Abstract
Glioblastomas (GBM) are very aggressive and malignant brain tumors, with frequent relapses despite an appropriate treatment combining surgery, chemotherapy and radiotherapy. In GBM, hypoxia is a characteristic feature and activation of Hypoxia Inducible Factors (HIF-1α and HIF-2α) has been associated with resistance to anti-cancer therapeutics. Int6, also named eIF3e, is the “e” subunit of the translation initiation factor eIF3, and was identified as novel regulator of HIF-2α. Eukaryotic initiation factors (eIFs) are key factors regulating total protein synthesis, which controls cell growth, size and proliferation. The functional significance of Int6 and the effect of Int6/EIF3E gene silencing on human brain GBM has not yet been described and its role on the HIFs is unknown in glioma cells. In the present study, we show that Int6/eIF3e suppression affects cell proliferation, cell cycle and apoptosis of various GBM cells. We highlight that Int6 inhibition induces a diminution of proliferation through cell cycle arrest and increased apoptosis. Surprisingly, these phenotypes are independent of global cell translation inhibition and are accompanied by decreased HIF expression when Int6 is silenced. In conclusion, we demonstrate here that Int6/eIF3e is essential for proliferation and survival of GBM cells, presumably through modulation of the HIFs.
Keywords: glioma, glioblastoma, proliferation, apoptosis, eukaryotic translation initiation factor, Int6, eIF3e, Hypoxia Inducible Factors, HIF-1α, HIF-2α
1. Introduction
Based on the World Health Organization (WHO) classification, Glioblastoma Multiforme (GBM) are grade IV astrocytic brain tumors [1]. They are the most common brain tumors in adults, representing the second cause of death in children and the third in adults. GBM are one of the most deadly human cancers and despite surgery, radiotherapy and chemotherapy, the median survival is approximately 12–14 months. Furthermore, these aggressive tumors are known to be hypoxic which could contribute for their resistance to anti-cancer therapeutics [2,3].
Indeed, cell response and adaptation to low oxygen conditions is primarily controlled by Hypoxia Inducible Factors (HIFs). These transcription factors act as a heterodimer, composed of two subunits: HIF-α and HIF-β. The two major HIF-α subunits are HIF-1α and HIF-2α and these factors mediate cellular adaptation to hypoxia [4]. The main regulation of the α subunits is the oxygen tension however their stabilization and consequent functions are also influenced by genetic alterations, cell metabolism, growth factors, cytokines and hormones [5]. HIFs promote adaptation to hypoxic conditions through the regulation of more than 150 genes involved mainly in angiogenesis, metabolism, proliferation and cell migration [4,6]. HIF-2α plays a particular role in GBM cells and glioma stem cells and is highly expressed in these cells [7,8]. HIF-2α can be active under non-hypoxic conditions creating a pseudo-hypoxic phenotype that influences cell behaviors. Its knockdown has been show to prevent glioma cell growth in vitro, which is associated with reduced levels of VEGF (Vascular Endothelial Growth Factor) as well as, poorly vascularized and highly necrotic tumors in vivo [7].
Recently, Int6, also known as eIF3e (“e” subunit of the eukaryotic translation Initiation Factor 3), has been described as a new regulator of HIF-2α [9–12]. Int6/eIF3e, through the Eukaryotic Initiation Factor 3 (eIF3), is mainly involved in protein synthesis, due to its direct binding to the 40S ribosome and facilitating ribosome recruitment to mRNA [13,14]. The core of eIF3 is composed of eIF3a, eIF3b, eIF3c, eIF3g and eIF3i, while eIF3e, eIF3f and eIF3h have been shown to stabilize the main core and modulate its activity [15,16]. Interestingly, it has been shown that some of these eIF3 subunits play a role in tumorigenesis [13,14]. Despite altered expression in different cancer types, eIF3e’s involvement in tumorigenesis is not yet clear. Of note, Int6 has other surprising functions such as contributing to the DNA damage response in HeLa cells through involvement of ATM and BRCA1 [17]. In breast carcinoma cells, Int6 depletion induces diminished proliferation, decreasing urokinase-type plasminogen activator (PLAU) and apoptotic regulator BCL-XL [18], and favors epithelial-to-mesenchymal transition increasing Snail and Zeb2 expression [19]. Finally, Int6 modulates HIF-2α expression and its target genes to control vascular remodeling and development [11,12].
To date, Int6/eIF3e expression in human glioma cells and its role in cell growth have not been studied. The aim of the present work was to determine the in vitro effect of Int6/EIF3E gene silencing by RNA interference on a panel of human GBM cell apoptosis and cell cycle and to elucidate its molecular mechanism potentially through HIF modulation.
