Abstract
Congenital heart disease (CHD) affects nearly 1 % of the population. It is a complex disease, which may be caused by multiple genetic and environmental factors. Studies in human genetics have led to the identification of more than 50 human genes, involved in isolated CHD or genetic syndromes, where CHD is part of the phenotype. Furthermore, mapping of genomic copy number variants and exome sequencing of CHD patients have led to the identification of a large number of candidate disease genes. Experiments in animal models, particularly in mice, have been used to verify human disease genes and to gain further insight into the molecular pathology behind CHD. The picture emerging from these studies suggest that genetic lesions associated with CHD affect a broad range of cellular signaling components, from ligands and receptors, across down-stream effector molecules to transcription factors and co-factors, including chromatin modifiers.
Keywords: Congenital heart disease, CHD, Disease genes, Copy number variants, CNVs
Introduction
Congenital heart disease (CHD) is the most prevalent birth defect, with a postnatal incidence of 0.8 % [1] and an approximately tenfold higher prenatal incidence [2]. CHD is a group of structural abnormalities of the heart, which include septal defects, valve defects and lesions affecting the outflow tract. The treatment of CHD has improved during the last 50 years, and today 95 % of CHD patients survive to adulthood, which has resulted in a growing population of adults living with CHD [3].
The etiology of CHD is complex and is associated with both environmental and genetic causes. Genetically, CHD is a very heterogeneous disease; 55 human disease genes have been identified so far (Table 1 and text below), however, experiments with targeted deletions in mice have revealed more than 500 genes which lead to heart defects when mutated (http://www.informatics.jax.org/). Thus it is likely that at least the same number of human CHD disease genes exist.
Table 1.
Genes associated with CHD via intragenic mutations
| HCNG gene symbol (alternative symbol) | Protein functiona | Type of CHDb | Reference |
|---|---|---|---|
| Genes encoding transription factors | |||
| CITED2 | Transcriptional co-activator | I | [18] |
| FOXH1 | Forkhead box TF | I | [29] |
| FOXP1 | Forkhead box TF | I | [246] |
| GATA4 | GATA-binding TF | I | [89–91] |
| GATA6 | GATA-binding TF | I | [247, 248] |
| IRX4 | Iroquois homeobox TF | I | [249] |
| MED13L | Multiprotein coactivator subunit | I | [250] |
| NKX2-5 | Homeobox TF | I | [69–71] |
| NKX2-6 | Homeobox TF | I | [251] |
| TBX1 | T-box TF | S (DiGeorge syndrome) | [156] |
| TBX5 | T-box TF | S (Holt–Oram syndrome) | [102, 103] |
| TBX20 | T-box TF | I | [104] |
| SALL4 | Zinc finger TF | I, S (Duane-radial ray syndrome) | [252–255] |
| TFAP2B | AP-2 TF | I, S (Char syndrome) | [110–112] |
| ZFPM2 | Zinc finger TF | I | [255, 256] |
| ZIC3 | Zinc finger TF | HTX | [19] |
| Genes involved in cell signaling | |||
| ACVR1 | Activin receptor, type 1 | I | [257] |
| ACVR2B | Activin receptor 2B | HTX | [14] |
| BRAF | Serine/threonine protein kinase | S (NS, LS, CFC) | [56, 64] |
| CBL | E3 ubiquitin ligase | S (NS-like) | [66] |
| CFC1 | Ligand (EGF family) | HTX | [17] |
| GDF1 | Ligand (BMP/TGFbeta family) | HTX | [16] |
| HRAS | RAS GTPase | S (Costello syndrome) | [63] |
| JAG1 | NOTCH ligand | S (Alagille syndrome) | [37, 38] |
| LEFTY2 (LEFTYA) | Ligand (BMP/TGFbeta family) | HTX | [14] |
| KRAS | RAS GTPase | S (NS, CFC) | [56, 57] |
| MAP2K1 (MEK1) | MAP kinase kinase | S (CFC) | [64] |
| MAP2K2 (MEK2) | MAP kinase kinase | S (CFC) | [64] |
| NF1 | Negative regulator of RAS-MAPK signalling | S (neurofibromatosis-NS) | [67] |
| NRAS | RAS GTPase | S (NS) | [62] |
| NODAL | Ligand (BMP/TGFbeta family) | HTX | [12, 13] |
| NOTCH1 | NOTCH receptor | I | [45–47] |
| NOTCH2 | NOTCH receptor | S (Alagille syndrome) | [41, 42] |
| PTGFRA | PTGFRα receptor | I | [258] |
| PTPN11 | Protein tyrosine phosphatase | S (NS) | [55] |
| RAF1 | MAP kinase kinase kinase | S (NS, LS) | [60, 61] |
| RIT1 | Ras-related GTPase | S (NS) | [259] |
| SHOC2 | RAS-MAPK modulator | S (NS) | [65] |
| SMAD6 | BMP/TGFbeta modulator | I | [260] |
| SOS1 | Guanine nucleotide exchange factor (RAS-MAPK pathway) | S (NS) | [58, 59] |
| TAB2 | Activator of MAP3K7 (TAK1) | I | [236] |
| TDGF1 | Co-receptor for TGF-β ligands | I | [29] |
| Genes encoding structural proteins | |||
| ACTC1 | Cardiac α-actinin | I | [124] |
| ELN | Elastin | Ic | [184] |
| MYH6 | Cardiac myosin HC | I | [118–120] |
| MYH7 | Cardiac myosin HC | I | [123] |
| MYH11 | Smooth muscle myosin HC | I | [125] |
| Genes encoding epigenetic regulators | |||
| CHD7 | Binding to H3K4Me3 | S (CHARGE syndrome) | [134, 135, 137] |
| KMT2D (MLL2) | H3K4 methyltransferase | S (Kabuki syndrome) | [132] |
| EP300 | Histone acetyltransferase | S (Rubinstein–Taybi syndrome) | [117] |
| CREBBP | Histone acetyltransferase | S (Rubinstein–Taybi syndrome) | [116] |
| EHMT1 | H3K9 methyltransferase | S (Kleefstra syndrome) | [205, 208] |
| Other genes | |||
| CRELD1 | Cell adhesion | I | [261] |
| MCTP2 d | Possible role in Ca2+ signalling | I | [262] |
| NPHP4 | Ciliary protein | I, HTX | [20] |
The list include genes, which have been associated with CHD by identification of mutations in two or more unrelated patient and/or genes where human genetic analyses are complemented with functional analyses
a TF transcription factor, HC heavy chain
b I isolated CHD, S syndromic CHD, HTX heterotaxy, NS Noonan syndrome, LS LEOPARD syndrome, CFC Cardiofaciocutaneous syndrome
cGenomic deletions, which include ELN cause Williams–Beuren syndrome
dIntragenic genomic duplication causing premature truncation at p.F697X
During the last two decades, linkage analysis has been used to successfully identify CHD disease genes in large families, segregating isolated CHD and genetic syndromes, where CHD is part of the phenotypic spectrum (syndromic CHD). Furthermore, fine-mapping of genomic copy number variants (CNVs) in patients with isolated CHD, or CHD in combination with additional birth defects, have been used to identify candidate disease genes. Follow-up studies of candidate genes in animal models, particularly in mice, have been very successful in validating the candidates, and to gain insight into the function of the gene products in heart development.
Identifying disease genes in CHD is critically important to understand the disease. Identification of a novel disease gene or a causative pathway will enhance current knowledge of the molecular biology involved in human cardiac development, and the molecular pathology underlying CHD. Such knowledge may lead to new preventive strategies and perhaps new treatments. Furthermore, such knowledge may also increase our understanding of the factors involved in cardiomyogenic stem cell differentiation, and may thus aid in the development of regenerative therapy for treatment of myocardial infarction. In familial cases of CHD, identification of disease genes will benefit the genetic counseling process for CHD-afflicted families. Such knowledge is particularly important for the growing population of adults with CHD, due to the high recurrence risk of certain forms of CHD [4].
In this review, we aim to summarize current knowledge in the molecular genetics of CHD, from the perspective of, disease gene identification efforts in humans, and functional analyses of disease genes in animal models.
Part I: genes associated with congenital heart disease
Below we discuss genes associated with syndromic and isolated CHD, juxtaposing studies from multiple model systems to clarify why errors in the underlying molecular machinery manifests themselves as congenital heart defects.
Genes associated with laterality defects
The heart is the first organ to break the bilateral symmetry of the developing embryo. During early embryogenesis, left–right asymmetry of the body-axis is established via intricate cross-talk amongst signaling pathways such as Notch, Nodal, Hedgehog, FGF and BMP, ultimately restricting NODAL signaling to the left side of the embryo. The nodal cilia model is the predominant model to elucidate induction of embryonic asymmetry in the developing embryo, although other models have been proposed [5]. Briefly, nodal cilia in the node of the primitive streak produce a directional fluid flow which induces left–right asymmetry by delivering morphogens to the left side of the embryo and/or by acting on mechanosensory cilia [6–9]. Ultimately, laterality cues are relayed downstream to ensure left-sided expression of the PITX2 transcription factor in the lateral plate mesoderm [10], a critical component in determining organ laterality [11]. NODAL [12, 13], LEFTY2 [14], ACVR2B [15], GDF1 [16] CFC1 [17], CITED2 [18] and ZIC3 [19] have all been localized to the laterality signaling pathway. Albeit human mutations in these genes show a wide range of heart malfunctions, many cluster around laterality defects such as heterotaxy and faulty looping of the heart. Interestingly, a recent study identified mutations in Nephronophthisis-4 (NPHP4), a cilia related gene, and linked them to a variety of cardiac laterality defects such as transposition of the great arteries (TGA), atrioventricular septal defects (AVSD), double outlet right ventricle (DORV), dextrocardia and abnormal pulmonary venous return. Laterality defects of the abdominal organs were also observed. Morpholino knock down of nphp4 in zebrafish resulted in reverse orientation or faulty looping of the heart [20]. Previous studies have also connected other NPHP family members to inborn heart defects and laterality deficiencies [21, 22].
The ZIC3 gene encodes a zinc finger transcription factor known to cause cardiovascular defects when mutated in humans. Mutations in ZIC3 cause X-linked familial heterotaxy but are also found in sporadic cases of heterotaxy and isolated CHDs [19, 23, 24]. Null and heterozygous Zic3 mice display a variety of cardiac defects such as TGA, interrupted aortic arch (IAA), atrial septum defect (ASD) and ventricular septal defect (VSD) in combination with various other developmental anomalies, thus resulting in a phenocopy of the clinical spectrum of malfunctions found in humans with heterotaxy [25]. How mutations in ZIC3 result in faulty heart looping is currently unknown. However, recent reports place ZIC3 upstream in the Nodal signal cascade [25, 26] with conditional loss-of-function studies showing that ZIC3 is required in the migrating mesoderm but not for heart progenitors and in the heart compartment [27].
Recently, the transcription factor FOXH1 was outlined as a possible signaling intersection between BMP and Nodal signaling to establish left/right asymmetry [28]. Mutations in FOXH1 have been linked to human heart defects [29], as well as the lack of outflow tract and right ventricle is seen in Foxh1 −/− mouse embryos [30].
In humans, mutations in the transcriptional co-activator CITED2 (Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain 2) are associated with laterality defects and cardiac anomalies such as septal defects and TGA [18]. Mice deficient in Cited2 die during gestation expressing partially penetrant laterality defects and fully penetrant heart defects [31, 32]. The heart anomalies include ASD, VSD, common atrioventricular canal (CAVC), DORV and IAA type B [32, 33]. Recently, Lopes et al. [31] showed that specific deletions of Cited2 in heart progenitors do not produce heart defects and that the cardiac malfunctions seen in Cited2 −/− embryos arise during the early phases of establishing the left–right body axis in close relation to NODAL signaling.
Genes encoding components of signaling pathways
Animal models have illustrated that cardiac development involves spatial and temporal coordination of a number of signaling pathways [34, 35]. The identification of disease genes in syndromic and isolated CHD has confirmed the involvement of a subgroup of these pathways in human heart development, and has further contributed new information about additional pathways.
The NOTCH signaling pathway acts locally as a cell-fate regulator, and as a patterning signal effector in many developmental processes. Its activity includes left–right axis partitioning and heart morphogenesis. For an extensive review on Notch signaling in cardiac development see [36].
Identification of mutations in JAG1, encoding a NOTCH signaling ligand, in patients with Alagille syndrome provided the first link between NOTCH signaling and human CHD [37, 38]. Alagille syndrome (AGS, OMIM #118450) is a multisystem disorder, which involves the liver, heart, eyes, face and skeleton [39]. Approximately 90 % of these patients have cardiovascular anomalies, often presenting in the form of stenosis in the pulmonary artery branch, valvular pulmonary stenosis (PS) and tetralogy of Fallot (TOF) [40]. The majority (>90 %) of AGS cases are caused by mutations in JAG1, however select (<1 %) cases are caused by mutation in NOTCH2 [41, 42]. Mice homozygous for targeted deletion of Jag1 die during embryonic development possibly due to vascular defects while heterozygous Jag1 −/+ mice display ocular defects [43].
In contrast, doubly heterozygous Jag1 +/−, Notch2 +/− mice exhibit multiorgan abnormalities characteristic of AGS, supporting a genetic interaction between JAG1 and NOTCH2 in AGS [44].
Mutations in NOTCH1 have been identified in patients with isolated CHD [45–47]. Patients with NOTCH1 mutations often present malfunctions of the aortic valve. NOTCH1 signaling has been linked to endothelial-to-mesenchymal transformation (EMT), a fundamental process in the early stages of cardiac valve formation, where endocardial cells detach to become a migratory mesenchyme that forms endocardial cushions, precursors of cardiac valves. Notch1 mutant mice develop hypoplastic endocardial cushions due to impaired EMT [48]. Recently, Luna-Zurita et al. and others [48–50] outlined that a NOTCH1, WNT4 and BMP2 signal interplay between the endocardium and myocardium underlie valve morphogenesis. Furthermore, Bosse et al. [51] showed that compound mutant Notch +/−; Nos3 −/− mice display an accelerated bicuspid aortic valve phenotype compared to Notch +/− and Nos3 −/− alone, suggesting an interaction between nitric oxide (NO) and NOTCH signaling in the development of the aortic valve. In the same paper, these data were further supported by in vitro data, suggesting that NO regulates Notch signaling in aortic valve interstitial cells.
Signal transduction through the RAS-mitogen activated protein kinase (MAPK) pathway can stimulate cell proliferation, differentiation, survival and metabolism. Identification of disease genes in Noonan syndrome (OMIM #163950), Costello syndrome (OMIM #218040), LEOPARD syndrome (OMIM #151100), Cardio-facio-cutaneous (CFC) syndrome (OMIM #115150) and a few other syndromes with distinct but overlapping phenotypes, collectively known as RASopathies (see [52] for review), have firmly established a link between the RAS-MAPK signal transduction pathway and human CHD. The RASopathies are manifested by a wide range of multisystem anomalies, including CHD. In Noonan syndrome approximately 85 % of patients have a variety of cardiac defects, most commonly including pulmonary valve stenosis, ASD and hypertrophic cardiomyopathy [53, 54].
Linkage analysis and mutation screening has identified mutations in PTPN11 as the cause of approximately half of cases with Noonan syndrome [55]. Subsequently, mutations in KRAS [56, 57], SOS1 [58, 59], RAF1 [60, 61], NRAS [62], HRAS [63], BRAF [56, 64], SHOC2 [65], MAP2K1 (MEK1) and MAP2K2 (MEK2) [64], CBL [66] and NF1 [67] have been associated with RASopathies in which CHD are observed (Table 1).
Genes encoding cardiac transcription factors
Cardiac developmental signals are conveyed to transcriptional circuits that regulate gene expression during normal heart development. At the heart of these transcriptional networks lie a set of core transcription factors many of which are associated with isolated CHD (Table 1). Transcriptional focal points include NKX2-5, GATA4 and TBX5. These transcription factors interact at cardiac promoters in synergistic fashions (see below). Their function and molecular signatures have been thoroughly described and excellently reviewed elsewhere, and will only be briefly mentioned here (see [68] for a recent review).
In humans, disease causing mutations in the homeodomain protein NKX2-5 result in a plethora of CHDs including ASD, VSD, TOF and DORV [69–71] Septal defects and atrio-ventricular conduction defects are commonly seen in patients with a mutated NKX2-5 gene [70]. Disruption of Tinman, the homologue of NKX2-5 in Drosophila melanogaster, results in a fruit fly devoid of the dorsal vessel, a structure analogous to the human heart [72]. Similarly, Nkx2-5 functionality is crucial in mice as homozygous mutations cause embryonic lethality due to faulty cardiac looping and insufficient myocardial differentiation during chamber formation [73, 74]. Mouse studies show that Nkx2-5 gene dosage is critically important for properly regulated development of the cardiac conduction system as Nkx2-5 null mice lack the primordium of the AV node and the conduction system of heterozygous mutant embryos only contain half the normal number of cells [75]. Additionally, Pashmforoush et al. [76] generated ventricular-restricted Nkx2-5 knockout mice that display progressive complete heart block and massive trabecular muscle overgrowth.
NKX2-5 ranks high in the cardiac regulatory hierarchy and is expressed in both the first- and second heart field (SHF) [77]. Its expression is closely coordinated through GATA factors, SMAD proteins and by self-autoregulation [78–81]. Proliferation of the SHF and outflow tract (OFT) morphology is regulated by Nkx2-5 feedback repression of BMP2/SMAD1 signaling [82]. It was recently shown that JARID2, which is also implicated in OFT development, is a direct target of NKX2-5 regulation [83]. Furthermore, it has been demonstrated that Nkx2-5 interacts with Gata4 within cardiac promoters, suggesting that the proteins cooperate in the transcriptional activation of cardiac target genes [84, 85]. Nkx2-5 interacts with Tbx5 in vitro and the two proteins were shown to activate a cardiac-specific Nppa promoter in a synergistic fashion [86] and Nkx2-5 cooperates with Tbx5 in development of the cardiac conduction system in vivo [87].
GATA binding protein 4 (GATA4) plays a pivotal role near the top of the transcriptional cascades that control heart development (see [88] for a recent review). In humans, the cardiac defects found in patients with interstitial deletions in 8p23.1 are attributed to haploinsufficiency of GATA4 (see below). Intragenic GATA4 mutations can also cause isolated CHDs, primarily cardiac septal defects, but PS, TOF and other defects have been reported [89–91]. In a recent paper human missense mutations in GATA4 were shown to disrupt GATA4–SMAD4 interactions in the BMP/TGF-β signaling pathway, likely causing AVSD and valve abnormalities in the affected patients [92]. Embryonic development in Gata4 deficient mice is arrested at E10.5 with incorrect ventral folding, endodermal malfunctions and an inability to establish a primitive heart tube [93, 94]. Correct Gata4 dosage is critically important for normal heart development, as mice homozygous for a hypomorphic allele develop CAVC, DORV and a hypoplastic ventricular myocardium [95]. Furthermore, it has also been shown that mice heterozygous for GATA4 mutations develop septation and endocardial cushion defects [90]. Heterozygous knock-in mice harbouring a Gata4 G296S mutation, previously identified in patients with septum defects and pulmonary valve stenosis, display ASD and semilunar valve stenosis [96].
GATA4 interacts with numerous transcription factors that promote cardiogenesis [88]. Direct downstream targets of GATA4 include HAND2 and MEF2C required for SHF development [97, 98]. GATA4 and TBX5 proteins physically interact and this interaction is disrupted by mutations in GATA4 [89]. Furthermore, Gata4 and Tbx5 double heterozygous mice develop cardiovascular defects, which point towards a genetic interaction between the two [99].
