synaptic outputs determine the sexual fate of postsynaptic targets. Curr Biol 20, 836–840, 10.1016/j.cub.2010.02.064 (2010). [DOI] [PubMed] [Google Scholar]
- Usui-Aoki K. et al. Formation of the male-specific muscle in female Drosophila by ectopic fruitless expression. Nat Cell Biol 2, 500–506, 10.1038/35019537 (2000). [DOI] [PubMed] [Google Scholar]
- Ryner L. C. et al. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell 87, 1079–1089 (1996). [DOI] [PubMed] [Google Scholar]
- Goodwin S. F. et al. Aberrant splicing and altered spatial expression patterns in fruitless mutants of Drosophila melanogaster. Genetics 154, 725–745 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G. et al. Spatial, temporal, and sexually dimorphic expression patterns of the fruitless gene in the Drosophila central nervous system. J Neurobiol 43, 404–426 (2000). [DOI] [PubMed] [Google Scholar]
- Billeter J. C. & Goodwin S. F. Characterization of Drosophila fruitless-gal4 transgenes reveals expression in male-specific fruitless neurons and innervation of male reproductive structures. J Comp Neurol 475, 270–287, 10.1002/cne.20177 (2004). [DOI] [PubMed] [Google Scholar]
- Lee G., Villella A., Taylor B. J. & Hall J. C. New reproductive anomalies in fruitless-mutant Drosophila males: extreme lengthening of mating durations and infertility correlated with defective serotonergic innervation of reproductive organs. J Neurobiol 47, 121–149, 10.1002/neu.1021 (2001). [DOI] [PubMed] [Google Scholar]
- Song H. J. et al. The fruitless gene is required for the proper formation of axonal tracts in the embryonic central nervous system of Drosophila. Genetics 162, 1703–1724 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter J. C. et al. Isoform-specific control of male neuronal differentiation and behavior in Drosophila by the fruitless gene. Curr Biol 16, 1063–1076, 10.1016/j.cub.2006.04.039 (2006). [DOI] [PubMed] [Google Scholar]
- Ito H. et al. Fruitless recruits two antagonistic chromatin factors to establish single-neuron sexual dimorphism. Cell 149, 1327–1338, 10.1016/j.cell.2012.04.025 (2012). [DOI] [PubMed] [Google Scholar]
- Ito H., Sato K. & Yamamoto D. Sex-switching of the Drosophila brain by two antagonistic chromatin factors. Fly (Austin) 7, 87–91, 10.4161/fly.24018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton J. E. et al. Male-specific fruitless isoforms have different regulatory roles conferred by distinct zinc finger DNA binding domains. BMC Genomics 14, 659, 10.1186/1471-2164-14-659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapeau M. D., Radovic A., Wittkopp P. J. & Long A. D. A gene necessary for normal male courtship, yellow, acts downstream of fruitless in the Drosophila melanogaster larval brain. J Neurobiol 55, 53–72, 10.1002/neu.10196 (2003). [DOI] [PubMed] [Google Scholar]
- Dauwalder B., Tsujimoto S., Moss J. & Mattox W. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev 16, 2879–2892, 10.1101/gad.1010302 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M. C. et al. Male-specific fruitless isoforms target neurodevelopmental genes to specify a sexually dimorphic nervous system. Curr Biol 24, 229–241, 10.1016/j.cub.2013.11.035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E. et al. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A 100, 7480–7485, 10.1073/pnas.1332608100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137, 10.1186/gb-2008-9-9-r137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. W., Peterson M. & Liu H. Essential role of postsynaptic NMDA receptors in developmental refinement of excitatory synapses. Proc Natl Acad Sci U S A 110, 1095–1100, 10.1073/pnas.1212971110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayyazuddin A., Zaheer M. A., Hiesinger P. R. & Bellen H. J. The nicotinic acetylcholine receptor Dalpha7 is required for an escape behavior in Drosophila. PLoS Biol 4, e63, 10.1371/journal.pbio.0040063 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura S. Y. et al. Cholinergic circuits integrate neighboring visual signals in a Drosophila motion detection pathway. Curr Biol 21, 2077–2084, 10.1016/j.cub.2011.10.053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer E. P., Iyer S. C., Sulkowski M. J. & Cox D. N. Isolation and purification of Drosophila peripheral neurons by magnetic bead sorting. J Vis Exp, 10.3791/1599 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddall N. A., Hime G. R., Pollock J. A. & Batterham P. Ttk69-dependent repression of lozenge prevents the ectopic development of R7 cells in the Drosophila larval eye disc. BMC Dev Biol 9, 64, 10.1186/1471-213X-9-64 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. J. Differentiation of a male-specific muscle in Drosophila melanogaster does not require the sex-determining genes doublesex or intersex. Genetics 132, 179–191 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol 27, 353–365 (1972). [PubMed] [Google Scholar]
- Zhang Y. et al. Expression in aneuploid Drosophila S2 cells. PLoS Biol 8, e1000320, 10.1371/journal.pbio.1000320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D. et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol Chapter 19, Unit 19 10 11–21, doi:10.1002/0471142727.mb1910s89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R. et al. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol 8, R129, 10.1186/gb-2007-8-7-r129 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes S. C. et al. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS genetics 7, e1002145, 10.1371/journal.pgen.1002145 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J. et al. DOG 1.0: illustrator of protein domain structures. Cell Res 19, 271–273, 10.1038/cr.2009.6 (2009). [DOI] [PubMed] [Google Scholar]
- Hulsen T., de Vlieg J. & Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488, 10.1186/1471-2164-9-488 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Supplementary Tables 1-3
Supplementary Tables 4-6
Supplementary Table 8-10
Supplementary Table 11
Supplementary Tables 12
Supplementary Table 14
Supplementary Tables 15
Supplementary Tables 16-18