2. Results and Discussion
2.1. Results
2.1.1. Int6/eIF3e Expression in Human Glioblastoma Cells
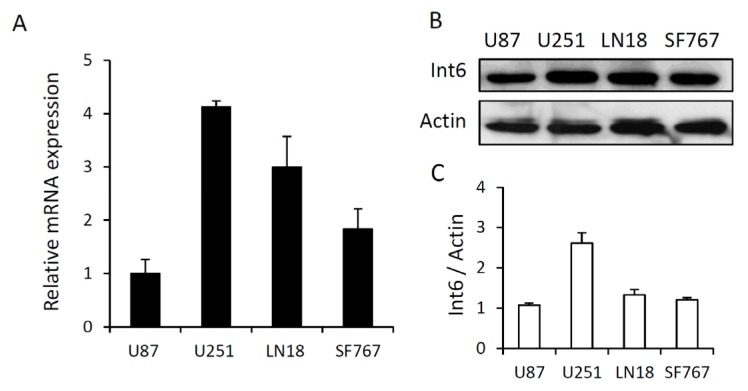
First, we analyzed Int6 expression in four different GBM cell lines (LN18, SF767, U87 and U251) by qRT-PCR and western blot analysis. qRT-PCR analyses revealed that Int6 mRNA is highly expressed in all glioma cell lines tested. U251 cells exhibit the highest mRNA expression and U87 cells the lowest (Figure 1A). In addition, basal Int6 protein expression was assessed by western blot and is partly correlated with Int6 mRNA expression. The U251 cells have the strongest Int6/eIF3e expression while the U87 cells have the lowest within the four different glioma cell lines (Figure 1B,C). These results show that Int6/eIF3e is well expressed in GBM cells and some differences between cell lines are observed.
Figure 1.
Basal Int6/eIF3e expression in four different glioblastoma cell lines. (A) Graph representing Int6 mRNA levels in LN18, SF767, U87 and U251 glioma cells analyzed by qRT-PCR (n = 4); (B) Western blot analysis showing basal Int6 protein expression in LN18, SF767, U87 and U251 glioma cells (n = 5); (C) Western blot quantifications showing the ratio Int6/eIF3e/Actin of at least 5 independent experiments.
2.1.2. RNA Interference Mediated Int6/EIF3E Silencing in Glioblastoma Cells
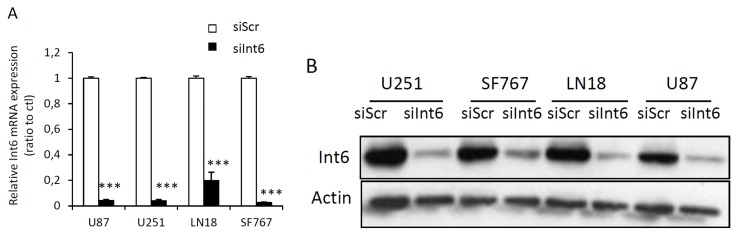
Using an RNA interference strategy, we tested different concentrations of control siRNA (siScr) or specific siRNA for Int6/EIF3E (siInt6) and performed a time course experiment in order to determine the efficiency of the siRNA over time. We show that siInt6 strongly and specifically inhibits Int6 mRNA and protein in all GBM cell lines compared to control siRNA (Figure 2 and Figure S1). The range of 1 nM to 50 nM of specific siRNA gave us a complete Int6 inhibition and 20 nM continued to inhibit Int6 seven days post transfection (Figure S1A,B). Of note, in order to be confident that the phenotypic results we observed were due to knockdown of the target of interest and not due to an off-target effect, we repeated some of the experiments described below with different and distinct siRNAs, targeting Int6/EIF3E, in parallel to confirm our results (see experimental section and Figure S1C–E and Figure S2A,B). These results demonstrate that siRNA transfection silenced Int6 gene and protein expression effectively.
Figure 2.
Efficient inhibition of Int6/eIF3e expression with siRNA. (A) qRT-PCR data showing strong Int6 mRNA depletion in GBM cells transfected with Int6 specific siRNA (siInt6) compared to control cells transfected with a scrambled sequence (siScr) (*** p < 0.001 siInt6 versus siScr, n > 4); (B) Western blots showing efficient inhibition of Int6/eIF3e protein expression in LN18, SF767, U87 and U251 GBM cells transfected with Int6 specific siRNA (siInt6) (n = 10).