The T-box transcription factors are important cardiac transcription factors. They are involved in fundamental cardiac developmental processes, including development of the chamber myocardium, outflow tract and the conduction system [100]. TBX1 regulates proliferation of cardiac progenitors in the SHF and haploinsufficiency of TBX1 is considered the primary cause of CHD in patients with DiGeorge syndrome (see below). TBX5 participates in regulation of gene expression in the developing chamber myocardium and conduction system [101]. Mutations in TBX5 cause Holt–Oram syndrome (OMIM #142900), a syndrome distinguished by upper limb defects and heart defects—primarily septal and conduction defects [102, 103]. Tbx5 null mice possess a deformed linear heart tube and underdeveloped atria while heterozygous Tbx5 mice model heart and limb abnormalities observed in Holt–Oram syndrome, potentially explaining cardiac conduction system defects seen in these patients [101].
Human mutations in TBX20 cause aberrant valvulogenesis, septal defects, TOF and cardiomyopathy [104]. Deletion of Tbx20 in mice generates a linear heart tube which fails to loop properly and exhibits insufficient chamber formation [105]. Heterozygous Tbx20 mice show onset of dilated cardiomyopathy recapitulating some of the human defects [106]. Recent papers by Cai and co-workers [107, 108] place TXB20 in the formation of the cardiac atrioventricular canal in a complex signaling network involving TBX20, TBX2 and BMP2.
In vitro, transcription factor AP-2gamma (TFAP2C) has been shown to bind the TBX20 promoter site and repress TBX20 expression [109]. Interestingly, mutations in TFAP2B causes Char syndrome (OMIM #169100) characterized by facial dysmorphism, anomalies of the fifth finger and patent ductus arteriosus (PDA) [110]. Isolated PDA has also been linked to mutations in TFAP2B [111, 112] and a recent Tfap2B knock out study in mice reported phenotypes resembling the characteristics of Char syndrome [113]. TFAP2 isoforms form a complex with CITED2, CREBBP and EP300 [114, 115]. Mutations in the transcriptional co-activators CREBBP and EP300 are associated with Rubinstein–Taybi syndrome (OMIM #180849) displaying mental retardation, broad thumbs and toes, facial abnormalities, and in some cases, CHD [116, 117].
Genes encoding components of the cardiac sarcomere
Mutations in genes encoding cardiac structural proteins have also been connected to CHD. Several studies link mutations in the cardiac sarcomeric protein MYH6 (myosin heavy chain 6) to ASD [118–120]. Morpholino knock down of myh6 in the developing chicken heart implies that its functionality is required in the formation of the atrial septum [120]. Molecular regulation of MYH6 expression involves transcription factors such as GATA4 [121], TBX5 and MEF2C [122].
Other members of the contractile units in cardiovascular muscle include MYH7 and ACTC1. A mutation in MYH7 encoding myosin heavy chain 7 was shown to cause CHDs such as Ebstein’s anomaly and septal defects [123]. Mutations in ACTC1 encoding the human α-cardiac muscle actin can cause ASD and morpholino knock down of Actc1 causes looping and atrial septal anomalies in chicken embryos [124].
Mutations in MYH11, encoding the major contractile protein of smooth muscle cells can cause thoracic aortic aneurysm and/or aortic dissection and PDA [125]. Mice homozygous for deletion of Myh11 show a delayed closure of the ductus arteriosus [126], which is connected to the shunting functions of smooth muscle cells upon birth [127, 128].
Genes encoding chromatin modifiers
Analysis of model organisms has shown that dynamic modification of chromatin structure serves as an important regulator of gene expression during heart development (reviewed in [129]). Genes that encode proteins which modify or bind to histones have been implicated as disease genes in syndromes causing heart defects. This evidence supports a functional link between chromatin modification and human heart development and defects.
Kabuki syndrome (OMIM #147920) is characterized by intellectual disability, craniofacial anomalies, skeletal and hand malformations. Abnormal organ development is also recurrent and includes CHD in approximately 50 % of the cases [130]. Heart defects usually present in the form of septal defects and CoA [131]. Recently, Ng et al. [132] used exome sequencing to identify mutation of KMT2D (MLL2) as a major cause of Kabuki syndrome. KMT2D encodes a histone methyltransferase involved in di- and tri-methylation of the Lys-4 position of histone H3, which marks actively transcribed genes [133].
CHARGE syndrome (OMIM #214800) is characterized by growth retardation and malformation of eyes, ears, genitals, choanae and heart defects—often in the form of outflow tract malformations [134–136]. Approximately two-thirds of the cases are caused by mutation of CHD7, which encodes a member of the chromodomain helicase DNA binding (CHD) family [134, 135, 137]. In vitro studies have shown that CHD7 binds DNA regions which correlate closely to regions of H3K4 methylation and regions with characteristics of enhancer elements. This hints that the protein is involved in transcriptional activation [138]. Recently, it was shown that CHD7 controls core components of the transcriptional circuit of neural crest cells and that CHD7 is essential for neural crest cell migration [139]. This function may explain the high frequency of outflow tract defects in CHARGE syndrome, as neural crest cells are known to play a crucial role in septation of the cardiac outflow tract [140].
In a recent study, Zaidi et al. [141] conducted a comprehensive screening of all protein coding genes in hundreds of patients with severe forms of CHD. In this study, whole-exomes of 362 children with CHD and their healthy parents were screened for de novo nucleotide variants by next-generation sequencing (NGS). De novo variants from these parent-offspring trios were compared to de novo variants identified in 264 healthy parent-offspring trios. The authors performed transcriptome profiling experiments to identify genes with a high expression in mouse embryonic hearts (HHE genes). In the trio datasets, they compared the number of de novo variants in genes, homologous to HHE genes. This comparison yielded a significant higher rate of de novo mutations in CHD patients compared to controls. When they compared the frequencies of damaging mutations (i.e. splice-site mutations, nonsense mutations and mutations introducing frameshift) between the two groups, the differences were even more pronounced, with an odds ratio of 7.5 (p = 0.001). Interestingly, GeneOntology analysis of 249 de novo mutations identified in CHD patients revealed significant enrichment for mutations in genes involved in H3K4 methylation. Moreover, in CHD patients 27 % of the damaging mutations within HHE genes were affecting proteins involved in H2K4 or H3K27 histone modification. These data suggest that genes involved in histone-modification are significant in the pathogenesis of isolated CHD.
Human genome analysis in combination with functional analysis of candidate genes in animal models has been instrumental in identifying the genes responsible for heart defects in several microdeletion syndromes (see below). Interestingly, several of these genes also encode chromatin modifying proteins, which support the potentially significant role of epigenetic mechanisms in both isolated and syndromic CHD.
Part II: chromosomal aberrations in congenital heart disease
Microscopically visible chromosomal aberrations are present in 8–18 % of CHD patients [142–144]. Furthermore, CHD is a characteristic part of the clinical spectrum in a significant number of syndromes caused by a chromosome abnormality. The most common chromosome syndrome associated with CHD is Down syndrome [145]. Congenital heart defects are seen in 45 % of individuals with Down syndrome, with the majority of cardiac defects being AVSD, ASD and VSD [146]. Cardiac defects are also found at a high frequency in other aneuploidy syndromes, including Turner syndrome (monosomy X), Edward syndrome (trisomy 18) and Patau syndrome (trisomy 13) [147–150].
CHD is a component of the clinical spectrum in a number of syndromes caused by submicroscopic chromosomal deletions or duplications (listed in Table 2). Some of these syndromes are well-studied microdeletion syndromes, for which the molecular defect has been known for many years. In addition, several novel microdeletion and microduplication syndromes associated with CHD have recently been discovered due to the widespread use of molecular cytogenetic methods.
Table 2.
Microdeletion and microduplication syndromes with genomic copy number variation and CHD
| Syndrome | Chromosome region | Frequency of CHD among patients (%) | CHD candidate gene(s) in regiona | Function of candidate gene(s)e | Reference (OMIM #) |
|---|---|---|---|---|---|
| Microdeletion syndromes | |||||
| 8p23.1 deletion syndrome | 8p23.1 | 94 | GATA4 | GATA-binding TF | [195] |
| 17q23 microdeletion syndrome | 17q23 | 86 | TBX2 | T-box TF | [216] (613355) |
| DiGeorge syndrome | 22q11.2 | 65–75 | TBX1; CRKL | T-box TF; Tyrosine kinase | [263] (188400) |
| 1p36 deletion syndrome | 1p36 | 71 | DVL1 | WNT signaling component | [264] (607872) |
| 2q31.1 microdeletion syndrome | 2q31.1 | 70 | SP3 | Sp TF | [265] |
| Kleefstra syndrome | 9q34 | 40 | EHMT1 | H3K9 methyl transferase | [205, 206, 208] (610253) |
| 16p12.2–p11.2 microdeletion syndrome | 16p12.2–p11.2 | 60 | N/A | [266] (613604) | |
| Jacobsen syndrome | 11q23-qter | 56 | N/A | [267] (147791) | |
| Wolf–Hirschhorn syndrome | 4p16.3 | 50 | WHSC1; FGFRL1 | H3K36 methyl transferase; fibroblast growth factor receptor | [268] (194190) |
| Williams–Beuren Syndrome | 7q11.23 | 53–85 | ELN; BAZ1B | Elastin; subunit of chromatin remodeling complex | [180] (194050) |
| Smith–Magenis syndrome | 17p11.2 | 40–45 | MAPK7 b | MAP kinase | [269, 270] (182290) |
| Koolen-De Vries syndrome | 17q21.31 | 27–36 | KANSL1 c | Subunit of NSL histone acetylation complex | [210, 212] (610443) |
| 1q21.1 deletion syndrome | 1q21 | 29 | GJA5 | Connexin 40 | [197, 271] |
| Miller–Dieker lissencephaly syndrome | 17p13.3 | 22 | N/Ad | [272, 273] (247200) | |
| Sotos syndrome | 5q35 | 21 | NSD1 | H3K36 methyl transferase | [274–276] (117550) |
| Brachydactyly-mental retardation syndrome | 2q37 | 20 | HDAC4 | Histone deacetylase | [277, 278] (600430) |
| 15q13 microdeletion syndrome | 15q13 | 15 | N/A | [279, 280] (612001) | |
| Microduplication syndromes | |||||
| 16p13.3 microduplication | 16p13.3 | 40 | CREBBP | Histone acetyltransferase | [281] (613458) |
| 16p13.11 microduplication | 16p13.11 | 20 | MYH11 | Smooth muscle myosin HC | [282] |
| Potocki–Lupski syndrome | 17p11.2 | 50 | MAPK7 | MAP kinase | [283, 284] (610883) |
| 22q11.2 duplication syndrome | 22q11.2 | 15 | TBX1 | T-box TF | [285] (608363) |
| Chromosomal aneuploidy | |||||
| Patau syndrome | 47, +13 (trisomy 13) | 86 | N/A | [150] | |
| Edward syndrome | 47, +18 (trisomy 18) | 61–94 | N/A | [147, 148] | |
| Down syndrome | 47, +21 (trisomy 21) | 50 | N/A | [146] (190685) | |
aGenes causing heart defects when deleted in mice (http://www.informatics.jax.org) (bold) and/or by point mutations in additional patients with CHD as part of the clinical spectrum (underlined)
bPoint mutations in RAI1 causes Smith–Magenis syndrome, but CHD have not been reported in patients with point mutations in RAI1 and cardiac defects are not observed in the Rai1 mouse model [286]
cOne out of four KDVS patients with point mutations in KANSL1 had CHD
dThe lissencephaly phenotype of MDLS is caused by haploinsufficiency of the PAFAH1B1 gene (also known as LIS1) [287, 288], but it is presently unknown which gene is responsible for heart defects in MDLS patients
e TF transcription factor
CHD candidate genes identified from microdeletion and microduplication syndromes
Genotype–phenotype comparisons in patients with microdeletion and microduplication syndromes have identified candidate CHD disease genes. Subsequent mutation screening of candidate genes in patients and studies of the genes and their product in animal models have substantially added to the understanding of CHD and cardiac developmental biology.
The majority of DiGeorge syndrome (DGS, OMIM #188400, also known as 22q11.2 deletion syndrome and velocardiofacial syndrome) are caused by a 3 Mb deletion in 22q11.2 [151]. 22q11.2 duplication syndrome (OMIM #608363) is caused by duplication of genomic material in 22q11.2. Most of the 22q11.2 duplications that have been reported are reciprocal to the common 3 Mb deletion involved in DGS [152].
The common 3 Mb deletion affects more than 50 genes, including the gene encoding the T-box transcription factor TBX1. It is generally accepted that haploinsufficiency of TBX1 significantly contributes to the CHD phenotype in DGS patients. Tbx1 −/− mice display similar cardiac phenotypes to individuals with 22q11.2 DS [153, 154]. Conditional knock-out experiments in mice have shown that Tbx1 is required for proliferation of cardiac progenitors in the SHF—a cell population which contributes to the development of the cardiac outflow tract [155]. Additionally, point mutations in TBX1 have been reported in patients without the 22q11.2 deletion, but exhibit a clinical presentation similar to DGS [156, 157].
Transgenic mice overexpressing Tbx1 display phenotypic similarities consistent with 22q11 duplication patients, including cardiac outflow tract defects. This suggests that correct gene-dosage of TBX1 is important for normal cardiac development [152, 158, 159].
Conversely, cases with cardiac defects carrying smaller (1.5 Mb) deletions within the common 3 Mb region, distal to TBX1 have also been reported [160–162], which implies that other genes in the 3 Mb region may contribute to the cardiac phenotype of DGS patients. An interesting candidate gene within this region is CRKL, encoding a protein kinase. Mice with targeted deletion of Crkl exhibit defective OFT development and VSDs [160, 163]. Furthermore, experiments with compound heterozygous Tbx1 +/−, Crkl +/− mice indicate a possible genetic interaction between the two genes, leading to the increased severity of the cardiac phenotypes in the double mutants [164]. A genetic interaction between Crkl and Fgf8 has also been shown [165], supporting a link among FGF8, TBX1 and CRKL in the pathogenesis of DGS (see below).
A high degree of phenotypic variability is a characteristic feature of DGS and 22q11.2 duplication syndrome. Parts of this variation may perhaps be explained by variations in genes located within 22q11.2, with TBX1 as the most likely candidate. An alternative explanation involves epistasis, as TBX1 has been shown to regulate or interact with several proteins and signaling networks. Gene expression profiling of tissues in the pharyngeal region from mouse models with targeted deletion of Tbx1, have identified several Tbx1 target genes, which include genes involved in homeostasis of retinoic acid (RA) [166–168]. Interestingly, RA regulates Tbx1 expression [169], thus there seems to be a dual relationship between TBX1 expression and RA signaling. During development of the pharyngeal arches TBX1 expression in the pharyngeal endoderm is regulated by the Hedgehog signaling pathway though the action of forkhead transcription factors [170, 171]. TBX1 itself regulates the expression of FGF8 in the SHF and in the pharyngeal endoderm [172, 173]. A genetic interaction among Tbx1, Six1/Eya1 and Fgf8 was recently demonstrated in mouse models [174]. Further, TBX1 can act as a negative modulator of BMP signaling by binding SMAD1 and hereby interfere with the SMAD1/SMAD4 interaction [175].
Williams–Beuren syndrome (WBS, OMIM #194050) is caused by deletion of genomic material in 7q11.23. Most patients with WBS are heterozygous for a 1.5–1.8 Mb deletion encompassing ~28 genes [176–179]. Cardiovascular abnormalities are present in 75 % of individuals with WBS, predominantly in the form of supravalvular aortic stenosis (SVAS) and pulmonary arterial stenosis [180]. In 6–10 % of cases aortic or mitral valve defects are also seen, and other so-called “atypical” cardiac defects in the form of ASD, VSD and TOF are observed in a significant fraction of the patients [180, 181].
The ELN gene, encoding elastin, is believed to be the gene responsible for SVAS in WBS. Patients with atypical deletions including only ELN and LIMK1 genes and SVAS have been reported [182, 183]. In addition, point mutations in ELN are associated with familial and sporadic SVAS [184, 185]. Targeted deletion of the Eln gene in mice results in reduced aortic lumen diameter due to subendothelial accumulation of smooth muscle cells [186].
However, deletion of ELN does not explain the occurrence of the atypical heart defects in a proportion of WBS patients. Results gained from a recently reported mouse model with targeted deletion of Baz1b, indicate that deletion of this gene may account for these defects. BAZ1B is located within the WBS common deleted region, and homozygous Baz1b −/− mice exhibit a range of cardiovascular defects, which include ASD, VSD, trabeculation defects, coarctation of the aorta (COA), hypoplastic pharyngeal arch artery and a low frequency of DORV [187]. BAZ1B (also known as WSTF) acts as a subunit in three ATP-dependent chromatin remodeling complexes; the WSTF including nucleosome assembly complex (WINAC) [188], the WICH complex (WSTF-ISWI chromatin remodeling complex) [189] and the B-WICH complex [190]. These complexes are important for gene regulation, DNA replication and DNA repair [189, 191]. Thus, the cardiac phenotypes of Baz1b knockout mice and the chromatin remodeling function of BAZ1B suggests that some of the phenotypes involved in WBS, including “atypical” heart defects, may be caused by epigenetic effects.
Wolf–Hirschhorn syndrome (WHS, OMIM #194190) is caused by microdeletions in 4p16.3. Genotype–phenotype comparisons in patients with submicroscopic deletions suggest that haploinsufficiency of the gene encoding the histone lysine methyl transferase WHSC1 (also known as NSD2) contributes significantly to the WHS phenotype [152, 192]. A recently published investigation of mice with targeted deletion of the H3K36me3-specific histone methyltransferase gene Whsc1 puts forth Whsc1as another component in heart development [193]. The Whsc1 −/− mutant mice displayed ASD and VSD, and co-immunoprecipitation experiments with nuclear extracts prepared from embryonic hearts showed that Whsc1 interacts with the cardiac transcription factor Nkx2-5. Furthermore, ChIP assays demonstrated that Whsc1 cooperates with Nkx2-5 in the transcriptional regulation of target genes. Cross-breeding experiments with Whsc1 −/+ and Nkx2-5 −/+ mice suggested a genetic interaction between the two genes during cardiac septal formation. Another candidate gene for heart defects in WHS is the FGFRL1 gene, which encode a member of the fibroblast growth factor receptor family. During mouse development Fgfrl1 is expressed in the brain, cranial placodes, pharyngeal arches, somites and heart [194]. Targeted deletion of Fgfrl1 in mice can result in a range of developmental defects, including heart defects in the form of VSD, and both semilunar and atrioventricular valve deformation [194].
Cardiac defects are observed in 94 % of cases with interstitial deletions in 8p23.1 [195]. The defects range from isolated septal defects to complex heart defects like TOF and hypoplastic left heart syndrome (HLHS). A proportion of the patients carry a ~3.7 Mb recurrent deletion flanked by low copy repeats, although some patients have larger deletions that may extend to the 8p telomere. The gene encoding the cardiac zinc finger transcription factor GATA4 is located within the recurrent deletion, and it is well documented that this gene is associated with congenital heart defects. Mutations in GATA4 cause human CHD, often in the form of septal defects, but other defects have been reported [89, 91, 196] Mice homozygous for targeted deletion of Gata4 display early defects in cardiogenesis [93, 94], and phenotypic characterization of mice homozygous for a hypomorphic allele of Gata4 supports that haploinsufficiency of GATA4 can cause CHD [95]. GATA4 interacts with several other transcription factors during cardiac development, including NKX2-5, TBX5, ZFPM2 (FOG2), SMAD4 and HAND2 [34, 89, 92, 121] (see above). Therefore it is possible that the complex cardiac phenotypes observed in a subset of 8p23 deletion patients are evoked by epistatic effects from genes encoding GATA4 binding partners.
