Abstract
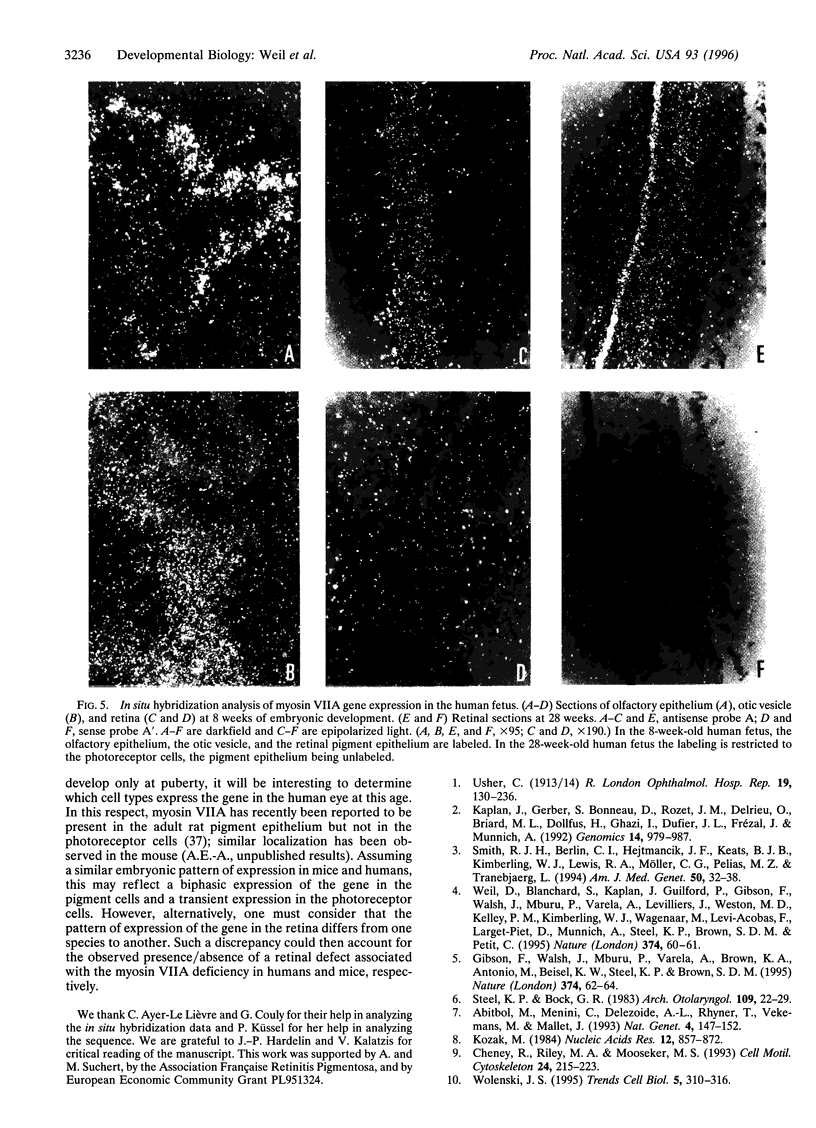
The gene encoding human myosin VIIA is responsible for Usher syndrome type III (USH1B), a disease which associates profound congenital sensorineural deafness, vestibular dysfunction, and retinitis pigmentosa. The reconstituted cDNA sequence presented here predicts a 2215 amino acid protein with a typical unconventional myosin structure. This protein is expected to dimerize into a two-headed molecule. The C terminus of its tail shares homology with the membrane-binding domain of the band 4.1 protein superfamily. The gene consists of 48 coding exons. It encodes several alternatively spliced forms. In situ hybridization analysis in human embryos demonstrates that the myosin VIIA gene is expressed in the pigment epithelium and the photoreceptor cells of the retina, thus indicating that both cell types may be involved in the USH1B retinal degenerative process. In addition, the gene is expressed in the human embryonic cochlear and vestibular neuroepithelia. We suggest that deafness and vestibular dysfunction in USH1B patients result from a defect in the morphogenesis of the inner ear sensory cell stereocilia.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abitbol M., Menini C., Delezoide A. L., Rhyner T., Vekemans M., Mallet J. Nucleus basalis magnocellularis and hippocampus are the major sites of FMR-1 expression in the human fetal brain. Nat Genet. 1993 Jun;4(2):147–153. doi: 10.1038/ng0693-147. [DOI] [PubMed] [Google Scholar]
- Adams R. J., Pollard T. D. Membrane-bound myosin-I provides new mechanisms in cell motility. Cell Motil Cytoskeleton. 1989;14(2):178–182. doi: 10.1002/cm.970140203. [DOI] [PubMed] [Google Scholar]
- Ashmore J. F., Kolston P. J. Hair cell based amplification in the cochlea. Curr Opin Neurobiol. 1994 Aug;4(4):503–508. doi: 10.1016/0959-4388(94)90050-7. [DOI] [PubMed] [Google Scholar]
- Cheney R. E., O'Shea M. K., Heuser J. E., Coelho M. V., Wolenski J. S., Espreafico E. M., Forscher P., Larson R. E., Mooseker M. S. Brain myosin-V is a two-headed unconventional myosin with motor activity. Cell. 1993 Oct 8;75(1):13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- Cheney R. E., Riley M. A., Mooseker M. S. Phylogenetic analysis of the myosin superfamily. Cell Motil Cytoskeleton. 1993;24(4):215–223. doi: 10.1002/cm.970240402. [DOI] [PubMed] [Google Scholar]
- Espreafico E. M., Cheney R. E., Matteoli M., Nascimento A. A., De Camilli P. V., Larson R. E., Mooseker M. S. Primary structure and cellular localization of chicken brain myosin-V (p190), an unconventional myosin with calmodulin light chains. J Cell Biol. 1992 Dec;119(6):1541–1557. doi: 10.1083/jcb.119.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George E. L., Ober M. B., Emerson C. P., Jr Functional domains of the Drosophila melanogaster muscle myosin heavy-chain gene are encoded by alternatively spliced exons. Mol Cell Biol. 1989 Jul;9(7):2957–2974. doi: 10.1128/mcb.9.7.