2.1.3. Int6 Inhibition Decreases Glioblastoma Cell Proliferation
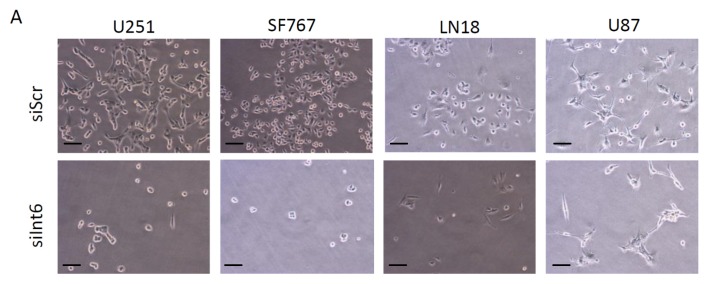
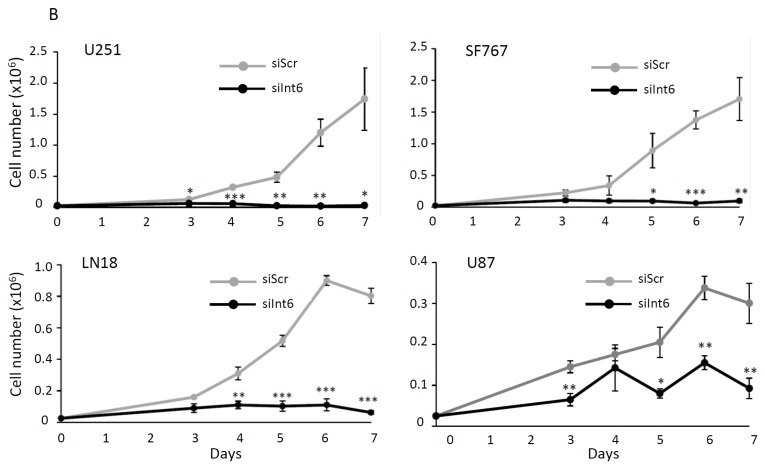
To investigate whether RNA interference-mediated Int6/EIF3E gene silencing affects glioblastoma cell growth and proliferation, we transfected LN18, SF767, U87 and U251 cells with control siRNA (siScr) or specific siRNA for Int6 (siInt6) and followed cell growth by counting cells daily for 7 days. Three days after transfection, we observed a significant and strong decrease in cell number when Int6 is silenced (siInt6) compared to controls in all GBM cells tested (Figure 3A,B and Figure S2). These results indicate that Int6 is essential for cell growth and proliferation of GBM cells.
Figure 3.
Int6/eIF3e inhibition suppresses GBM cell proliferation. (A) Photographs taken during the proliferation assay and representing LN18, SF767, U87 and U251 GBM cells 72 h after transfection with siInt6 or siScr (scale bars: 20 μm); (B) Proliferation assay performed with each GBM cell lines (U251, SF767, LN18 and U87) showing a significant decrease in cell number when cells are transfected with siInt6 (grey curve: control siScr, black curve: siInt6) (* p < 0.05, ** p < 0.01, *** p < 0.001 siInt6 versus siScr, n > 4).
2.1.4. Int6 Inhibition Induces Cell Cycle Arrest and G0 Transition
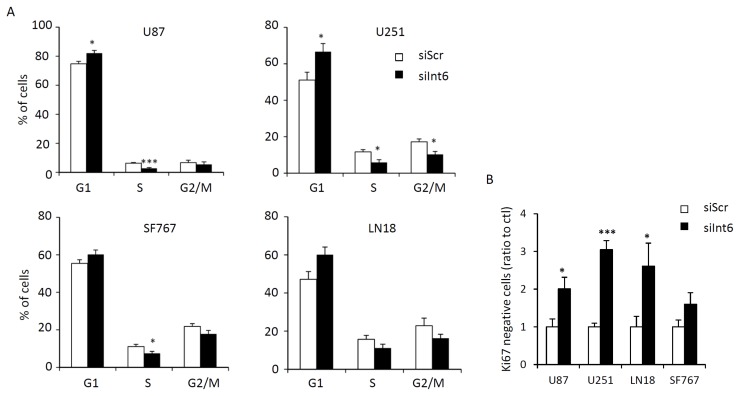
In order to explain the decreased proliferation with Int6 inhibition, we first looked at cell cycle and cell cycle arrest in human GBM cells. Using PI/Ki67 staining and flow cytometry, we analyzed cell cycle and cells in G0. We demonstrate that Int6 inhibition leads to a modest but significant block in the G1 phase (Figure 4A and Figure S3A). Cells transfected with siInt6 accumulated more in G1 (siInt6: 82.10% for U87, 66.66% for U251, 60.22% for SF767 and 60.10 for LN18) compared to cells transfected with control siRNA (siScr: 74.74% for U87, 55.00% for U251, 54.41% for SF767 and 47.18% for LN18). In correlation with this G1 block, we also observed an increase in the number of cells in G0 when Int6 was inhibited. The percentage of Ki67 negative cells was at least twice higher in response to Int6 silencing (Figure 4B and Figure S3B). These results indicate that inhibition of GBM cell growth by Int6 gene silencing may induce G1 phase cell cycle arrest and transition in G0.