Other microdeletion and microduplication syndromes which comprise CHD
The widespread use of molecular cytogenetic methods like fluorescent in situ hybridization (FISH) and especially array comparative genome hybridization (array CGH) in clinical genetics laboratories has led to the recent delineation of a number of microdeletion and microduplication syndromes, which incorporate CHD as a component of their clinical spectrum (Table 2).
The minimal deleted region in 1q21.1 deletion syndrome contains the GJA5 gene [197]. A recent screen of 807 TOF cases revealed significant enrichment of small duplications encompassing GJA5, thus providing convincing evidence for a link between GJA5 and CHD [152, 192, 198]. Cardiac defects have been reported in a proportion of mice with targeted deletion of Gja5 [199, 200], suggesting that haploinsufficiency of GJA5 may be responsible for cardiac defects in some individuals with 1q21.1 deletions. GJA5 encodes the cardiac gap junction subunit Connexin 40, which is expressed in the atrial myocardium and the atrioventricular conduction system [201, 202]. Gap junctions are cell membrane channels that interconnect the cytoplasm of neighboring cells. In the heart, these channels contribute to the atrioventricular conduction [203, 204], but at present there is no proposed mechanism describing how GJA5 haploinsufficiency results in structural heart defects.
Molecular delineation of 9q34 microdeletions and mapping of the chromosomal breakpoints in a patient with a t(X;9) translocation suggested that the EHMT1 gene is responsible for Kleefstra syndrome (KS, OMIM #610253) [205–207]. Mutation screening in patients without deletions in 9q34 subsequently confirmed that haploinsufficiency of EHMT1 causes KS [205, 208]. Approximately 40 % of patients with KS and deletion of 9q34 have CHD, and the presence of CHD in five out of eleven KS patients with point mutations in EHMT1 confirm that this gene is responsible for CHD in KS. EHMT1 encodes euchromatic histone-lysine N-methyltransferase 1, which regulates transcription by methylation of histone H3 lysine 9 (H3K9Me2) in euchromatic DNA [209].
Koolen-De Vries syndrome (KDVS, OMIM #610443) is caused by recurrent deletions in 17q21.31. Between 27 and 36 % of KDVS patients have CHD [210, 211]. Recent delineation of the critical region of 17q21.31 and mutation screening of KDVS patients without deletion of 17q21 revealed that KDVS is caused by haploinsufficiency of KANSL1 [212, 213]. One out of four patients with point mutations in KDVS has CHD, hinting that KANSL1 is a CHD disease gene, although further patient data is needed to confirm this link. KANSL1 encodes a member of the male specific lethal (MSL) complex initially described in Drosophila (reviewed in [214]). Within the MSL complex KANSL1 interacts with KAT8, a histone acetyltransferase which regulates gene expression through acetylation of H4 lysine 16 (H4K16) [215].
Another interesting CHD candidate gene is TBX2, which is located within the deleted region in 17q23 deletion syndrome [216]. TBX2 is expressed in non-chamber myocardium of the developing heart, and mice with targeted mutation in Tbx2 have defects in the development of the atrioventricular canal (AVC) and the OFT [217]. It has been hypothesized that TBX2 is involved in cardiac chamber development and functions as a local repressor of the chamber-specific gene program in non-chamber regions like the AVC and OFT [218, 219].
Pathogenic copy number variants identified in cohorts of CHD patients
Array CGH and similar methods have been used to screen cohorts of CHD patients for pathogenic CNVs in the form of duplications and deletions. Since 2007, 14 whole-genome CNV screening studies have been reported, comprising more than 5,000 patients (Table 3, [220–234]). The reported studies show large differences, which include size and phenotypic composition of patient cohorts and the experimental and analytical setup, thus it is somewhat difficult to compare the results. Nevertheless, we find it safe to conclude that pathogenic CNVs are found among a significant portion of CHD patients.
Table 3.
CNV screens in patients with heart defects
| Patients | Phenotype of patientsd | Microarray type | Main results | Candidate genes in CNVsg | Reference |
|---|---|---|---|---|---|
| 60 | Congenital heart disease and extracardiac abnormalities | In-house-made microarray containing BAC/PACe clones. Average genomic distance of probes were 1 Mbp | CNVs considered to be causal were identified in 10 (17 %) patients | EHMT1, NKX2-5, NOTCH1, NSD1 | [233] |
| 105 | Congenital heart disease with and without extracardiac abnormalities. Subjects with documented syndromes were excluded | In-house-made microarray containing 32 k overlapping BAC clones | Rare de novo or inherited CNVs (0.34–13.9 Mb in size) were detected in 18 (17 %) patients | GJA5, LTBP1, TBX1 | [222] |
| 40 | Congenital heart disease with and without extracardiac abnormalities | NimbleGen Systems, Inc. whole-genome 385 K oligo array | Seven large CNVs were identified in 5 (12.5 %) patients | N/A | [230] |
| 114 | Tetralogy of Fallot (TOF) | Affymetrix Genome-Wide Human SNP Array 6.0 | Eleven (9.6 %) rare de novo CNVs (>20 kb) were identified in 114 TOF trios | JAG1, NOTCH1, RAB10, RAF1, TBX1 | [225] |
| 150a | Congenital heart disease and extracardiac abnormalities | In-house-made microarray containing BAC/PAC clones. Average genomic distance of probes were 1 Mbp | CNVs considered to be causal were identified in 26 (17.3 %) patients | ATRX, CREBBP, EHMT1, FOXC1, GATA4, NOTCH1, RAI, TBX1 | [220] |
| 46 | Isolated congenital heart disease | Affymetrix Genome-Wide Human SNP Array 6.0 | De novo CNVs were identified in two (4 %) patients | GJA5, NOTCH1, PDGFRA, TBX1 | [221] |
| 58 | Congenital heart disease and extracardiac abnormalities | Affymetrix GeneChip 100 K microarray | Potentially pathogenic CNVs (0.2–9.6 Mb in size) were detected in 12 (20.7 %) patients | ADAM19, HAND1, MESP1, NRP1, NTRK | [224] |
| 53 | Hypoplastic left heart syndrome (HLHS) | Agilent customized genome-wide 400 K array | Thirty-three rare non-polymorphic CNVs (2–1,554 kb in size) were detected in 25 (47 %) patients | BMPR2, ZNF423 | [227] |
| 262 | Heterotaxy (patients with D-transposition of the great arteries were also included in the sample) | Illumina 610Quad Beadchip platform | Forty-five previously unrecorded genic CNVs (0.27–25 Mb in size) were identified in 39 (14.5 %) patients. A significant (p = 1.5e − 4) burden of rare genic CNVs were found in HTX cases (14.5 %) compared to controls (7.4 %) | GALNT11, NEK2, NUP188, ROCK2, TGFBR2 | [223] |
| 43 | HLHS | NimbleGen Systems, Inc. whole-genome 385 K oligo array | A significant (p < 0.03) burden of CNVs were found in patients (4.6/subject) compared to controls (2.94/subject). The burden of unique CNVs in CHD patients was not found to be significant | N/A | [229] |
| 67b | Left-sided congenital heart disease (BAV, AS, COA, HLHS) | Affymetrix Human Genome-Wide SNP Array 6.0 | A total of 73 unique inherited or de novo CNVs (>20 kb) were identified in 54 individuals | ADORA2B, ANG, CACNA1C, COPS3, CRMP1, CTHRC1, ERCC5, EVC2, FLII, GRPEL1, HSD17B10, ITGA10, LIMS1, MAPK7, MFAP4, MSX1, MTHFD2, NCOR1, NGEF, PLA2G12A, PRPSAP2, RASD1, SBEBF1, SMC1A, ULK2 | [226] |
| 2,539 | Isolated congenital heart disease (808 TOF and 1,448 other CHDs). Subjects with documented syndromes known to cause CHD were excluded | Illumina 660 W-Quad SNP platform | A significant (p = 0.008) burden of rare genic CNVs were found in CHD cases (7.8 %) compared to controls (4.4 %) | CNOT6, EDIL3, GATA4, GJA5, HAND2, PPM1K and 13 genes in the WNT-signaling pathway (CDH18, CDH2, CTBP1, CTNNB1, FAT1, LRP5L, NFATC1, PCDH15, PCDHB7, PCDHB8, PRKCB, PRKCQ, WNT7B) | [232] |
| 203 + 511c | Congenital heart disease and extracardiac abnormalities. | Customized 105 k oligonucleotide arrays manufactured by Agilent. Average resolution of 30 kb, with denser coverage at disease loci | A total of 55 rare CNVs (>50 kb) were identified in patients from the discovery cohort. Sixteen of these CNVs were identified in the second cohort | PDE1A, NALCN, ANKRD11, SOX7, GATA4, CRK, CAMTA2, CECR1 | [228] |
| 433 | Tetralogy of Fallot-pulmonary atresia or pulmonary atresia and ventricular septal defect. Subjects with documented syndromes were excluded | Affymetrix Genome-Wide Human SNP Array 6.0 | 47 large (>500 kb) rare CNVs were found in 43 (9.9 %) patients | ANGPT2, ARHGEF10, ARHGEF4, BARD1, BBS9, C12oerf66, CASP1, CASP12, CASP4, CASP5, CCDC148, CDH19, CHL1, CHRM3, CHST8, CNDP2, CNN2, CRKL, DISP1, DNAH11, EDIL3, FGF10, FOXO3B, FSTL3, FSTL4, GJA5, GMDS, GNA11, HIRA, HNF1B, HRIP3, IDS, KCNB2, KIAA1609, LBH, MAPK3, NBEA, NFATC1, NXN, PARD6G, PDS5B, PLXNA2, PPM1K, PPP4C, PTBP1, RAF1, S1PR4, SEMA3D, SEMA3E, SFPQ, SLC25A46, SNX8, SOX4, SPG20, TBX1, TBX6, TNFSF11, VCAN, WDR18, WNK3, ZNF347 | [231] |
| 945 | Congenital heart disease with and without extracardiac abnormalities | Affymetrix Genome-Wide Human SNP Array 6.0 | Known CHD-related chromosomal abnormalitiesf were identified in 135 (14.3 %) patients. Large, rare CNVs (0.22–32.1 Mb in size) were identified in 35 (3.7 %) patients | FKBP6, ELN, GTF2IRD1, GATA4, CRKL, TBX1, ATRX, GPC3, BCOR, ZIC3, FLNA, MID1, PRKAB2, FMO5, CHD1L, BCL9, ACP6, GJA5, HRAS, GATA6, RUNX1 | [234] |
Genes known to cause CHD in humans are underlined, genes with reported cardiovascular system involvement (e.g., from targeted deletion in mice) are bold
aIncludes 60 patients from Thienpont et al. [233]
bA total of 174 patients from 67 families
cA discovery cohort of 203 patients and a second independent cohort of 511 patients were analyzed
d AS aortic stenosis, BAV bicuspid aortic valve, COA coarctation of the aorta, HLHS hypoplastic left heart syndrome, TOF tetralogy of Fallot
e BAC bacterial artificial chromosome, PAC P1-derived artificial chromosome
fTrisomy 21 (n = 80), trisomy 18 (n = 1), 22qDS (n = 42), Turner syndrome (n = 8), William’s syndrome (n = 3), and Triple X syndrome (n = 1)
gCandidate genes suggested by the authors
The highest frequency of pathogenic CNVs is found among patients with CHD and extra-cardiac anomalies. Based on the current reports [220, 224, 228, 230, 233, 234] we estimate that pathogenic CNVs are present in 15–20 % of patients with CHD and extra-cardiac anomalies.
Among patients with isolated CHD, the frequency of pathogenic CNVs is significantly lower. Here, we estimate the frequency to be between 4 and 14 % [221, 222, 225, 231, 232]. However, this estimate should be treated with caution, due to the aforementioned large differences in study design.
Identification of disease genes and pathways from CNVs detected in cohorts of CHD patients
In principle, CNVs identified as pathogenic in CHD patients should contain one or more dosage sensitive cardiac developmental genes. Thus, each pathogenic CNV, or at least overlapping CNVs should define a disease locus for CHD and it therefore should be possible to use CNVs to detect CHD disease genes. Several groups have reported identification of CNVs spanning genes, which span genes previously recognized to cause CHD in animal models (Table 3), thereby providing a plausible link between these genes and CHD in humans.
However, many of the identified CNVs do not contain a well-established cardiac developmental gene. These CNVs often contain several genes, although only one is likely to be the gene responsible for CHD. Three approaches have been utilized to identify the causal genes in such cases: (1) narrowing of the locus by comparison of multiple samples with overlapping CNVs, (2) in silico gene prioritization and (3) functional investigations of candidate genes within the CNVs.
Hitz et al. [226] used Endeavour [235] to test for enrichment of angiogenesis-associated genes within 73 CNVs identified in patients with left-sided CHD. They also searched for genes with expression in the developing heart in serial analysis of gene expression (SAGE) and public databases. By combining these prioritization methods, they identified 25 CHD candidate genes (Table 3). Soemedi et al. [232] performed genomic region annotation enrichment analysis on rare deletions and duplications identified in 2,256 CHD cases. They found enrichment of 13 genes encoding proteins involved in the WNT signaling pathway (Table 3). Silversides et al. [231] performed a systematic review of genes within rare CNVs identified among 433 cases with TOF and identified 62 CHD candidate genes (Table 3). They also assessed whether genes, in predefined gene-sets derived from GeneOntology (GO) annotations and pathway and protein domain databases, were significantly overrepresented in CNVs detected in TOF cases compared to controls. They found enrichment of gene-sets belonging to five functional clusters: vasculature development, chromosome organization, cell motility, chemotaxis and neuron projection and development. Lalani et al. [228] identified eight candidate genes in CNVs identified in patients with CHD and extracardiac anomalies (Table 3). They grouped genes within enriched CNVs based on GO categories and found enrichment for genes encoding proteins involved in G-protein coupled receptor internalization, hemopoiesis and cytoskeleton organization. Furthermore they analyzed protein–protein interactions between proteins encoded by candidate genes in CNVs identified in patients and a set of 276 proteins from GO cardiac development categories. They identified 11 proteins with at least one connection with a human cardiac-specific protein (significant at p = 0.03).
Thienpont et al. [236] identified TAB2 as a dosage sensitive CHD disease gene by comparing overlapping CNVs within 6q25. The overlapping region of seven CNVs identified in CHD patients revealed a CHD locus containing 11 candidate genes, including TAB2. For prioritization of the candidate genes in the locus and surrounding genomic region, the authors performed in silico analyses of 105 genes in 6q24–25, using an adapted version of Endeavour [235]. This analysis predicted TAB2 as the highest-ranking candidate gene in 6q24–25. Further functional analyses of TAB2 in human embryonic heart tissues and zebrafish suggested that TAB2 is a cardiac developmental gene. Point mutations localized within TAB2 in two unrelated CHD patients and mapping of a translocation breakpoints within TAB2 in a CHD family segregating a t(2;6) translocation, further verified TAB2 as a CHD disease gene.
Fakhro et al. [223] performed whole-genome CNV screening of 262 patients with Heterotaxy and isolated TGA. They identified 45 unrecorded gene-containing CNVs, including two different CNVs affecting TGFBR2. Evaluation of candidate genes using in situ hybridization and Morpholino-based gene knock-down in X. tropicalis showed that the genes tgfbr2, rock2, galnt11, nek2 and nup188 are involved in left–right patterning of the heart (Table 3). ROCK2 and NEK2 are ciliary proteins, thus this study confirmed the importance of cilia and TGF-β signaling in LR patterning [237]. In addition, this study identified two novel genes (GALNT11 and NUP188) with unknown functions in LR development.
The molecular pathology of congenital heart disease
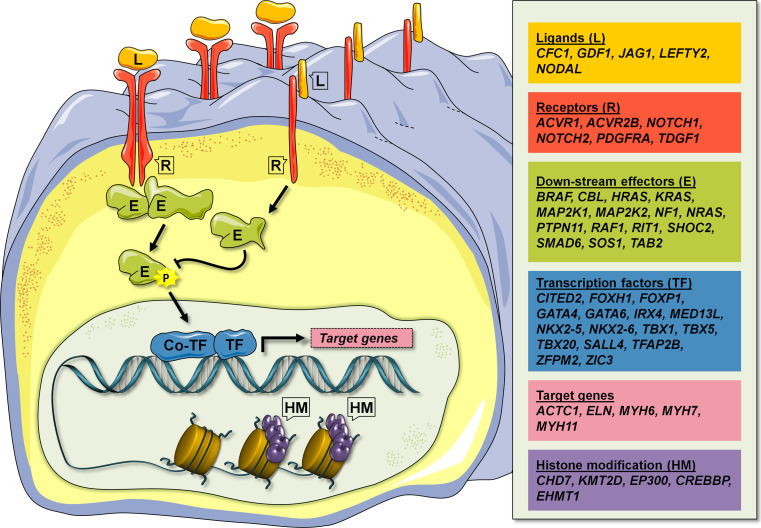
Cardiac development is controlled by a large number of signaling pathways, which are tightly regulated in time and space, and interact in complex developmental networks [34]. The CHD disease genes, which have been identified to date, suggest that all aspects of developmental signaling pathways may be involved in human CHD: from ligands (e.g. JAG1) and receptors (e.g. NOTCH, PDGFRA), across down-stream signaling effectors (e.g. PTPN11, SMAD6), to transcription factors (e.g. GATA4, NKX2-5) and targets (e.g. ACTC1, MYH6) (Fig. 1). Moreover, discoveries of disease genes encoding histone-modifying proteins (e.g. CHD7, KMT2D), suggest that epigenetic regulation of an unknown number of target genes, may add an additional layer of regulation on consensus cardiac developmental networks.
Fig. 1.
Schematic representation of the different cell signaling components affected by mutations in human CHD disease genes. These include ligands (L), receptors (R), down-stream effectors (E), transcription regulators, which include transcription factors (TF), transcription co-factors (co-TF) and histone modifying proteins (HM), and target genes. Known human CHD disease genes within the six groups are shown in the panel at the right. Colored figure are shown in the on-line version of the article
Lage et al. [238] have recently shown that a wide range of CHD risk factors, functionally converge in complex, yet discrete, protein networks driving heart development. These findings, combined with the potentially hundreds of CHD disease genes [141], suggest that CHD may be caused by a very large number of combinations of mutations and environmental risk factors.
Reduced penetrance of CHD are often observed in human pedigrees (e.g. [239], unpublished observations in Danish pedigrees) and in carriers of CNVs known to cause CHD (see Table 2 and discussions in text). Some of the reduced penetrance may simply be due to unidentified asymptomatic heart defects in some carriers, but may also be caused by epistasis. Several examples of epistasis in mouse models are mentioned in the text above. Winston et al. [240] performed a systematic study of the influence of genetic background on the expression of heart defects in Nkx2-5 +/− heterozygous mice. The authors compared Nkx2-5 +/− heterozygous C57Bl/6 mice with Nkx2-5 +/− heterozygous F1 progeny from crosses with two other mouse strains. The data showed that the F1 hybrid mice presented with a significantly lower incidence of septal defects compared to mice with the original C57Bl/6 background. The authors suggest that modifying alleles can either direct the manifestation of a cardiac developmental defect or buffer the effect from perturbations. The latter situation, which was the case in the study, may ensure robustness of normal heart development.