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson F., Walsh J., Mburu P., Varela A., Brown K. A., Antonio M., Beisel K. W., Steel K. P., Brown S. D. A type VII myosin encoded by the mouse deafness gene shaker-1. Nature. 1995 Mar 2;374(6517):62–64. doi: 10.1038/374062a0. [DOI] [PubMed] [Google Scholar]
- Halsall D. J., Hammer J. A., 3rd A second isoform of chicken brush border myosin I contains a 29-residue inserted sequence that binds calmodulin. FEBS Lett. 1990 Jul 2;267(1):126–130. doi: 10.1016/0014-5793(90)80305-3. [DOI] [PubMed] [Google Scholar]
- Hasson T., Heintzelman M. B., Santos-Sacchi J., Corey D. P., Mooseker M. S. Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc Natl Acad Sci U S A. 1995 Oct 10;92(21):9815–9819. doi: 10.1073/pnas.92.21.9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson T., Mooseker M. S. Porcine myosin-VI: characterization of a new mammalian unconventional myosin. J Cell Biol. 1994 Oct;127(2):425–440. doi: 10.1083/jcb.127.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden S. M., Wolenski J. S., Mooseker M. S. Binding of brush border myosin I to phospholipid vesicles. J Cell Biol. 1990 Aug;111(2):443–451. doi: 10.1083/jcb.111.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz J. A., Hammer J. A., 3rd A new Acanthamoeba myosin heavy chain. Cloning of the gene and immunological identification of the polypeptide. J Biol Chem. 1990 Nov 25;265(33):20646–20652. [PubMed] [Google Scholar]
- Houdusse A., Cohen C. Target sequence recognition by the calmodulin superfamily: implications from light chain binding to the regulatory domain of scallop myosin. Proc Natl Acad Sci U S A. 1995 Nov 7;92(23):10644–10647. doi: 10.1073/pnas.92.23.10644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudspeth A. J., Gillespie P. G. Pulling springs to tune transduction: adaptation by hair cells. Neuron. 1994 Jan;12(1):1–9. doi: 10.1016/0896-6273(94)90147-3. [DOI] [PubMed] [Google Scholar]
- Kaplan J., Gerber S., Bonneau D., Rozet J. M., Delrieu O., Briard M. L., Dollfus H., Ghazi I., Dufier J. L., Frézal J. A gene for Usher syndrome type I (USH1A) maps to chromosome 14q. Genomics. 1992 Dec;14(4):979–987. doi: 10.1016/s0888-7543(05)80120-x. [DOI] [PubMed] [Google Scholar]
- Kellerman K. A., Miller K. G. An unconventional myosin heavy chain gene from Drosophila melanogaster. J Cell Biol. 1992 Nov;119(4):823–834. doi: 10.1083/jcb.119.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991 May 24;252(5009):1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mercer J. A., Seperack P. K., Strobel M. C., Copeland N. G., Jenkins N. A. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature. 1991 Feb 21;349(6311):709–713. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- Mullen R. J., LaVail M. M. Inherited retinal dystrophy: primary defect in pigment epithelium determined with experimental rat chimeras. Science. 1976 May 21;192(4241):799–801. doi: 10.1126/science.1265483. [DOI] [PubMed] [Google Scholar]