How the large heterogeneity in CHD and potentially large epistatic effects translates into lesions in the personal genome of the individual patient remains to be investigated. One possible scenario could be that individual combinations of several risk alleles may be the cause of CHD in part of the patients.
Future perspectives
It was recently demonstrated that exome-sequencing is a powerful tool for identification of de novo mutations in CHD [141]. It is very likely that more studies based on exome-sequencing will reveal new CHD disease genes in the near future. The high number of variants identified in exome-sequencing experiments is a big challenge in very heterogeneous disorders like CHD. Thus, it is likely that such studies will be performed on large numbers of parent-offspring trios or on families with dominant or recessive inherited CHD.
The past 5 years have shown that analysis of genome rearrangements in the form of CNVs, translocations or inversions can lead to detection of new CHD disease genes or loci. The technology for mapping such rearrangements is continuously improving, and breakpoints in balanced translocations and inversions can now be mapped within days using NGS [241].
Untreated, CHD is a disorder with a high mortality rate, therefore a large part of the disease causing mutations are likely rare in populations due to negative selection. However, it is also possible that some variants associated with CHD may escape negative selection. Such variants may be discovered through genome wide association studies, as has recently been demonstrated [242–244].
Interesting therapeutic opportunities could arise from the current knowledge of the molecular pathogenesis of CHD. A significant part of CHD seems to be caused by mutations which perturb complex developmental networks. These networks are characterized by extensive communication within and between specific signaling pathways, and with the environment. Thus, given the apparent epistatic effects observed in patients and animal models, it should be possible to manipulate the signaling pathways in the developmental networks with synthetic agonists or antagonists, and thereby alleviate effects from mutations or redirect signaling events towards normal heart development. A recent study suggests that this could be possible someday. Tian et al. [245] showed that defects in the cardiac inflow tract and AV canal (resembling complete CAVC in humans) in Wnt2 −/− mice, could be rescued by transient pharmacological activation of Wnt signaling with LiCl.
Naturally, such therapeutic opportunities are presently very hypothetical, and to become reality, much more knowledge about the molecular genetics and the molecular pathology of CHD are needed. Combining human genetics/genomics with functional studies in cell models or animal models like zebrafish, Xenopus frogs, chicken or mice are likely to have the greatest impact on our understanding of the molecular pathology in human CHD.
Acknowledgments
We thank Josie Bodle for language editing of the manuscript. LAL and TAA were supported by The Danish Council for Independent Research. KDT was supported by the Lundbeck foundation. Wilhelm Johannsen Centre for Functional Genome Research is established by the Danish National Research Foundation.
References
- 1.Reller MD, Strickland MJ, Riehle-Colarusso T, Mahle WT, Correa A. Prevalence of congenital heart defects in metropolitan Atlanta, 1998–2005. J Pediatr. 2008;153(6):807–813. doi: 10.1016/j.jpeds.2008.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman JI. Incidence of congenital heart disease: II. Prenatal incidence. Pediatr Cardiol. 1995;16(4):155–165. doi: 10.1007/BF00794186. [DOI] [PubMed] [Google Scholar]
- 3.Warnes CA, Williams RG, Bashore TM, Child JS, Connolly HM, Dearani JA, del Nido P, Fasules JW, Graham TP, Jr, Hijazi ZM, Hunt SA, King ME, Landzberg MJ, Miner PD, Radford MJ, Walsh EP, Webb GD. ACC/AHA 2008 guidelines for the management of adults with congenital heart disease: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (writing committee to develop guidelines on the management of adults with congenital heart disease) Circulation. 2008;118(23):e714–e833. doi: 10.1161/CIRCULATIONAHA.108.190690. [DOI] [PubMed] [Google Scholar]
- 4.Parrott A, Ware SM. The role of the geneticist and genetic counselor in an ACHD clinic. Prog Pediatr Cardiol. 2012;34(1):15–20. doi: 10.1016/j.ppedcard.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vandenberg LN, Levin M. A unified model for left-right asymmetry? Comparison and synthesis of molecular models of embryonic laterality. Dev Biol. 2013;379(1):1–15. doi: 10.1016/j.ydbio.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka Y, Okada Y, Hirokawa N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature. 2005;435(7039):172–177. doi: 10.1038/nature03494. [DOI] [PubMed] [Google Scholar]
- 7.McGrath J, Somlo S, Makova S, Tian X, Brueckner M. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell. 2003;114(1):61–73. doi: 10.1016/s0092-8674(03)00511-7. [DOI] [PubMed] [Google Scholar]
- 8.Nonaka S, Shiratori H, Saijoh Y, Hamada H. Determination of left-right patterning of the mouse embryo by artificial nodal flow. Nature. 2002;418(6893):96–99. doi: 10.1038/nature00849. [DOI] [PubMed] [Google Scholar]
- 9.Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95(6):829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 10.Ramsdell AF. Left-right asymmetry and congenital cardiac defects: getting to the heart of the matter in vertebrate left-right axis determination. Dev Biol. 2005;288(1):1–20. doi: 10.1016/j.ydbio.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 11.Yoshioka H, Meno C, Koshiba K, Sugihara M, Itoh H, Ishimaru Y, Inoue T, Ohuchi H, Semina EV, Murray JC, Hamada H, Noji S. Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell. 1998;94(3):299–305. doi: 10.1016/s0092-8674(00)81473-7. [DOI] [PubMed] [Google Scholar]
- 12.De Luca A, Sarkozy A, Consoli F, Ferese R, Guida V, Dentici ML, Mingarelli R, Bellacchio E, Tuo G, Limongelli G, Digilio MC, Marino B, Dallapiccola B. Familial transposition of the great arteries caused by multiple mutations in laterality genes. Heart. 2010;96(9):673–677. doi: 10.1136/hrt.2009.181685. [DOI] [PubMed] [Google Scholar]
- 13.Mohapatra B, Casey B, Li H, Ho-Dawson T, Smith L, Fernbach SD, Molinari L, Niesh SR, Jefferies JL, Craigen WJ, Towbin JA, Belmont JW, Ware SM. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum Mol Genet. 2009;18(5):861–871. doi: 10.1093/hmg/ddn411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosaki K, Bassi MT, Kosaki R, Lewin M, Belmont J, Schauer G, Casey B. Characterization and mutation analysis of human LEFTY A and LEFTY B, homologues of murine genes implicated in left-right axis development. Am J Hum Genet. 1999;64(3):712–721. doi: 10.1086/302289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosaki R, Gebbia M, Kosaki K, Lewin M, Bowers P, Towbin JA, Casey B. Left-right axis malformations associated with mutations in ACVR2B, the gene for human activin receptor type IIB. Am J Med Genet. 1999;82(1):70–76. doi: 10.1002/(sici)1096-8628(19990101)82:1<70::aid-ajmg14>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 16.Karkera JD, Lee JS, Roessler E, Banerjee-Basu S, Ouspenskaia MV, Mez J, Goldmuntz E, Bowers P, Towbin J, Belmont JW, Baxevanis AD, Schier AF, Muenke M. Loss-of-function mutations in growth differentiation factor-1 (GDF1) are associated with congenital heart defects in humans. Am J Hum Genet. 2007;81(5):987–994. doi: 10.1086/522890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldmuntz E, Bamford R, Karkera JD, dela Cruz J, Roessler E, Muenke M. CFC1 mutations in patients with transposition of the great arteries and double-outlet right ventricle. Am J Hum Genet. 2002;70(3):776–780. doi: 10.1086/339079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sperling S, Grimm CH, Dunkel I, Mebus S, Sperling HP, Ebner A, Galli R, Lehrach H, Fusch C, Berger F, Hammer S. Identification and functional analysis of CITED2 mutations in patients with congenital heart defects. Hum Mutat. 2005;26(6):575–582. doi: 10.1002/humu.20262. [DOI] [PubMed] [Google Scholar]
- 19.Ware SM, Peng J, Zhu L, Fernbach S, Colicos S, Casey B, Towbin J, Belmont JW. Identification and functional analysis of ZIC3 mutations in heterotaxy and related congenital heart defects. Am J Hum Genet. 2004;74(1):93–105. doi: 10.1086/380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.French VM, van de Laar IM, Wessels MW, Rohe C, Roos-Hesselink JW, Wang G, Frohn-Mulder IM, Severijnen LA, de Graaf BM, Schot R, Breedveld G, Mientjes E, van Tienhoven M, Jadot E, Jiang Z, Verkerk A, Swagemakers S, Venselaar H, Rahimi Z, Najmabadi H, Meijers-Heijboer H, de Graaff E, Helbing WA, Willemsen R, Devriendt K, Belmont JW, Oostra BA, Amack JD, Bertoli-Avella AM. NPHP4 variants are associated with pleiotropic heart malformations. Circ Res. 2012;110(12):1564–1574. doi: 10.1161/CIRCRESAHA.112.269795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto EA, Schermer B, Obara T, O’Toole JF, Hiller KS, Mueller AM, Ruf RG, Hoefele J, Beekmann F, Landau D, Foreman JW, Goodship JA, Strachan T, Kispert A, Wolf MT, Gagnadoux MF, Nivet H, Antignac C, Walz G, Drummond IA, Benzing T, Hildebrandt F. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat Genet. 2003;34(4):413–420. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaki M, Hoefele J, Allen SJ, Ramaswami G, Janssen S, Bergmann C, Heckenlively JR, Otto EA, Hildebrandt F. Genotype-phenotype correlation in 440 patients with NPHP-related ciliopathies. Kidney Int. 2011;80(11):1239–1245. doi: 10.1038/ki.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gebbia M, Ferrero GB, Pilia G, Bassi MT, Aylsworth A, Penman-Splitt M, Bird LM, Bamforth JS, Burn J, Schlessinger D, Nelson DL, Casey B. X-linked situs abnormalities result from mutations in ZIC3. Nat Genet. 1997;17(3):305–308. doi: 10.1038/ng1197-305. [DOI] [PubMed] [Google Scholar]
- 24.Megarbane A, Salem N, Stephan E, Ashoush R, Lenoir D, Delague V, Kassab R, Loiselet J, Bouvagnet P. X-linked transposition of the great arteries and incomplete penetrance among males with a nonsense mutation in ZIC3. Eur J Hum Genet. 2000;8(9):704–708. doi: 10.1038/sj.ejhg.5200526. [DOI] [PubMed] [Google Scholar]
- 25.Purandare SM, Ware SM, Kwan KM, Gebbia M, Bassi MT, Deng JM, Vogel H, Behringer RR, Belmont JW, Casey B. A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development. 2002;129(9):2293–2302. doi: 10.1242/dev.129.9.2293. [DOI] [PubMed] [Google Scholar]
- 26.Ware SM, Harutyunyan KG, Belmont JW. Heart defects in X-linked heterotaxy: evidence for a genetic interaction of Zic3 with the nodal signaling pathway. Dev Dyn. 2006;235(6):1631–1637. doi: 10.1002/dvdy.20719. [DOI] [PubMed] [Google Scholar]
- 27.Sutherland MJ, Wang S, Quinn ME, Haaning A, Ware SM. Zic3 is required in the migrating primitive streak for node morphogenesis and left-right patterning. Hum Mol Genet. 2013;22(10):1913–1923. doi: 10.1093/hmg/ddt001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenhart KF, Holtzman NG, Williams JR, Burdine RD. Integration of nodal and BMP signals in the heart requires FoxH1 to create left-right differences in cell migration rates that direct cardiac asymmetry. PLoS Genet. 2013;9(1):e1003109. doi: 10.1371/journal.pgen.1003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roessler E, Ouspenskaia MV, Karkera JD, Velez JI, Kantipong A, Lacbawan F, Bowers P, Belmont JW, Towbin JA, Goldmuntz E, Feldman B, Muenke M. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am J Hum Genet. 2008;83(1):18–29. doi: 10.1016/j.ajhg.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Both I, Silvestri C, Erdemir T, Lickert H, Walls JR, Henkelman RM, Rossant J, Harvey RP, Attisano L, Wrana JL. Foxh1 is essential for development of the anterior heart field. Dev Cell. 2004;7(3):331–345. doi: 10.1016/j.devcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 31.Lopes FK, Artap ST, Preis JI, Fatkin D, Chapman G, Furtado MB, Harvey RP, Hamada H, Sparrow DB, Dunwoodie SL. Loss of Cited2 causes congenital heart disease by perturbing left-right patterning of the body axis. Hum Mol Genet. 2011;20(6):1097–1110. doi: 10.1093/hmg/ddq554. [DOI] [PubMed] [Google Scholar]
- 32.Weninger WJ, Lopes FK, Bennett MB, Withington SL, Preis JI, Barbera JP, Mohun TJ, Dunwoodie SL. Cited2 is required both for heart morphogenesis and establishment of the left-right axis in mouse development. Development. 2005;132(6):1337–1348. doi: 10.1242/dev.01696. [DOI] [PubMed] [Google Scholar]
- 33.Bamforth SD, Braganca J, Farthing CR, Schneider JE, Broadbent C, Michell AC, Clarke K, Neubauer S, Norris D, Brown NA, Anderson RH, Bhattacharya S. Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat Genet. 2004;36(11):1189–1196. doi: 10.1038/ng1446. [DOI] [PubMed] [Google Scholar]
- 34.Lage K, Mollgard K, Greenway S, Wakimoto H, Gorham JM, Workman CT, Bendsen E, Hansen NT, Rigina O, Roque FS, Wiese C, Christoffels VM, Roberts AE, Smoot LB, Pu WT, Donahoe PK, Tommerup N, Brunak S, Seidman CE, Seidman JG, Larsen LA. Dissecting spatio-temporal protein networks driving human heart development and related disorders. Mol Syst Biol. 2010;6:381. doi: 10.1038/msb.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srivastava D. Genetic assembly of the heart: implications for congenital heart disease. Annu Rev Physiol. 2001;63:451–469. doi: 10.1146/annurev.physiol.63.1.451. [DOI] [PubMed] [Google Scholar]
- 36.de la Pompa JL, Epstein JA. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22(2):244–254. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, Costa T, Pierpont ME, Rand EB, Piccoli DA, Hood L, Spinner NB. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16(3):243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 38.Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, Chandrasekharappa SC. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16(3):235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 39.Emerick KM, Rand EB, Goldmuntz E, Krantz ID, Spinner NB, Piccoli DA. Features of Alagille syndrome in 92 patients: frequency and relation to prognosis. Hepatology. 1999;29(3):822–829. doi: 10.1002/hep.510290331. [DOI] [PubMed] [Google Scholar]
- 40.McElhinney DB, Krantz ID, Bason L, Piccoli DA, Emerick KM, Spinner NB, Goldmuntz E. Analysis of cardiovascular phenotype and genotype-phenotype correlation in individuals with a JAG1 mutation and/or Alagille syndrome. Circulation. 2002;106(20):2567–2574. doi: 10.1161/01.cir.0000037221.45902.69. [DOI] [PubMed] [Google Scholar]
- 41.Kamath BM, Bauer RC, Loomes KM, Chao G, Gerfen J, Hutchinson A, Hardikar W, Hirschfield G, Jara P, Krantz ID, Lapunzina P, Leonard L, Ling S, Ng VL, Hoang PL, Piccoli DA, Spinner NB. NOTCH2 mutations in Alagille syndrome. J Med Genet. 2012;49(2):138–144. doi: 10.1136/jmedgenet-2011-100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet. 2006;79(1):169–173. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue Y, Gao X, Lindsell CE, Norton CR, Chang B, Hicks C, Gendron-Maguire M, Rand EB, Weinmaster G, Gridley T. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet. 1999;8(5):723–730. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]
- 44.McCright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 2002;129(4):1075–1082. doi: 10.1242/dev.129.4.1075. [DOI] [PubMed] [Google Scholar]
- 45.McKellar SH, Tester DJ, Yagubyan M, Majumdar R, Ackerman MJ, Sundt TM., III Novel NOTCH1 mutations in patients with bicuspid aortic valve disease and thoracic aortic aneurysms. J Thorac Cardiovasc Surg. 2007;134(2):290–296. doi: 10.1016/j.jtcvs.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 46.McBride KL, Riley MF, Zender GA, Fitzgerald-Butt SM, Towbin JA, Belmont JW, Cole SE. NOTCH1 mutations in individuals with left ventricular outflow tract malformations reduce ligand-induced signaling. Hum Mol Genet. 2008;17(18):2886–2893. doi: 10.1093/hmg/ddn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437(7056):270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 48.Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, de la Pompa JL. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18(1):99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Wu B, Chamberlain AA, Lui W, Koirala P, Susztak K, Klein D, Taylor V, Zhou B. Endocardial to myocardial notch-wnt-bmp axis regulates early heart valve development. PLoS ONE. 2013;8(4):e60244. doi: 10.1371/journal.pone.0060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luna-Zurita L, Prados B, Grego-Bessa J, Luxan G, del Monte G, Benguria A, Adams RH, Perez-Pomares JM, de la Pompa JL. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J Clin Invest. 2010;120(10):3493–3507. doi: 10.1172/JCI42666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bosse K, Hans CP, Zhao N, Koenig SN, Huang N, Guggilam A, Lahaye S, Tao G, Lucchesi PA, Lincoln J, Lilly B, Garg V. Endothelial nitric oxide signaling regulates Notch1 in aortic valve disease. J Mol Cell Cardiol. 2013;60:27–35. doi: 10.1016/j.yjmcc.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19(3):230–236. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sznajer Y, Keren B, Baumann C, Pereira S, Alberti C, Elion J, Cave H, Verloes A. The spectrum of cardiac anomalies in Noonan syndrome as a result of mutations in the PTPN11 gene. Pediatrics. 2007;119(6):e1325–e1331. doi: 10.1542/peds.2006-0211. [DOI] [PubMed] [Google Scholar]
- 54.Marino B, Digilio MC, Toscano A, Giannotti A, Dallapiccola B. Congenital heart diseases in children with Noonan syndrome: an expanded cardiac spectrum with high prevalence of atrioventricular canal. J Pediatr. 1999;135(6):703–706. doi: 10.1016/s0022-3476(99)70088-0. [DOI] [PubMed] [Google Scholar]
- 55.Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29(4):465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 56.Niihori T, Aoki Y, Narumi Y, Neri G, Cave H, Verloes A, Okamoto N, Hennekam RC, Gillessen-Kaesbach G, Wieczorek D, Kavamura MI, Kurosawa K, Ohashi H, Wilson L, Heron D, Bonneau D, Corona G, Kaname T, Naritomi K, Baumann C, Matsumoto N, Kato K, Kure S, Matsubara Y. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38(3):294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- 57.Schubbert S, Zenker M, Rowe SL, Boll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner LE, Nguyen H, West B, Zhang KY, Sistermans E, Rauch A, Niemeyer CM, Shannon K, Kratz CP. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38(3):331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 58.Tartaglia M, Pennacchio LA, Zhao C, Yadav KK, Fodale V, Sarkozy A, Pandit B, Oishi K, Martinelli S, Schackwitz W, Ustaszewska A, Martin J, Bristow J, Carta C, Lepri F, Neri C, Vasta I, Gibson K, Curry CJ, Siguero JP, Digilio MC, Zampino G, Dallapiccola B, Bar-Sagi D, Gelb BD. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39(1):75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 59.Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, Li L, Yassin Y, Tamburino AM, Neel BG, Kucherlapati RS. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007;39(1):70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- 60.Razzaque MA, Nishizawa T, Komoike Y, Yagi H, Furutani M, Amo R, Kamisago M, Momma K, Katayama H, Nakagawa M, Fujiwara Y, Matsushima M, Mizuno K, Tokuyama M, Hirota H, Muneuchi J, Higashinakagawa T, Matsuoka R. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet. 2007;39(8):1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- 61.Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, Pogna EA, Schackwitz W, Ustaszewska A, Landstrom A, Bos JM, Ommen SR, Esposito G, Lepri F, Faul C, Mundel P, Lopez Siguero JP, Tenconi R, Selicorni A, Rossi C, Mazzanti L, Torrente I, Marino B, Digilio MC, Zampino G, Ackerman MJ, Dallapiccola B, Tartaglia M, Gelb BD. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007;39(8):1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 62.Cirstea IC, Kutsche K, Dvorsky R, Gremer L, Carta C, Horn D, Roberts AE, Lepri F, Merbitz-Zahradnik T, Konig R, Kratz CP, Pantaleoni F, Dentici ML, Joshi VA, Kucherlapati RS, Mazzanti L, Mundlos S, Patton MA, Silengo MC, Rossi C, Zampino G, Digilio C, Stuppia L, Seemanova E, Pennacchio LA, Gelb BD, Dallapiccola B, Wittinghofer A, Ahmadian MR, Tartaglia M, Zenker M. A restricted spectrum of NRAS mutations causes Noonan syndrome. Nat Genet. 2010;42(1):27–29. doi: 10.1038/ng.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aoki Y, Niihori T, Kawame H, Kurosawa K, Ohashi H, Tanaka Y, Filocamo M, Kato K, Suzuki Y, Kure S, Matsubara Y. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37(10):1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 64.Rodriguez-Viciana P, Tetsu O, Tidyman WE, Estep AL, Conger BA, Cruz MS, McCormick F, Rauen KA. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311(5765):1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- 65.Cordeddu V, Di SE, Pennacchio LA, Ma’ayan A, Sarkozy A, Fodale V, Cecchetti S, Cardinale A, Martin J, Schackwitz W, Lipzen A, Zampino G, Mazzanti L, Digilio MC, Martinelli S, Flex E, Lepri F, Bartholdi D, Kutsche K, Ferrero GB, Anichini C, Selicorni A, Rossi C, Tenconi R, Zenker M, Merlo D, Dallapiccola B, Iyengar R, Bazzicalupo P, Gelb BD, Tartaglia M. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat Genet. 2009;41(9):1022–1026. doi: 10.1038/ng.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martinelli S, de Luca A, Stellacci E, Rossi C, Checquolo S, Lepri F, Caputo V, Silvano M, Buscherini F, Consoli F, Ferrara G, Digilio MC, Cavaliere ML, van Hagen JM, Zampino G, van der Burgt I, Ferrero GB, Mazzanti L, Screpanti I, Yntema HG, Nillesen WM, Savarirayan R, Zenker M, Dallapiccola B, Gelb BD, Tartaglia M. Heterozygous germline mutations in the CBL tumor-suppressor gene cause a Noonan syndrome-like phenotype. Am J Hum Genet. 2010;87(2):250–257. doi: 10.1016/j.ajhg.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Luca A, Bottillo I, Sarkozy A, Carta C, Neri C, Bellacchio E, Schirinzi A, Conti E, Zampino G, Battaglia A, Majore S, Rinaldi MM, Carella M, Marino B, Pizzuti A, Digilio MC, Tartaglia M, Dallapiccola B. NF1 gene mutations represent the major molecular event underlying neurofibromatosis-Noonan syndrome. Am J Hum Genet. 2005;77(6):1092–1101. doi: 10.1086/498454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCulley DJ, Black BL. Transcription factor pathways and congenital heart disease. Curr Top Dev Biol. 2012;100:253–277. doi: 10.1016/B978-0-12-387786-4.00008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281(5373):108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 70.Benson DW, Silberbach GM, Kavanaugh-McHugh A, Cottrill C, Zhang Y, Riggs S, Smalls O, Johnson MC, Watson MS, Seidman JG, Seidman CE, Plowden J, Kugler JD. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J Clin Invest. 1999;104(11):1567–1573. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goldmuntz E, Geiger E, Benson DW. NKX2.5 mutations in patients with tetralogy of Fallot. Circulation. 2001;104(21):2565–2568. doi: 10.1161/hc4601.098427. [DOI] [PubMed] [Google Scholar]
- 72.Azpiazu N, Frasch M. Tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7(7B):1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- 73.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126(6):1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 74.Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nk2–5. Genes Dev. 1995;9(13):1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 75.Jay PY, Harris BS, Maguire CT, Buerger A, Wakimoto H, Tanaka M, Kupershmidt S, Roden DM, Schultheiss TM, O’Brien TX, Gourdie RG, Berul CI, Izumo S. Nk2–5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest. 2004;113(8):1130–1137. doi: 10.1172/JCI19846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, Evans SM, Clark B, Feramisco JR, Giles W, Ho SY, Benson DW, Silberbach M, Shou W, Chien KR. Nk2–5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117(3):373–386. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 77.Zaffran S, Kelly RG. New developments in the second heart field. Differentiation. 2012;84(1):17–24. doi: 10.1016/j.diff.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 78.Searcy RD, Vincent EB, Liberatore CM, Yutzey KE. A GATA-dependent nkx-2.5 regulatory element activates early cardiac gene expression in transgenic mice. Development. 1998;125(22):4461–4470. doi: 10.1242/dev.125.22.4461. [DOI] [PubMed] [Google Scholar]
- 79.Lien CL, McAnally J, Richardson JA, Olson EN. Cardiac-specific activity of an Nk2–5 enhancer requires an evolutionarily conserved Smad binding site. Dev Biol. 2002;244(2):257–266. doi: 10.1006/dbio.2002.0603. [DOI] [PubMed] [Google Scholar]
- 80.Lien CL, Wu C, Mercer B, Webb R, Richardson JA, Olson EN. Control of early cardiac-specific transcription of Nk2–5 by a GATA-dependent enhancer. Development. 1999;126(1):75–84. doi: 10.1242/dev.126.1.75. [DOI] [PubMed] [Google Scholar]
- 81.Liberatore CM, Searcy-Schrick RD, Vincent EB, Yutzey KE. Nkx-2.5 gene induction in mice is mediated by a Smad consensus regulatory region. Dev Biol. 2002;244(2):243–256. doi: 10.1006/dbio.2002.0604. [DOI] [PubMed] [Google Scholar]
- 82.Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, Stennard FA, Wise N, Schaft D, Wolstein O, Furtado MB, Shiratori H, Chien KR, Hamada H, Black BL, Saga Y, Robertson EJ, Buckingham ME, Harvey RP. An Nk2–5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128(5):947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barth JL, Clark CD, Fresco VM, Knoll EP, Lee B, Argraves WS, Lee KH. Jarid2 is among a set of genes differentially regulated by Nkx2.5 during outflow tract morphogenesis. Dev Dyn. 2010;239(7):2024–2033. doi: 10.1002/dvdy.22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nk2–5 and GATA-4 are mutual cofactors. EMBO J. 1997;16(18):5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tonjes M, Dunkel I, Sperling SR. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet. 2011;7(2):e1001313. doi: 10.1371/journal.pgen.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I. Tbx5 associates with Nk2–5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28(3):276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- 87.Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, Seidman JG, Seidman CE. A molecular pathway including Id2, Tbx5, and Nk2–5 required for cardiac conduction system development. Cell. 2007;129(7):1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 88.Zhou P, He A, Pu WT. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr Top Dev Biol. 2012;100:143–169. doi: 10.1016/B978-0-12-387786-4.00005-1. [DOI] [PubMed] [Google Scholar]
- 89.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, Srivastava D. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424(6947):443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 90.Rajagopal SK, Ma Q, Obler D, Shen J, Manichaikul A, Tomita-Mitchell A, Boardman K, Briggs C, Garg V, Srivastava D, Goldmuntz E, Broman KW, Benson DW, Smoot LB, Pu WT. Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol. 2007;43(6):677–685. doi: 10.1016/j.yjmcc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tomita-Mitchell A, Maslen CL, Morris CD, Garg V, Goldmuntz E. GATA4 sequence variants in patients with congenital heart disease. J Med Genet. 2007;44(12):779–783. doi: 10.1136/jmg.2007.052183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moskowitz IP, Wang J, Peterson MA, Pu WT, Mackinnon AC, Oxburgh L, Chu GC, Sarkar M, Berul C, Smoot L, Robertson EJ, Schwartz R, Seidman JG, Seidman CE. Transcription factor genes Smad4 and Gata4 cooperatively regulate cardiac valve development (corrected) Proc Natl Acad Sci USA. 2011;108(10):4006–4011. doi: 10.1073/pnas.1019025108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11(8):1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 94.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11(8):1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 95.Pu WT, Ishiwata T, Juraszek AL, Ma Q, Izumo S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol. 2004;275(1):235–244. doi: 10.1016/j.ydbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 96.Misra C, Sachan N, McNally CR, Koenig SN, Nichols HA, Guggilam A, Lucchesi PA, Pu WT, Srivastava D, Garg V. Congenital heart disease-causing Gata4 mutation displays functional deficits in vivo. PLoS Genet. 2012;8(5):e1002690. doi: 10.1371/journal.pgen.1002690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McFadden DG, Charite J, Richardson JA, Srivastava D, Firulli AB, Olson EN. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127(24):5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- 98.Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131(16):3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- 99.Maitra M, Schluterman MK, Nichols HA, Richardson JA, Lo CW, Srivastava D, Garg V. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev Biol. 2009;326(2):368–377. doi: 10.1016/j.ydbio.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Greulich F, Rudat C, Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc Res. 2011;91(2):212–222. doi: 10.1093/cvr/cvr112. [DOI] [PubMed] [Google Scholar]
- 101.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG. A murine model of Holt–Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106(6):709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 102.Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, Bonnet D, Lyonnet S, Young ID, Raeburn JA, Buckler AJ, Law DJ, Brook JD. Holt–Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15(1):21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 103.Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, Renault B, Kucherlapati R, Seidman JG, Seidman CE. Mutations in human TBX5 (corrected) cause limb and cardiac malformation in Holt–Oram syndrome. Nat Genet. 1997;15(1):30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 104.Kirk EP, Sunde M, Costa MW, Rankin SA, Wolstein O, Castro ML, Butler TL, Hyun C, Guo G, Otway R, Mackay JP, Waddell LB, Cole AD, Hayward C, Keogh A, Macdonald P, Griffiths L, Fatkin D, Sholler GF, Zorn AM, Feneley MP, Winlaw DS, Harvey RP. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet. 2007;81(2):280–291. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Singh MK, Christoffels VM, Dias JM, Trowe MO, Petry M, Schuster-Gossler K, Burger A, Ericson J, Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005;132(12):2697–2707. doi: 10.1242/dev.01854. [DOI] [PubMed] [Google Scholar]
- 106.Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL, Elliott DE, Prall OW, Black BL, Fatkin D, Harvey RP. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132(10):2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- 107.Cai CL, Zhou W, Yang L, Bu L, Qyang Y, Zhang X, Li X, Rosenfeld MG, Chen J, Evans S. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132(10):2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cai X, Nomura-Kitabayashi A, Cai W, Yan J, Christoffels VM, Cai CL. Myocardial Tbx20 regulates early atrioventricular canal formation and endocardial epithelial-mesenchymal transition via Bmp2. Dev Biol. 2011;360(2):381–390. doi: 10.1016/j.ydbio.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hammer S, Toenjes M, Lange M, Fischer JJ, Dunkel I, Mebus S, Grimm CH, Hetzer R, Berger F, Sperling S. Characterization of TBX20 in human hearts and its regulation by TFAP2. J Cell Biochem. 2008;104(3):1022–1033. doi: 10.1002/jcb.21686. [DOI] [PubMed] [Google Scholar]
- 110.Satoda M, Zhao F, Diaz GA, Burn J, Goodship J, Davidson HR, Pierpont ME, Gelb BD. Mutations in TFAP2B cause Char syndrome, a familial form of patent ductus arteriosus. Nat Genet. 2000;25(1):42–46. doi: 10.1038/75578. [DOI] [PubMed] [Google Scholar]
- 111.Chen YW, Zhao W, Zhang ZF, Fu Q, Shen J, Zhang Z, Ji W, Wang J, Li F. Familial nonsyndromic patent ductus arteriosus caused by mutations in TFAP2B. Pediatr Cardiol. 2011;32(7):958–965. doi: 10.1007/s00246-011-0024-7. [DOI] [PubMed] [Google Scholar]
- 112.Khetyar M, Syrris P, Tinworth L, Abushaban L, Carter N. Novel TFAP2B mutation in nonsyndromic patent ductus arteriosus. Genet Test. 2008;12(3):457–459. doi: 10.1089/gte.2008.0015. [DOI] [PubMed] [Google Scholar]
- 113.Zhao F, Bosserhoff AK, Buettner R, Moser M. A heart-hand syndrome gene: Tfap2b plays a critical role in the development and remodeling of mouse ductus arteriosus and limb patterning. PLoS One. 2011;6(7):e22908. doi: 10.1371/journal.pone.0022908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Braganca J, Swingler T, Marques FI, Jones T, Eloranta JJ, Hurst HC, Shioda T, Bhattacharya S. Human CREB-binding protein/p300-interacting transactivator with ED-rich tail (CITED) 4, a new member of the CITED family, functions as a co-activator for transcription factor AP-2. J Biol Chem. 2002;277(10):8559–8565. doi: 10.1074/jbc.M110850200. [DOI] [PubMed] [Google Scholar]
- 115.Braganca J, Eloranta JJ, Bamforth SD, Ibbitt JC, Hurst HC, Bhattacharya S. Physical and functional interactions among AP-2 transcription factors, p300/CREB-binding protein, and CITED2. J Biol Chem. 2003;278(18):16021–16029. doi: 10.1074/jbc.M208144200. [DOI] [PubMed] [Google Scholar]
- 116.Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ. Rubinstein–Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376(6538):348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 117.Roelfsema JH, White SJ, Ariyurek Y, Bartholdi D, Niedrist D, Papadia F, Bacino CA, den Dunnen JT, van Ommen GJ, Breuning MH, Hennekam RC, Peters DJ. Genetic heterogeneity in Rubinstein–Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet. 2005;76(4):572–580. doi: 10.1086/429130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Posch MG, Waldmuller S, Muller M, Scheffold T, Fournier D, Andrade-Navarro MA, De GB, Guillaumont S, Dauphin C, Yousseff D, Schmitt KR, Perrot A, Berger F, Hetzer R, Bouvagnet P, Ozcelik C. Cardiac alpha-myosin (MYH6) is the predominant sarcomeric disease gene for familial atrial septal defects. PLoS One. 2011;6(12):e28872. doi: 10.1371/journal.pone.0028872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Granados-Riveron JT, Ghosh TK, Pope M, Bu’Lock F, Thornborough C, Eason J, Kirk EP, Fatkin D, Feneley MP, Harvey RP, Armour JA, David BJ. Alpha-cardiac myosin heavy chain (MYH6) mutations affecting myofibril formation are associated with congenital heart defects. Hum Mol Genet. 2010;19(20):4007–4016. doi: 10.1093/hmg/ddq315. [DOI] [PubMed] [Google Scholar]
- 120.Ching YH, Ghosh TK, Cross SJ, Packham EA, Honeyman L, Loughna S, Robinson TE, Dearlove AM, Ribas G, Bonser AJ, Thomas NR, Scotter AJ, Caves LS, Tyrrell GP, Newbury-Ecob RA, Munnich A, Bonnet D, Brook JD. Mutation in myosin heavy chain 6 causes atrial septal defect. Nat Genet. 2005;37(4):423–428. doi: 10.1038/ng1526. [DOI] [PubMed] [Google Scholar]
- 121.Dai YS, Cserjesi P, Markham BE, Molkentin JD. The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J Biol Chem. 2002;277(27):24390–24398. doi: 10.1074/jbc.M202490200. [DOI] [PubMed] [Google Scholar]
- 122.Ghosh TK, Song FF, Packham EA, Buxton S, Robinson TE, Ronksley J, Self T, Bonser AJ, Brook JD. Physical interaction between TBX5 and MEF2C is required for early heart development. Mol Cell Biol. 2009;29(8):2205–2218. doi: 10.1128/MCB.01923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Postma AV, van Engelen K, van de Meerakker J, Rahman T, Probst S, Baars MJ, Bauer U, Pickardt T, Sperling SR, Berger F, Moorman AF, Mulder BJ, Thierfelder L, Keavney B, Goodship J, Klaassen S. Mutations in the sarcomere gene MYH7 in Ebstein anomaly. Circ Cardiovasc Genet. 2011;4(1):43–50. doi: 10.1161/CIRCGENETICS.110.957985. [DOI] [PubMed] [Google Scholar]