- Rayment I., Holden H. M., Whittaker M., Yohn C. B., Lorenz M., Holmes K. C., Milligan R. A. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993 Jul 2;261(5117):58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- Rayment I., Rypniewski W. R., Schmidt-Bäse K., Smith R., Tomchick D. R., Benning M. M., Winkelmann D. A., Wesenberg G., Holden H. M. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993 Jul 2;261(5117):50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Rees D. J., Ades S. E., Singer S. J., Hynes R. O. Sequence and domain structure of talin. Nature. 1990 Oct 18;347(6294):685–689. doi: 10.1038/347685a0. [DOI] [PubMed] [Google Scholar]
- Reizes O., Barylko B., Li C., Südhof T. C., Albanesi J. P. Domain structure of a mammalian myosin I beta. Proc Natl Acad Sci U S A. 1994 Jul 5;91(14):6349–6353. doi: 10.1073/pnas.91.14.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert C., Kroschewski R., Bähler M. Identification, characterization and cloning of myr 1, a mammalian myosin-I. J Cell Biol. 1993 Mar;120(6):1393–1403. doi: 10.1083/jcb.120.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Smith R. J., Berlin C. I., Hejtmancik J. F., Keats B. J., Kimberling W. J., Lewis R. A., Möller C. G., Pelias M. Z., Tranebjaerg L. Clinical diagnosis of the Usher syndromes. Usher Syndrome Consortium. Am J Med Genet. 1994 Mar 1;50(1):32–38. doi: 10.1002/ajmg.1320500107. [DOI] [PubMed] [Google Scholar]
- Steel K. P., Bock G. R. Hereditary inner-ear abnormalities in animals. Relationships with human abnormalities. Arch Otolaryngol. 1983 Jan;109(1):22–29. doi: 10.1001/archotol.1983.00800150026005. [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Kawashima A., Nagafuchi A., Tsukita S. Structural diversity of band 4.1 superfamily members. J Cell Sci. 1994 Jul;107(Pt 7):1921–1928. doi: 10.1242/jcs.107.7.1921. [DOI] [PubMed] [Google Scholar]
- Tsukita S., Oishi K., Sato N., Sagara J., Kawai A., Tsukita S. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J Cell Biol. 1994 Jul;126(2):391–401. doi: 10.1083/jcb.126.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolenski J. S. Regulation of calmodulin-binding myosins. Trends Cell Biol. 1995 Aug;5(8):310–316. doi: 10.1016/s0962-8924(00)89053-4. [DOI] [PubMed] [Google Scholar]
- Xie X., Harrison D. H., Schlichting I., Sweet R. M., Kalabokis V. N., Szent-Györgyi A. G., Cohen C. Structure of the regulatory domain of scallop myosin at 2.8 A resolution. Nature. 1994 Mar 24;368(6469):306–312. doi: 10.1038/368306a0. [DOI] [PubMed] [Google Scholar]