- 124.Matsson H, Eason J, Bookwalter CS, Klar J, Gustavsson P, Sunnegardh J, Enell H, Jonzon A, Vikkula M, Gutierrez I, Granados-Riveron J, Pope M, Bu’Lock F, Cox J, Robinson TE, Song F, Brook DJ, Marston S, Trybus KM, Dahl N. Alpha-cardiac actin mutations produce atrial septal defects. Hum Mol Genet. 2008;17(2):256–265. doi: 10.1093/hmg/ddm302. [DOI] [PubMed] [Google Scholar]
- 125.Zhu L, Vranckx R, Van Khau KP, Lalande A, Boisset N, Mathieu F, Wegman M, Glancy L, Gasc JM, Brunotte F, Bruneval P, Wolf JE, Michel JB, Jeunemaitre X. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat Genet. 2006;38(3):343–349. doi: 10.1038/ng1721. [DOI] [PubMed] [Google Scholar]
- 126.Morano I, Chai GX, Baltas LG, Lamounier-Zepter V, Lutsch G, Kott M, Haase H, Bader M. Smooth-muscle contraction without smooth-muscle myosin. Nat Cell Biol. 2000;2(6):371–375. doi: 10.1038/35014065. [DOI] [PubMed] [Google Scholar]
- 127.Bokenkamp R, DeRuiter MC, van Munsteren C, Gittenberger-de-Groot AC. Insights into the pathogenesis and genetic background of patency of the ductus arteriosus. Neonatology. 2010;98(1):6–17. doi: 10.1159/000262481. [DOI] [PubMed] [Google Scholar]
- 128.Slomp J, Gittenberger-de Groot AC, Glukhova MA, van Conny MJ, Kockx MM, Schwartz SM, Koteliansky VE. Differentiation, dedifferentiation, and apoptosis of smooth muscle cells during the development of the human ductus arteriosus. Arterioscler Thromb Vasc Biol. 1997;17(5):1003–1009. doi: 10.1161/01.atv.17.5.1003. [DOI] [PubMed] [Google Scholar]
- 129.Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu Rev Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- 130.Hannibal MC, Buckingham KJ, Ng SB, Ming JE, Beck AE, McMillin MJ, Gildersleeve HI, Bigham AW, Tabor HK, Mefford HC, Cook J, Yoshiura K, Matsumoto T, Matsumoto N, Miyake N, Tonoki H, Naritomi K, Kaname T, Nagai T, Ohashi H, Kurosawa K, Hou JW, Ohta T, Liang D, Sudo A, Morris CA, Banka S, Black GC, Clayton-Smith J, Nickerson DA, Zackai EH, Shaikh TH, Donnai D, Niikawa N, Shendure J, Bamshad MJ. Spectrum of MLL2 (ALR) mutations in 110 cases of Kabuki syndrome. Am J Med Genet A. 2011;155A(7):1511–1516. doi: 10.1002/ajmg.a.34074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Digilio MC, Marino B, Toscano A, Giannotti A, Dallapiccola B. Congenital heart defects in Kabuki syndrome. Am J Med Genet. 2001;100(4):269–274. doi: 10.1002/ajmg.1265. [DOI] [PubMed] [Google Scholar]
- 132.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42(9):790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20(3):341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lalani SR, Safiullah AM, Fernbach SD, Harutyunyan KG, Thaller C, Peterson LE, McPherson JD, Gibbs RA, White LD, Hefner M, Davenport SL, Graham JM, Bacino CA, Glass NL, Towbin JA, Craigen WJ, Neish SR, Lin AE, Belmont JW. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am J Hum Genet. 2006;78(2):303–314. doi: 10.1086/500273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jongmans MC, Admiraal RJ, van der Donk KP, Vissers LE, Baas AF, Kapusta L, van Hagen JM, Donnai D, de Ravel TJ, Veltman JA, van Geurts KA, de Vries BB, Brunner HG, Hoefsloot LH, van Ravenswaaij CM. CHARGE syndrome: the phenotypic spectrum of mutations in the CHD7 gene. J Med Genet. 2006;43(4):306–314. doi: 10.1136/jmg.2005.036061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wyse RK, al-Mahdawi S, Burn J, Blake K. Congenital heart disease in CHARGE association. Pediatr Cardiol. 1993;14(2):75–81. doi: 10.1007/BF00796983. [DOI] [PubMed] [Google Scholar]
- 137.Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, Schoenmakers EF, Brunner HG, Veltman JA, van Kessel AG. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36(9):955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 138.Schnetz MP, Bartels CF, Shastri K, Balasubramanian D, Zentner GE, Balaji R, Zhang X, Song L, Wang Z, Laframboise T, Crawford GE, Scacheri PC. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19(4):590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463(7283):958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Keyte A, Hutson MR. The neural crest in cardiac congenital anomalies. Differentiation. 2012;84(1):25–40. doi: 10.1016/j.diff.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, Carriero NJ, Cheung YH, Deanfield J, Depalma S, Fakhro KA, Glessner J, Hakonarson H, Italia MJ, Kaltman JR, Kaski J, Kim R, Kline JK, Lee T, Leipzig J, Lopez A, Mane SM, Mitchell LE, Newburger JW, Parfenov M, Pe’er I, Porter G, Roberts AE, Sachidanandam R, Sanders SJ, Seiden HS, State MW, Subramanian S, Tikhonova IR, Wang W, Warburton D, White PS, Williams IA, Zhao H, Seidman JG, Brueckner M, Chung WK, Gelb BD, Goldmuntz E, Seidman CE, Lifton RP. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Harris JA, Francannet C, Pradat P, Robert E. The epidemiology of cardiovascular defects, part 2: a study based on data from three large registries of congenital malformations. Pediatr Cardiol. 2003;24(3):222–235. doi: 10.1007/s00246-002-9402-5. [DOI] [PubMed] [Google Scholar]
- 143.Meberg A, Hals J, Thaulow E. Congenital heart defects–chromosomal anomalies, syndromes and extracardiac malformations. Acta Paediatr. 2007;96(8):1142–1145. doi: 10.1111/j.1651-2227.2007.00381.x. [DOI] [PubMed] [Google Scholar]
- 144.Schellberg R, Schwanitz G, Gravinghoff L, Kallenberg R, Trost D, Raff R, Wiebe W. New trends in chromosomal investigation in children with cardiovascular malformations. Cardiol Young. 2004;14(6):622–629. doi: 10.1017/S1047951104006079. [DOI] [PubMed] [Google Scholar]
- 145.Centers for Disease Control and Prevention (CDC) Improved national prevalence estimates for 18 selected major birth defects–United States, 1999–2001. MMWR Morb Mortal Wkly Rep. 2006;54(51):1301–1305. [PubMed] [Google Scholar]
- 146.Freeman SB, Bean LH, Allen EG, Tinker SW, Locke AE, Druschel C, Hobbs CA, Romitti PA, Royle MH, Torfs CP, Dooley KJ, Sherman SL. Ethnicity, sex, and the incidence of congenital heart defects: a report from the national Down syndrome project. Genet Med. 2008;10(3):173–180. doi: 10.1097/GIM.0b013e3181634867. [DOI] [PubMed] [Google Scholar]
- 147.Embleton ND, Wyllie JP, Wright MJ, Burn J, Hunter S. Natural history of trisomy 18. Arch Dis Child Fetal Neonatal Ed. 1996;75(1):F38–F41. doi: 10.1136/fn.75.1.f38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lin HY, Lin SP, Chen YJ, Hung HY, Kao HA, Hsu CH, Chen MR, Chang JH, Ho CS, Huang FY, Shyur SD, Lin DS, Lee HC. Clinical characteristics and survival of trisomy 18 in a medical center in Taipei, 1988–2004. Am J Med Genet A. 2006;140(9):945–951. doi: 10.1002/ajmg.a.31173. [DOI] [PubMed] [Google Scholar]
- 149.Mazzanti L, Cacciari E. Congenital heart disease in patients with Turner’s syndrome. Italian study group for Turner syndrome (ISGTS) J Pediatr. 1998;133(5):688–692. doi: 10.1016/s0022-3476(98)70119-2. [DOI] [PubMed] [Google Scholar]
- 150.Wyllie JP, Wright MJ, Burn J, Hunter S. Natural history of trisomy 13. Arch Dis Child. 1994;71(4):343–345. doi: 10.1136/adc.71.4.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Donald-McGinn DM, Zackai EH. Genetic counseling for the 22q11.2 deletion. Dev Disabil Res Rev. 2008;14(1):69–74. doi: 10.1002/ddrr.10. [DOI] [PubMed] [Google Scholar]
- 152.Ou Z, Berg JS, Yonath H, Enciso VB, Miller DT, Picker J, Lenzi T, Keegan CE, Sutton VR, Belmont J, Chinault AC, Lupski JR, Cheung SW, Roeder E, Patel A. Microduplications of 22q11.2 are frequently inherited and are associated with variable phenotypes. Genet Med. 2008;10(4):267–277. doi: 10.1097/GIM.0b013e31816b64c2. [DOI] [PubMed] [Google Scholar]
- 153.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27(3):286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 154.Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haploinsufficiency in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410(6824):97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 155.Xu H, Morishima M, Wylie JN, Schwartz RJ, Bruneau BG, Lindsay EA, Baldini A. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;131(13):3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- 156.Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, Ichida F, Joo K, Kimura M, Imamura S, Kamatani N, Momma K, Takao A, Nakazawa M, Shimizu N, Matsuoka R. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362(9393):1366–1373. doi: 10.1016/s0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- 157.Zweier C, Sticht H, Aydin-Yaylagul I, Campbell CE, Rauch A. Human TBX1 missense mutations cause gain of function resulting in the same phenotype as 22q11.2 deletions. Am J Hum Genet. 2007;80(3):510–517. doi: 10.1086/511993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Funke B, Epstein JA, Kochilas LK, Lu MM, Pandita RK, Liao J, Bauerndistel R, Schuler T, Schorle H, Brown MC, Adams J, Morrow BE. Mice overexpressing genes from the 22q11 region deleted in velo-cardio-facial syndrome/DiGeorge syndrome have middle and inner ear defects. Hum Mol Genet. 2001;10(22):2549–2556. doi: 10.1093/hmg/10.22.2549. [DOI] [PubMed] [Google Scholar]
- 159.Liao J, Kochilas L, Nowotschin S, Arnold JS, Aggarwal VS, Epstein JA, Brown MC, Adams J, Morrow BE. Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Hum Mol Genet. 2004;13(15):1577–1585. doi: 10.1093/hmg/ddh176. [DOI] [PubMed] [Google Scholar]
- 160.Garcia-Minaur S, Fantes J, Murray RS, Porteous ME, Strain L, Burns JE, Stephen J, Warner JP. A novel atypical 22q11.2 distal deletion in father and son. J Med Genet. 2002;39(10):E62. doi: 10.1136/jmg.39.10.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rauch A, Zink S, Zweier C, Thiel CT, Koch A, Rauch R, Lascorz J, Huffmeier U, Weyand M, Singer H, Hofbeck M. Systematic assessment of atypical deletions reveals genotype-phenotype correlation in 22q11.2. J Med Genet. 2005;42(11):871–876. doi: 10.1136/jmg.2004.030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sorensen KM, El-Segaier M, Fernlund E, Errami A, Bouvagnet P, Nehme N, Steensberg J, Hjortdal V, Soller M, Behjati M, Werge T, Kirchoff M, Schouten J, Tommerup N, Andersen PS, Larsen LA. Screening of congenital heart disease patients using multiplex ligation-dependent probe amplification: early diagnosis of syndromic patients. Am J Med Genet A. 2012;158A(4):720–725. doi: 10.1002/ajmg.a.35214. [DOI] [PubMed] [Google Scholar]
- 163.Guris DL, Fantes J, Tara D, Druker BJ, Imamoto A. Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat Genet. 2001;27(3):293–298. doi: 10.1038/85855. [DOI] [PubMed] [Google Scholar]
- 164.Guris DL, Duester G, Papaioannou VE, Imamoto A. Dose-dependent interaction of Tbx1 and Crkl and locally aberrant RA signaling in a model of del22q11 syndrome. Dev Cell. 2006;10(1):81–92. doi: 10.1016/j.devcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 165.Moon AM, Guris DL, Seo JH, Li L, Hammond J, Talbot A, Imamoto A. Crkl deficiency disrupts Fgf8 signaling in a mouse model of 22q11 deletion syndromes. Dev Cell. 2006;10(1):71–80. doi: 10.1016/j.devcel.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liao J, Aggarwal VS, Nowotschin S, Bondarev A, Lipner S, Morrow BE. Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev Biol. 2008;316(2):524–537. doi: 10.1016/j.ydbio.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Roberts C, Ivins S, Cook AC, Baldini A, Scambler PJ. Cyp26 genes a1, b1 and c1 are down-regulated in Tbx1 null mice and inhibition of Cyp26 enzyme function produces a phenocopy of DiGeorge Syndrome in the chick. Hum Mol Genet. 2006;15(23):3394–3410. doi: 10.1093/hmg/ddl416. [DOI] [PubMed] [Google Scholar]
- 168.Ivins S, van Lammerts BK, Roberts C, James C, Lindsay E, Baldini A, Ataliotis P, Scambler PJ. Microarray analysis detects differentially expressed genes in the pharyngeal region of mice lacking Tbx1. Dev Biol. 2005;285(2):554–569. doi: 10.1016/j.ydbio.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 169.Roberts C, Ivins SM, James CT, Scambler PJ. Retinoic acid down-regulates Tbx1 expression in vivo and in vitro. Dev Dyn. 2005;232(4):928–938. doi: 10.1002/dvdy.20268. [DOI] [PubMed] [Google Scholar]
- 170.Garg V, Yamagishi C, Hu T, Kathiriya IS, Yamagishi H, Srivastava D. Tbx1, a DiGeorge syndrome candidate gene, is regulated by sonic hedgehog during pharyngeal arch development. Dev Biol. 2001;235(1):62–73. doi: 10.1006/dbio.2001.0283. [DOI] [PubMed] [Google Scholar]
- 171.Yamagishi H, Maeda J, Hu T, McAnally J, Conway SJ, Kume T, Meyers EN, Yamagishi C, Srivastava D. Tbx1 is regulated by tissue-specific forkhead proteins through a common Sonic hedgehog-responsive enhancer. Genes Dev. 2003;17(2):269–281. doi: 10.1101/gad.1048903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hu T, Yamagishi H, Maeda J, McAnally J, Yamagishi C, Srivastava D. Tbx1 regulates fibroblast growth factors in the anterior heart field through a reinforcing autoregulatory loop involving forkhead transcription factors. Development. 2004;131(21):5491–5502. doi: 10.1242/dev.01399. [DOI] [PubMed] [Google Scholar]
- 173.Vitelli F, Taddei I, Morishima M, Meyers EN, Lindsay EA, Baldini A. A genetic link between Tbx1 and fibroblast growth factor signaling. Development. 2002;129(19):4605–4611. doi: 10.1242/dev.129.19.4605. [DOI] [PubMed] [Google Scholar]
- 174.Guo C, Sun Y, Zhou B, Adam RM, Li X, Pu WT, Morrow BE, Moon A, Li X. A Tbx1-Six1/Eya1-Fgf8 genetic pathway controls mammalian cardiovascular and craniofacial morphogenesis. J Clin Invest. 2011;121(4):1585–1595. doi: 10.1172/JCI44630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fulcoli FG, Huynh T, Scambler PJ, Baldini A. Tbx1 regulates the BMP-Smad1 pathway in a transcription independent manner. PLoS One. 2009;4(6):e6049. doi: 10.1371/journal.pone.0006049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 176.Ewart AK, Morris CA, Atkinson D, Jin W, Sternes K, Spallone P, Stock AD, Leppert M, Keating MT. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 1993;5(1):11–16. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- 177.Lowery MC, Morris CA, Ewart A, Brothman LJ, Zhu XL, Leonard CO, Carey JC, Keating M, Brothman AR. Strong correlation of elastin deletions, detected by FISH, with Williams syndrome: evaluation of 235 patients. Am J Hum Genet. 1995;57(1):49–53. [PMC free article] [PubMed] [Google Scholar]
- 178.Perez Jurado LA, Peoples R, Kaplan P, Hamel BC, Francke U. Molecular definition of the chromosome 7 deletion in Williams syndrome and parent-of-origin effects on growth. Am J Hum Genet. 1996;59(4):781–792. [PMC free article] [PubMed] [Google Scholar]
- 179.Pober BR. Williams–Beuren syndrome. N Engl J Med. 2010;362(3):239–252. doi: 10.1056/NEJMra0903074. [DOI] [PubMed] [Google Scholar]
- 180.del Pasqua A, Rinelli G, Toscano A, Iacobelli R, Digilio C, Marino B, Saffirio C, Mondillo S, Pasquini L, Sanders SP, de Zorzi A. New findings concerning cardiovascular manifestations emerging from long-term follow-up of 150 patients with the Williams–Beuren–Beuren syndrome. Cardiol Young. 2009;19(6):563–567. doi: 10.1017/S1047951109990837. [DOI] [PubMed] [Google Scholar]
- 181.Eronen M, Peippo M, Hiippala A, Raatikka M, Arvio M, Johansson R, Kahkonen M. Cardiovascular manifestations in 75 patients with Williams syndrome. J Med Genet. 2002;39(8):554–558. doi: 10.1136/jmg.39.8.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Frangiskakis JM, Ewart AK, Morris CA, Mervis CB, Bertrand J, Robinson BF, Klein BP, Ensing GJ, Everett LA, Green ED, Proschel C, Gutowski NJ, Noble M, Atkinson DL, Odelberg SJ, Keating MT. LIM-kinase1 hemizygosity implicated in impaired visuospatial constructive cognition. Cell. 1996;86(1):59–69. doi: 10.1016/s0092-8674(00)80077-x. [DOI] [PubMed] [Google Scholar]
- 183.Tassabehji M, Metcalfe K, Karmiloff-Smith A, Carette MJ, Grant J, Dennis N, Reardon W, Splitt M, Read AP, Donnai D. Williams syndrome: use of chromosomal microdeletions as a tool to dissect cognitive and physical phenotypes. Am J Hum Genet. 1999;64(1):118–125. doi: 10.1086/302214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li DY, Toland AE, Boak BB, Atkinson DL, Ensing GJ, Morris CA, Keating MT. Elastin point mutations cause an obstructive vascular disease, supravalvular aortic stenosis. Hum Mol Genet. 1997;6(7):1021–1028. doi: 10.1093/hmg/6.7.1021. [DOI] [PubMed] [Google Scholar]
- 185.Metcalfe K, Rucka AK, Smoot L, Hofstadler G, Tuzler G, McKeown P, Siu V, Rauch A, Dean J, Dennis N, Ellis I, Reardon W, Cytrynbaum C, Osborne L, Yates JR, Read AP, Donnai D, Tassabehji M. Elastin: mutational spectrum in supravalvular aortic stenosis. Eur J Hum Genet. 2000;8(12):955–963. doi: 10.1038/sj.ejhg.5200564. [DOI] [PubMed] [Google Scholar]
- 186.Li DY, Faury G, Taylor DG, Davis EC, Boyle WA, Mecham RP, Stenzel P, Boak B, Keating MT. Novel arterial pathology in mice and humans hemizygous for elastin. J Clin Invest. 1998;102(10):1783–1787. doi: 10.1172/JCI4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yoshimura K, Kitagawa H, Fujiki R, Tanabe M, Takezawa S, Takada I, Yamaoka I, Yonezawa M, Kondo T, Furutani Y, Yagi H, Yoshinaga S, Masuda T, Fukuda T, Yamamoto Y, Ebihara K, Li DY, Matsuoka R, Takeuchi JK, Matsumoto T, Kato S. Distinct function of 2 chromatin remodeling complexes that share a common subunit, Williams syndrome transcription factor (WSTF) Proc Natl Acad Sci USA. 2009;106(23):9280–9285. doi: 10.1073/pnas.0901184106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 188.Kitagawa H, Fujiki R, Yoshimura K, Mezaki Y, Uematsu Y, Matsui D, Ogawa S, Unno K, Okubo M, Tokita A, Nakagawa T, Ito T, Ishimi Y, Nagasawa H, Matsumoto T, Yanagisawa J, Kato S. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell. 2003;113(7):905–917. doi: 10.1016/s0092-8674(03)00436-7. [DOI] [PubMed] [Google Scholar]
- 189.Bozhenok L, Wade PA, Varga-Weisz P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 2002;21(9):2231–2241. doi: 10.1093/emboj/21.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cavellan E, Asp P, Percipalle P, Farrants AK. The WSTF-SNF2 h chromatin remodeling complex interacts with several nuclear proteins in transcription. J Biol Chem. 2006;281(24):16264–16271. doi: 10.1074/jbc.M600233200. [DOI] [PubMed] [Google Scholar]
- 191.Poot RA, Bozhenok L, van den Berg DL, Steffensen S, Ferreira F, Grimaldi M, Gilbert N, Ferreira J, Varga-Weisz PD. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat Cell Biol. 2004;6(12):1236–1244. doi: 10.1038/ncb1196. [DOI] [PubMed] [Google Scholar]
- 192.Maas NM, Van BG, Hannes F, Thienpont B, Sanlaville D, Kok K, Midro A, Andrieux J, Anderlid BM, Schoumans J, Hordijk R, Devriendt K, Fryns JP, Vermeesch JR. Genotype-phenotype correlation in 21 patients with Wolf-Hirschhorn syndrome using high resolution array comparative genome hybridisation (CGH) J Med Genet. 2008;45(2):71–80. doi: 10.1136/jmg.2007.052910. [DOI] [PubMed] [Google Scholar]
- 193.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone H3 lysine 36 trimethyltransferase links Nk2–5 to Wolf-Hirschhorn syndrome. Nature. 2009;460(7252):287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 194.Catela C, Bilbao-Cortes D, Slonimsky E, Kratsios P, Rosenthal N, Te WP. Multiple congenital malformations of Wolf–Hirschhorn syndrome are recapitulated in Fgfrl1 null mice. Dis Model Mech. 2009;2(5–6):283–294. doi: 10.1242/dmm.002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wat MJ, Shchelochkov OA, Holder AM, Breman AM, Dagli A, Bacino C, Scaglia F, Zori RT, Cheung SW, Scott DA, Kang SH. Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am J Med Genet A. 2009;149A(8):1661–1677. doi: 10.1002/ajmg.a.32896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhang W, Li X, Shen A, Jiao W, Guan X, Li Z. GATA4 mutations in 486 Chinese patients with congenital heart disease. Eur J Med Genet. 2008;51(6):527–535. doi: 10.1016/j.ejmg.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 197.Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, Collins A, Mercer C, Norga K, de Ravel T, Devriendt K, Bongers EM, de Leeuw N, Reardon W, Gimelli S, Bena F, Hennekam RC, Male A, Gaunt L, Clayton-Smith J, Simonic I, Park SM, Mehta SG, Nik-Zainal S, Woods CG, Firth HV, Parkin G, Fichera M, Reitano S, Lo GM, Li KE, Casuga I, Broomer A, Conrad B, Schwerzmann M, Raber L, Gallati S, Striano P, Coppola A, Tolmie JL, Tobias ES, Lilley C, Armengol L, Spysschaert Y, Verloo P, De Coene A, Goossens L, Mortier G, Speleman F, van Binsbergen E, Nelen MR, Hochstenbach R, Poot M, Gallagher L, Gill M, McClellan J, King MC, Regan R, Skinner C, Stevenson RE, Antonarakis SE, Chen C, Estivill X, Menten B, Gimelli G, Gribble S, Schwartz S, Sutcliffe JS, Walsh T, Knight SJ, Sebat J, Romano C, Schwartz CE, Veltman JA, de Vries BB, Vermeesch JR, Barber JC, Willatt L, Tassabehji M, Eichler EE. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359(16):1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Soemedi R, Topf A, Wilson IJ, Darlay R, Rahman T, Glen E, Hall D, Huang N, Bentham J, Bhattacharya S, Cosgrove C, Brook JD, Granados-Riveron J, Setchfield K, Bu’Lock F, Thornborough C, Devriendt K, Breckpot J, Hofbeck M, Lathrop M, Rauch A, Blue GM, Winlaw DS, Hurles M, Santibanez-Koref M, Cordell HJ, Goodship JA, Keavney BD. Phenotype-specific effect of chromosome 1q21.1 rearrangements and GJA5 duplications in 2436 congenital heart disease patients and 6760 controls. Hum Mol Genet. 2012;21(7):1513–1520. doi: 10.1093/hmg/ddr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gu H, Smith FC, Taffet SM, Delmar M. High incidence of cardiac malformations in connexin40-deficient mice. Circ Res. 2003;93(3):201–206. doi: 10.1161/01.RES.0000084852.65396.70. [DOI] [PubMed] [Google Scholar]
- 200.Kirchhoff S, Kim JS, Hagendorff A, Thonnissen E, Kruger O, Lamers WH, Willecke K. Abnormal cardiac conduction and morphogenesis in connexin40 and connexin43 double-deficient mice. Circ Res. 2000;87(5):399–405. doi: 10.1161/01.res.87.5.399. [DOI] [PubMed] [Google Scholar]
- 201.Gourdie RG, Severs NJ, Green CR, Rothery S, Germroth P, Thompson RP. The spatial distribution and relative abundance of gap-junctional connexin40 and connexin43 correlate to functional properties of components of the cardiac atrioventricular conduction system. J Cell Sci. 1993;105(Pt 4):985–991. doi: 10.1242/jcs.105.4.985. [DOI] [PubMed] [Google Scholar]
- 202.Gros D, Jarry-Guichard T, Ten Velde I, de Maziere A, van Kempen MJ, Davoust J, Briand JP, Moorman AF, Jongsma HJ. Restricted distribution of connexin40, a gap junctional protein, in mammalian heart. Circ Res. 1994;74(5):839–851. doi: 10.1161/01.res.74.5.839. [DOI] [PubMed] [Google Scholar]
- 203.Simon AM, Goodenough DA, Paul DL. Mice lacking connexin40 have cardiac conduction abnormalities characteristic of atrioventricular block and bundle branch block. Curr Biol. 1998;8(5):295–298. doi: 10.1016/s0960-9822(98)70113-7. [DOI] [PubMed] [Google Scholar]
- 204.Kirchhoff S, Nelles E, Hagendorff A, Kruger O, Traub O, Willecke K. Reduced cardiac conduction velocity and predisposition to arrhythmias in connexin40-deficient mice. Curr Biol. 1998;8(5):299–302. doi: 10.1016/s0960-9822(98)70114-9. [DOI] [PubMed] [Google Scholar]
- 205.Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, Magee A, Genevieve D, Cormier-Daire V, Van EH, Fryns JP, Hamel BC, Sistermans EA, de Vries BB, van Bokhoven H. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet. 2006;79(2):370–377. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kleefstra T, Smidt M, Banning MJ, Oudakker AR, Van Esch H, de Brouwer AP, Nillesen W, Sistermans EA, Hamel BC, de Bruijn D, Fryns JP, Yntema HG, Brunner HG, de Vries BB, van Bokhoven H. Disruption of the gene euchromatin histone methyl transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J Med Genet. 2005;42(4):299–306. doi: 10.1136/jmg.2004.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Stewart DR, Huang A, Faravelli F, Anderlid BM, Medne L, Ciprero K, Kaur M, Rossi E, Tenconi R, Nordenskjold M, Gripp KW, Nicholson L, Meschino WS, Capua E, Quarrell OW, Flint J, Irons M, Giampietro PF, Schowalter DB, Zaleski CA, Malacarne M, Zackai EH, Spinner NB, Krantz ID. Subtelomeric deletions of chromosome 9q: a novel microdeletion syndrome. Am J Med Genet A. 2004;128A(4):340–351. doi: 10.1002/ajmg.a.30136. [DOI] [PubMed] [Google Scholar]
- 208.Willemsen MH, Vulto-van Silfhout AT, Nillesen WM, Wissink-Lindhout WM, van Bokhoven H, Philip N, Berry-Kravis EM, Kini U, van Ravenswaaij-Arts CM, Delle CB, Innes AM, Houge G, Kosonen T, Cremer K, Fannemel M, Stray-Pedersen A, Reardon W, Ignatius J, Lachlan K, Mircher C, Helderman van den Enden PT, Mastebroek M, Cohn-Hokke PE, Yntema HG, Drunat S, Kleefstra T (2012) Update on Kleefstra syndrome. Mol Syndromol 2(3–5):202–212 [DOI] [PMC free article] [PubMed]
- 209.Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19(7):815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Dubourg C, Sanlaville D, Doco-Fenzy M, Le CC, Missirian C, Jaillard S, Schluth-Bolard C, Landais E, Boute O, Philip N, Toutain A, David A, Edery P, Moncla A, Martin-Coignard D, Vincent-Delorme C, Mortemousque I, Duban-Bedu B, Drunat S, Beri M, Mosser J, Odent S, David V, Andrieux J. Clinical and molecular characterization of 17q21.31 microdeletion syndrome in 14 French patients with mental retardation. Eur J Med Genet. 2011;54(2):144–151. doi: 10.1016/j.ejmg.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 211.Koolen DA, Sharp AJ, Hurst JA, Firth HV, Knight SJ, Goldenberg A, Saugier-Veber P, Pfundt R, Vissers LE, Destree A, Grisart B, Rooms L, Aa N, Field M, Hackett A, Bell K, Nowaczyk MJ, Mancini GM, Poddighe PJ, Schwartz CE, Rossi E, Gregori M, Antonacci-Fulton LL, McLellan MD, Garrett JM, Wiechert MA, Miner TL, Crosby S, Ciccone R, Willatt L, Rauch A, Zenker M, Aradhya S, Manning MA, Strom TM, Wagenstaller J, Krepischi-Santos AC, Vianna-Morgante AM, Rosenberg C, Price SM, Stewart H, Shaw-Smith C, Brunner HG, Wilkie AO, Veltman JA, Zuffardi O, Eichler EE, Vries BB. Clinical and molecular delineation of the 17q21.31 microdeletion syndrome. J Med Genet. 2008;45(11):710–720. doi: 10.1136/jmg.2008.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Koolen DA, Kramer JM, Neveling K, Nillesen WM, Moore-Barton HL, Elmslie FV, Toutain A, Amiel J, Malan V, Tsai AC, Cheung SW, Gilissen C, Verwiel ET, Martens S, Feuth T, Bongers EM, de Vries P, Scheffer H, Vissers LE, Brouwer AP, Brunner HG, Veltman JA, Schenck A, Yntema HG, de Vries BB. Mutations in the chromatin modifier gene KANSL1 cause the 17q21.31 microdeletion syndrome. Nat Genet. 2012;44(6):639–641. doi: 10.1038/ng.2262. [DOI] [PubMed] [Google Scholar]
- 213.Zollino M, Orteschi D, Murdolo M, Lattante S, Battaglia D, Stefanini C, Mercuri E, Chiurazzi P, Neri G, Marangi G. Mutations in KANSL1 cause the 17q21.31 microdeletion syndrome phenotype. Nat Genet. 2012;44(6):636–638. doi: 10.1038/ng.2257. [DOI] [PubMed] [Google Scholar]
- 214.Laverty C, Lucci J, Akhtar A. The MSL complex: X chromosome and beyond. Curr Opin Genet Dev. 2010;20(2):171–178. doi: 10.1016/j.gde.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 215.Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25(21):9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ballif BC, Theisen A, Rosenfeld JA, Traylor RN, Gastier-Foster J, Thrush DL, Astbury C, Bartholomew D, McBride KL, Pyatt RE, Shane K, Smith WE, Banks V, Gallentine WB, Brock P, Rudd MK, Adam MP, Keene JA, Phillips JA, III, Pfotenhauer JP, Gowans GC, Stankiewicz P, Bejjani BA, Shaffer LG (2010) Identification of a recurrent microdeletion at 17q23.1q23.2 flanked by segmental duplications associated with heart defects and limb abnormalities. Am J Hum Genet 86(3):454–461 [DOI] [PMC free article] [PubMed]
- 217.Harrelson Z, Kelly RG, Goldin SN, Gibson-Brown JJ, Bollag RJ, Silver LM, Papaioannou VE. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004;131(20):5041–5052. doi: 10.1242/dev.01378. [DOI] [PubMed] [Google Scholar]
- 218.Christoffels VM, Hoogaars WM, Tessari A, Clout DE, Moorman AF, Campione M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev Dyn. 2004;229(4):763–770. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- 219.Habets PE, Moorman AF, Clout DE, Roon MA, Lingbeek M, Lohuizen M, Campione M, Christoffels VM. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16(10):1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Breckpot J, Thienpont B, Peeters H, Ravel T, Singer A, Rayyan M, Allegaert K, Vanhole C, Eyskens B, Vermeesch JR, Gewillig M, Devriendt K. Array comparative genomic hybridization as a diagnostic tool for syndromic heart defects. J Pediatr. 2010;156(5):810–817. doi: 10.1016/j.jpeds.2009.11.049. [DOI] [PubMed] [Google Scholar]
- 221.Breckpot J, Thienpont B, Arens Y, Tranchevent LC, Vermeesch JR, Moreau Y, Gewillig M, Devriendt K. Challenges of interpreting copy number variation in syndromic and non-syndromic congenital heart defects. Cytogenet Genome Res. 2011;135(3–4):251–259. doi: 10.1159/000331272. [DOI] [PubMed] [Google Scholar]
- 222.Erdogan F, Larsen LA, Zhang L, Tumer Z, Tommerup N, Chen W, Jacobsen JR, Schubert M, Jurkatis J, Tzschach A, Ropers HH, Ullmann R. High frequency of submicroscopic genomic aberrations detected by tiling path array comparative genome hybridisation in patients with isolated congenital heart disease. J Med Genet. 2008;45(11):704–709. doi: 10.1136/jmg.2008.058776. [DOI] [PubMed] [Google Scholar]
- 223.Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci USA. 2011;108(7):2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Goldmuntz E, Paluru P, Glessner J, Hakonarson H, Biegel JA, White PS, Gai X, Shaikh TH. Microdeletions and microduplications in patients with congenital heart disease and multiple congenital anomalies. Congenit Heart Dis. 2011;6(6):592–602. doi: 10.1111/j.1747-0803.2011.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, Ergul E, Conta JH, Korn JM, McCarroll SA, Gorham JM, Gabriel S, Altshuler DM, Quintanilla-Dieck ML, Artunduaga MA, Eavey RD, Plenge RM, Shadick NA, Weinblatt ME, De Jager PL, Hafler DA, Breitbart RE, Seidman JG, Seidman CE. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41(8):931–935. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Hitz MP, Lemieux-Perreault LP, Marshall C, Feroz-Zada Y, Davies R, Yang SW, Lionel AC, D’Amours G, Lemyre E, Cullum R, Bigras JL, Thibeault M, Chetaille P, Montpetit A, Khairy P, Overduin B, Klaassen S, Hoodless P, Nemer M, Stewart AF, Boerkoel C, Scherer SW, Richter A, Dube MP, Andelfinger G. Rare copy number variants contribute to congenital left-sided heart disease. PLoS Genet. 2012;8(9):e1002903. doi: 10.1371/journal.pgen.1002903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Iascone M, Ciccone R, Galletti L, Marchetti D, Seddio F, Lincesso AR, Pezzoli L, Vetro A, Barachetti D, Boni L, Federici D, Soto AM, Comas JV, Ferrazzi P, Zuffardi O. Identification of de novo mutations and rare variants in hypoplastic left heart syndrome. Clin Genet. 2012;81(6):542–554. doi: 10.1111/j.1399-0004.2011.01674.x. [DOI] [PubMed] [Google Scholar]
- 228.Lalani SR, Shaw C, Wang X, Patel A, Patterson LW, Kolodziejska K, Szafranski P, Ou Z, Tian Q, Kang SH, Jinnah A, Ali S, Malik A, Hixson P, Potocki L, Lupski JR, Stankiewicz P, Bacino CA, Dawson B, Beaudet AL, Boricha FM, Whittaker R, Li C, Ware SM, Cheung SW, Penny DJ, Jefferies JL, Belmont JW. Rare DNA copy number variants in cardiovascular malformations with extracardiac abnormalities. Eur J Hum Genet. 2013;21(2):173–181. doi: 10.1038/ejhg.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Payne AR, Chang SW, Koenig SN, Zinn AR, Garg V. Submicroscopic chromosomal copy number variations identified in children with hypoplastic left heart syndrome. Pediatr Cardiol. 2012;33(5):757–763. doi: 10.1007/s00246-012-0208-9. [DOI] [PubMed] [Google Scholar]
- 230.Richards AA, Santos LJ, Nichols HA, Crider BP, Elder FF, Hauser NS, Zinn AR, Garg V. Cryptic chromosomal abnormalities identified in children with congenital heart disease. Pediatr Res. 2008;64(4):358–363. doi: 10.1203/PDR.0b013e31818095d0. [DOI] [PubMed] [Google Scholar]
- 231.Silversides CK, Lionel AC, Costain G, Merico D, Migita O, Liu B, Yuen T, Rickaby J, Thiruvahindrapuram B, Marshall CR, Scherer SW, Bassett AS. Rare copy number variations in adults with tetralogy of Fallot implicate novel risk gene pathways. PLoS Genet. 2012;8(8):e1002843. doi: 10.1371/journal.pgen.1002843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Soemedi R, Wilson IJ, Bentham J, Darlay R, Topf A, Zelenika D, Cosgrove C, Setchfield K, Thornborough C, Granados-Riveron J, Blue GM, Breckpot J, Hellens S, Zwolinkski S, Glen E, Mamasoula C, Rahman TJ, Hall D, Rauch A, Devriendt K, Gewillig M, O’Sullivan J, Winlaw DS, Bu’Lock F, Brook JD, Bhattacharya S, Lathrop M, Santibanez-Koref M, Cordell HJ, Goodship JA, Keavney BD. Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. Am J Hum Genet. 2012;91(3):489–501. doi: 10.1016/j.ajhg.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Thienpont B, Mertens L, Ravel T, Eyskens B, Boshoff D, Maas N, Fryns JP, Gewillig M, Vermeesch JR, Devriendt K. Submicroscopic chromosomal imbalances detected by array-CGH are a frequent cause of congenital heart defects in selected patients. Eur Heart J. 2007;28(22):2778–2784. doi: 10.1093/eurheartj/ehl560. [DOI] [PubMed] [Google Scholar]
- 234.Tomita-Mitchell A, Mahnke DK, Struble CA, Tuffnell ME, Stamm KD, Hidestrand M, Harris SE, Goetsch MA, Simpson PM, Bick DP, Broeckel U, Pelech AN, Tweddell JS, Mitchell ME. Human gene copy number spectra analysis in congenital heart malformations. Physiol Genomics. 2012;44(9):518–541. doi: 10.1152/physiolgenomics.00013.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Aerts S, Lambrechts D, Maity S, Loo P, Coessens B, Smet F, Tranchevent LC, Moor B, Marynen P, Hassan B, Carmeliet P, Moreau Y. Gene prioritization through genomic data fusion. Nat Biotechnol. 2006;24(5):537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 236.Thienpont B, Zhang L, Postma AV, Breckpot J, Tranchevent LC, Loo P, Mollgard K, Tommerup N, Bache I, Tumer Z, Engelen K, Menten B, Mortier G, Waggoner D, Gewillig M, Moreau Y, Devriendt K, Larsen LA. Haploinsufficiency of TAB2 causes congenital heart defects in humans. Am J Hum Genet. 2010;86(6):839–849. doi: 10.1016/j.ajhg.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Shiratori H, Hamada H. The left-right axis in the mouse: from origin to morphology. Development. 2006;133(11):2095–2104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- 238.Lage K, Greenway SC, Rosenfeld JA, Wakimoto H, Gorham JM, Segre AV, Roberts AE, Smoot LB, Pu WT, Pereira AC, Mesquita SM, Tommerup N, Brunak S, Ballif BC, Shaffer LG, Donahoe PK, Daly MJ, Seidman JG, Seidman CE, Larsen LA. Genetic and environmental risk factors in congenital heart disease functionally converge in protein networks driving heart development. Proc Natl Acad Sci USA. 2012;109(35):14035–14040. doi: 10.1073/pnas.1210730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.McBride KL, Zender GA, Fitzgerald-Butt SM, Koehler D, Menesses-Diaz A, Fernbach S, Lee K, Towbin JA, Leal S, Belmont JW. Linkage analysis of left ventricular outflow tract malformations (aortic valve stenosis, coarctation of the aorta, and hypoplastic left heart syndrome) Eur J Hum Genet. 2009;17(6):811–819. doi: 10.1038/ejhg.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Winston JB, Erlich JM, Green CA, Aluko A, Kaiser KA, Takematsu M, Barlow RS, Sureka AO, LaPage MJ, Janss LL, Jay PY. Heterogeneity of genetic modifiers ensures normal cardiac development. Circulation. 2010;121(11):1313–1321. doi: 10.1161/CIRCULATIONAHA.109.887687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, Ernst C, Hanscom C, Rossin E, Lindgren AM, Pereira S, Ruderfer D, Kirby A, Ripke S, Harris DJ, Lee JH, Ha K, Kim HG, Solomon BD, Gropman AL, Lucente D, Sims K, Ohsumi TK, Borowsky ML, Loranger S, Quade B, Lage K, Miles J, Wu BL, Shen Y, Neale B, Shaffer LG, Daly MJ, Morton CC, Gusella JF. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149(3):525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Hu Z, Shi Y, Mo X, Xu J, Zhao B, Lin Y, Yang S, Xu Z, Dai J, Pan S, Da M, Wang X, Qian B, Wen Y, Wen J, Xing J, Guo X, Xia Y, Ma H, Jin G, Yu S, Liu J, Zhou Z, Wang X, Chen Y, Sha J, Shen H. A genome-wide association study identifies two risk loci for congenital heart malformations in Han Chinese populations. Nat Genet. 2013;45(7):818–821. doi: 10.1038/ng.2636. [DOI] [PubMed] [Google Scholar]
- 243.Cordell HJ, Bentham J, Topf A, Zelenika D, Heath S, Mamasoula C, Cosgrove C, Blue G, Granados-Riveron J, Setchfield K, Thornborough C, Breckpot J, Soemedi R, Martin R, Rahman TJ, Hall D, Engelen K, Moorman AF, Zwinderman AH, Barnett P, Koopmann TT, Adriaens ME, Varro A, George AL, Dos RC, Bishopric NH, Bezzina CR, O’Sullivan J, Gewillig M, Bu’Lock FA, Winlaw D, Bhattacharya S, Devriendt K, Brook JD, Mulder BJ, Mital S, Postma AV, Lathrop GM, Farrall M, Goodship JA, Keavney BD. Genome-wide association study of multiple congenital heart disease phenotypes identifies a susceptibility locus for atrial septal defect at chromosome 4p16. Nat Genet. 2013;45(7):822–824. doi: 10.1038/ng.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Cordell HJ, Topf A, Mamasoula C, Postma AV, Bentham J, Zelenika D, Heath S, Blue G, Cosgrove C, Granados RJ, Darlay R, Soemedi R, Wilson IJ, Ayers KL, Rahman TJ, Hall D, Mulder BJ, Zwinderman AH, Engelen K, Brook JD, Setchfield K, Bu’Lock FA, Thornborough C, O’Sullivan J, Stuart AG, Parsons J, Bhattacharya S, Winlaw D, Mital S, Gewillig M, Breckpot J, Devriendt K, Moorman AF, Rauch A, Lathrop GM, Keavney BD, Goodship JA. Genome-wide association study identifies loci on 12q24 and 13q32 associated with tetralogy of Fallot. Hum Mol Genet. 2013;22(7):1473–1481. doi: 10.1093/hmg/dds552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Tian Y, Yuan L, Goss AM, Wang T, Yang J, Lepore JJ, Zhou D, Schwartz RJ, Patel V, Cohen ED, Morrisey EE. Characterization and in vivo pharmacological rescue of a Wnt2-Gata6 pathway required for cardiac inflow tract development. Dev Cell. 2010;18(2):275–287. doi: 10.1016/j.devcel.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Chang SW, Mislankar M, Misra C, Huang N, Dajusta DG, Harrison SM, McBride KL, Baker LA, Garg V (2013) Genetic abnormalities in FOXP1 are associated with congenital heart defects. Hum Mutat (Epub ahead of print) [DOI] [PMC free article] [PubMed]
- 247.Kodo K, Nishizawa T, Furutani M, Arai S, Yamamura E, Joo K, Takahashi T, Matsuoka R, Yamagishi H. GATA6 mutations cause human cardiac outflow tract defects by disrupting semaphorin-plexin signaling. Proc Natl Acad Sci USA. 2009;106(33):13933–13938. doi: 10.1073/pnas.0904744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Maitra M, Koenig SN, Srivastava D, Garg V. Identification of GATA6 sequence variants in patients with congenital heart defects. Pediatr Res. 2010;68(4):281–285. doi: 10.1203/PDR.0b013e3181ed17e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Cheng Z, Wang J, Su D, Pan H, Huang G, Li X, Li Z, Shen A, Xie X, Wang B, Ma X. Two novel mutations of the IRX4 gene in patients with congenital heart disease. Hum Genet. 2011;130(5):657–662. doi: 10.1007/s00439-011-0996-7. [DOI] [PubMed] [Google Scholar]
- 250.Muncke N, Jung C, Rudiger H, Ulmer H, Roeth R, Hubert A, Goldmuntz E, Driscoll D, Goodship J, Schon K, Rappold G. Missense mutations and gene interruption in PROSIT240, a novel TRAP240-like gene, in patients with congenital heart defect (transposition of the great arteries) Circulation. 2003;108(23):2843–2850. doi: 10.1161/01.CIR.0000103684.77636.CD. [DOI] [PubMed] [Google Scholar]
- 251.Heathcote K, Braybrook C, Abushaban L, Guy M, Khetyar ME, Patton MA, Carter ND, Scambler PJ, Syrris P. Common arterial trunk associated with a homeodomain mutation of NKX2.6. Hum Mol Genet. 2005;14(5):585–593. doi: 10.1093/hmg/ddi055. [DOI] [PubMed] [Google Scholar]
- 252.Kohlhase J, Heinrich M, Schubert L, Liebers M, Kispert A, Laccone F, Turnpenny P, Winter RM, Reardon W. Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet. 2002;11(23):2979–2987. doi: 10.1093/hmg/11.23.2979. [DOI] [PubMed] [Google Scholar]
- 253.Al-Baradie R, Yamada K, St HC, Chan WM, Andrews C, McIntosh N, Nakano M, Martonyi EJ, Raymond WR, Okumura S, Okihiro MM, Engle EC. Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and results from mutations in SALL4, a new member of the SAL family. Am J Hum Genet. 2002;71(5):1195–1199. doi: 10.1086/343821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Wang B, Li L, Xie X, Wang J, Yan J, Mu Y, Ma X. Genetic variation of SAL-Like 4 (SALL4) in ventricular septal defect. Int J Cardiol. 2010;145(2):224–226. doi: 10.1016/j.ijcard.2009.05.067. [DOI] [PubMed] [Google Scholar]
- 255.Pizzuti A, Sarkozy A, Newton AL, Conti E, Flex E, Digilio MC, Amati F, Gianni D, Tandoi C, Marino B, Crossley M, Dallapiccola B. Mutations of ZFPM2/FOG2 gene in sporadic cases of tetralogy of Fallot. Hum Mutat. 2003;22(5):372–377. doi: 10.1002/humu.10261. [DOI] [PubMed] [Google Scholar]
- 256.Nemer G, Fadlalah F, Usta J, Nemer M, Dbaibo G, Obeid M, Bitar F. A novel mutation in the GATA4 gene in patients with Tetralogy of Fallot. Hum Mutat. 2006;27(3):293–294. doi: 10.1002/humu.9410. [DOI] [PubMed] [Google Scholar]
- 257.Smith KA, Joziasse IC, Chocron S, Dinther M, Guryev V, Verhoeven MC, Rehmann H, Smagt JJ, Doevendans PA, Cuppen E, Mulder BJ, Ten DP, Bakkers J. Dominant-negative ALK2 allele associates with congenital heart defects. Circulation. 2009;119(24):3062–3069. doi: 10.1161/CIRCULATIONAHA.108.843714. [DOI] [PubMed] [Google Scholar]
- 258.Bleyl SB, Saijoh Y, Bax NA, de Groot Gittenberger AC, Wisse LJ, Chapman SC, Hunter J, Shiratori H, Hamada H, Yamada S, Shiota K, Klewer SE, Leppert MF, Schoenwolf GC. Dysregulation of the PDGFRA gene causes inflow tract anomalies including TAPVR: integrating evidence from human genetics and model organisms. Hum Mol Genet. 2010;19(7):1286–1301. doi: 10.1093/hmg/ddq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Aoki Y, Niihori T, Banjo T, Okamoto N, Mizuno S, Kurosawa K, Ogata T, Takada F, Yano M, Ando T, Hoshika T, Barnett C, Ohashi H, Kawame H, Hasegawa T, Okutani T, Nagashima T, Hasegawa S, Funayama R, Nagashima T, Nakayama K, Inoue SI, Watanabe Y, Ogura T, Matsubara Y. Gain-of-Function Mutations in RIT1 Cause Noonan Syndrome, a RAS/MAPK Pathway Syndrome. Am J Hum Genet. 2013;93(1):173–180. doi: 10.1016/j.ajhg.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Tan HL, Glen E, Topf A, Hall D, O’Sullivan JJ, Sneddon L, Wren C, Avery P, Lewis RJ, Ten DP, Arthur HM, Goodship JA, Keavney BD. Nonsynonymous variants in the SMAD6 gene predispose to congenital cardiovascular malformation. Hum Mutat. 2012;33(4):720–727. doi: 10.1002/humu.22030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Robinson SW, Morris CD, Goldmuntz E, Reller MD, Jones MA, Steiner RD, Maslen CL. Missense mutations in CRELD1 are associated with cardiac atrioventricular septal defects. Am J Hum Genet. 2003;72(4):1047–1052. doi: 10.1086/374319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lalani SR, Ware SM, Wang X, Zapata G, Tian Q, Franco LM, Jiang Z, Bucasas K, Scott DA, Campeau PM, Hanchard N, Umana L, Cast A, Patel A, Cheung SW, McBride KL, Bray M, Craig CA, Boggs BA, Huang M, Baker MR, Hamilton S, Towbin J, Jefferies JL, Fernbach SD, Potocki L, Belmont JW (2013) MCTP2 is a dosage-sensitive gene required for cardiac outflow tract development. Hum Mol Genet (Epub ahead of print) [DOI] [PMC free article] [PubMed]
- 263.Ryan AK, Goodship JA, Wilson DI, Philip N, Levy A, Seidel H, Schuffenhauer S, Oechsler H, Belohradsky B, Prieur M, Aurias A, Raymond FL, Clayton-Smith J, Hatchwell E, McKeown C, Beemer FA, Dallapiccola B, Novelli G, Hurst JA, Ignatius J, Green AJ, Winter RM, Brueton L, Brondum-Nielsen K, Scambler PJ. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet. 1997;34(10):798–804. doi: 10.1136/jmg.34.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Battaglia A, Hoyme HE, Dallapiccola B, Zackai E, Hudgins L, Donald-McGinn D, Bahi-Buisson N, Romano C, Williams CA, Brailey LL, Zuberi SM, Carey JC. Further delineation of deletion 1p36 syndrome in 60 patients: a recognizable phenotype and common cause of developmental delay and mental retardation. Pediatrics. 2008;121(2):404–410. doi: 10.1542/peds.2007-0929. [DOI] [PubMed] [Google Scholar]
- 265.Mitter D, Chiaie BD, Ludecke HJ, Gillessen-Kaesbach G, Bohring A, Kohlhase J, Caliebe A, Siebert R, Roepke A, Ramos-Arroyo MA, Nieva B, Menten B, Loeys B, Mortier G, Wieczorek D. Genotype-phenotype correlation in eight new patients with a deletion encompassing 2q31.1. Am J Med Genet A. 2010;152A(5):1213–1224. doi: 10.1002/ajmg.a.33344. [DOI] [PubMed] [Google Scholar]
- 266.Ballif BC, Hornor SA, Jenkins E, Madan-Khetarpal S, Surti U, Jackson KE, Asamoah A, Brock PL, Gowans GC, Conway RL, Graham JM, Medne L, Zackai EH, Shaikh TH, Geoghegan J, Selzer RR, Eis PS, Bejjani BA, Shaffer LG. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2–p12.2. Nat Genet. 2007;39(9):1071–1073. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- 267.Grossfeld PD, Mattina T, Lai Z, Favier R, Jones KL, Cotter F, Jones C. The 11q terminal deletion disorder: a prospective study of 110 cases. Am J Med Genet A. 2004;129A(1):51–61. doi: 10.1002/ajmg.a.30090. [DOI] [PubMed] [Google Scholar]
- 268.Battaglia A, Filippi T, Carey JC. Update on the clinical features and natural history of Wolf-Hirschhorn (4p-) syndrome: experience with 87 patients and recommendations for routine health supervision. Am J Med Genet C Semin Med Genet. 2008;148C(4):246–251. doi: 10.1002/ajmg.c.30187. [DOI] [PubMed] [Google Scholar]
- 269.Girirajan S, Vlangos CN, Szomju BB, Edelman E, Trevors CD, Dupuis L, Nezarati M, Bunyan DJ, Elsea SH. Genotype-phenotype correlation in Smith–Magenis syndrome: evidence that multiple genes in 17p11.2 contribute to the clinical spectrum. Genet Med. 2006;8(7):417–427. doi: 10.1097/01.gim.0000228215.32110.89. [DOI] [PubMed] [Google Scholar]
- 270.Potocki L, Shaw CJ, Stankiewicz P, Lupski JR. Variability in clinical phenotype despite common chromosomal deletion in Smith–Magenis syndrome [del(17)(p11.2p11.2)] Genet Med. 2003;5(6):430–434. doi: 10.1097/01.gim.0000095625.14160.ab. [DOI] [PubMed] [Google Scholar]
- 271.Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, Shen J, Kang SH, Pursley A, Lotze T, Kennedy G, Lansky-Shafer S, Weaver C, Roeder ER, Grebe TA, Arnold GL, Hutchison T, Reimschisel T, Amato S, Geragthy MT, Innis JW, Obersztyn E, Nowakowska B, Rosengren SS, Bader PI, Grange DK, Naqvi S, Garnica AD, Bernes SM, Fong CT, Summers A, Walters WD, Lupski JR, Stankiewicz P, Cheung SW, Patel A. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40(12):1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Cardoso C, Leventer RJ, Ward HL, Toyo-Oka K, Chung J, Gross A, Martin CL, Allanson J, Pilz DT, Olney AH, Mutchinick OM, Hirotsune S, Wynshaw-Boris A, Dobyns WB, Ledbetter DH. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller–Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am J Hum Genet. 2003;72(4):918–930. doi: 10.1086/374320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Dobyns WB, Curry CJ, Hoyme HE, Turlington L, Ledbetter DH. Clinical and molecular diagnosis of Miller–Dieker syndrome. Am J Hum Genet. 1991;48(3):584–594. [PMC free article] [PubMed] [Google Scholar]
- 274.Nagai T, Matsumoto N, Kurotaki N, Harada N, Niikawa N, Ogata T, Imaizumi K, Kurosawa K, Kondoh T, Ohashi H, Tsukahara M, Makita Y, Sugimoto T, Sonoda T, Yokoyama T, Uetake K, Sakazume S, Fukushima Y, Naritomi K. Sotos syndrome and haploinsufficiency of NSD1: clinical features of intragenic mutations and submicroscopic deletions. J Med Genet. 2003;40(4):285–289. doi: 10.1136/jmg.40.4.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Saugier-Veber P, Bonnet C, Afenjar A, Drouin-Garraud V, Coubes C, Fehrenbach S, Holder-Espinasse M, Roume J, Malan V, Portnoi MF, Jeanne N, Baumann C, Heron D, David A, Gerard M, Bonneau D, Lacombe D, Cormier-Daire V, Billette de Villemeur T, Frebourg T, Burglen L. Heterogeneity of NSD1 alterations in 116 patients with Sotos syndrome. Hum Mutat. 2007;28(11):1098–1107. doi: 10.1002/humu.20568. [DOI] [PubMed] [Google Scholar]
- 276.Tatton-Brown K, Douglas J, Coleman K, Baujat G, Cole TR, Das S, Horn D, Hughes HE, Temple IK, Faravelli F, Waggoner D, Turkmen S, Cormier-Daire V, Irrthum A, Rahman N. Genotype-phenotype associations in Sotos syndrome: an analysis of 266 individuals with NSD1 aberrations. Am J Hum Genet. 2005;77(2):193–204. doi: 10.1086/432082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Aldred MA, Sanford RO, Thomas NS, Barrow MA, Wilson LC, Brueton LA, Bonaglia MC, Hennekam RC, Eng C, Dennis NR, Trembath RC. Molecular analysis of 20 patients with 2q37.3 monosomy: definition of minimum deletion intervals for key phenotypes. J Med Genet. 2004;41(6):433–439. doi: 10.1136/jmg.2003.017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Williams SR, Aldred MA, Der Kaloustian VM, Halal F, Gowans G, McLeod DR, Zondag S, Toriello HV, Magenis RE, Elsea SH. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am J Hum Genet. 2010;87(2):219–228. doi: 10.1016/j.ajhg.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, Schroer RJ, Novara F, De GM, Ciccone R, Broomer A, Casuga I, Wang Y, Xiao C, Barbacioru C, Gimelli G, Bernardina BD, Torniero C, Giorda R, Regan R, Murday V, Mansour S, Fichera M, Castiglia L, Failla P, Ventura M, Jiang Z, Cooper GM, Knight SJ, Romano C, Zuffardi O, Chen C, Schwartz CE, Eichler EE. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40(3):322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Bon BW, Mefford HC, Menten B, Koolen DA, Sharp AJ, Nillesen WM, Innis JW, Ravel TJ, Mercer CL, Fichera M, Stewart H, Connell LE, Ounap K, Lachlan K, Castle B, Aa N, Ravenswaaij C, Nobrega MA, Serra-Juhe C, Simonic I, Leeuw N, Pfundt R, Bongers EM, Baker C, Finnemore P, Huang S, Maloney VK, Crolla JA, Kalmthout M, Elia M, Vandeweyer G, Fryns JP, Janssens S, Foulds N, Reitano S, Smith K, Parkel S, Loeys B, Woods CG, Oostra A, Speleman F, Pereira AC, Kurg A, Willatt L, Knight SJ, Vermeesch JR, Romano C, Barber JC, Mortier G, Perez-Jurado LA, Kooy F, Brunner HG, Eichler EE, Kleefstra T, de Vries BB. Further delineation of the 15q13 microdeletion and duplication syndromes. J Med Genet. 2009;46(8):511–523. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Thienpont B, Bena F, Breckpot J, Philip N, Menten B, Van EH, Scalais E, Salamone JM, Fong CT, Kussmann JL, Grange DK, Gorski JL, Zahir F, Yong SL, Morris MM, Gimelli S, Fryns JP, Mortier G, Friedman JM, Villard L, Bottani A, Vermeesch JR, Cheung SW, Devriendt K. Duplications of the critical Rubinstein-Taybi deletion region on chromosome 16p13.3 cause a novel recognisable syndrome. J Med Genet. 2010;47(3):155–161. doi: 10.1136/jmg.2009.070573. [DOI] [PubMed] [Google Scholar]
- 282.Nagamani SC, Erez A, Bader P, Lalani SR, Scott DA, Scaglia F, Plon SE, Tsai CH, Reimschisel T, Roeder E, Malphrus AD, Eng PA, Hixson PM, Kang SH, Stankiewicz P, Patel A, Cheung SW. Phenotypic manifestations of copy number variation in chromosome 16p13.11. Eur J Hum Genet. 2011;19(3):280–286. doi: 10.1038/ejhg.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Potocki L, Bi W, Treadwell-Deering D, Carvalho CM, Eifert A, Friedman EM, Glaze D, Krull K, Lee JA, Lewis RA, Mendoza-Londono R, Robbins-Furman P, Shaw C, Shi X, Weissenberger G, Withers M, Yatsenko SA, Zackai EH, Stankiewicz P, Lupski JR. Characterization of Potocki–Lupski syndrome [dup(17)(p11.2p11.2)] and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am J Hum Genet. 2007;80(4):633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Soler-Alfonso C, Motil KJ, Turk CL, Robbins-Furman P, Friedman EM, Zhang F, Lupski JR, Fraley JK, Potocki L. Potocki–Lupski syndrome: a microduplication syndrome associated with oropharyngeal dysphagia and failure to thrive. J Pediatr. 2011;158(4):655–659. doi: 10.1016/j.jpeds.2010.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Portnoi MF. Microduplication 22q11.2: a new chromosomal syndrome. Eur J Med Genet. 2009;52(2–3):88–93. doi: 10.1016/j.ejmg.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 286.Bi W, Ohyama T, Nakamura H, Yan J, Visvanathan J, Justice MJ, Lupski JR. Inactivation of Rai1 in mice recapitulates phenotypes observed in chromosome engineered mouse models for Smith–Magenis syndrome. Hum Mol Genet. 2005;14(8):983–995. doi: 10.1093/hmg/ddi085. [DOI] [PubMed] [Google Scholar]
- 287.Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, Clark GD, Ledbetter DH, McBain CJ, Wynshaw-Boris A. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19(4):333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- 288.Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller–Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364(6439):717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]