Figure 4.
Int6/eIF3e inhibition halts GBM cell cycle. (A) Quantification of cell cycle distribution of GBM cells using flow cytometry and PI staining. Int6 knockdown (siInt6) induces decreased cell number in S phase and increased cell number in G1 phase compared to control cells (siScr); (B) Quantification of GBM cells in G0 phase using flow cytometry and Ki67/PI staining. Int6 knockdown significantly increases the number of cells in G0 phase (* p < 0.05, *** p < 0.001 compared with the negative control, siScr, n = 4).
2.1.5. Int6 Inhibition Increases Glioblastoma Cell Apoptosis
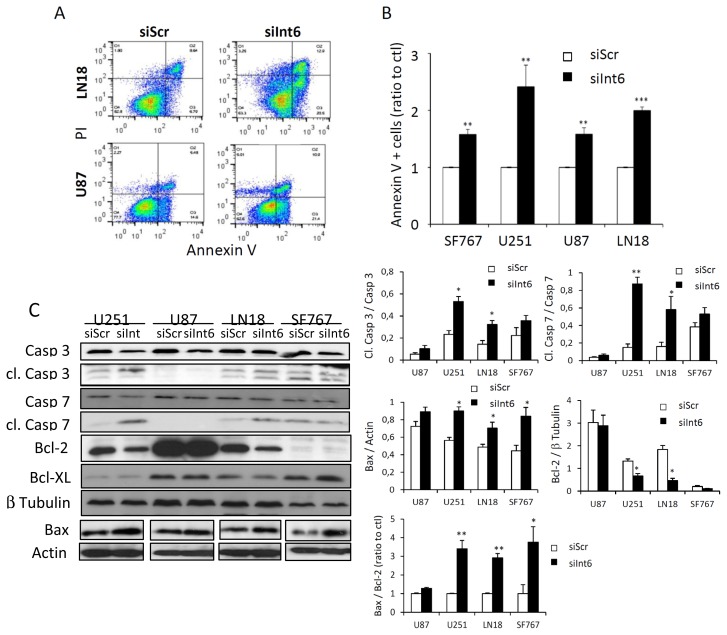
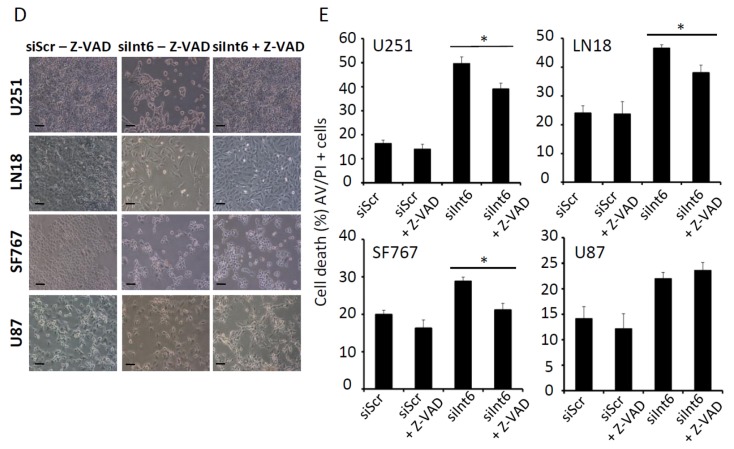
Apoptosis often is a consequence of the G0/G1 arrest and could be another explanation for the effect of Int6 inhibition on glioma cell proliferation. We decided to assess apoptosis rate by flow cytometry and western blot in the four different GBM cell lines using Annexin-V/PI staining and apoptosis-related proteins such as caspase 3, caspase 7, PARP, Bcl-XL and Bax, respectively. Three days following RNA interference-mediated Int6 gene silencing (siInt6), the apoptosis rate, reflected by the percentage of AnnexinV positive cells, was significantly increased compared to control cells (siScr) (Figure 5A,B and Figure S4A). In addition, we confirmed the apoptotic process by studying caspase 3, caspase 7, PARP, Bcl-XL and Bax expression in our cell lines. All cell lines exhibit a slight decrease in total caspase 3, caspase 7 and PARP expression and a slight increase in expression of the pro-apoptotic protein Bax (Figure 5C and Figure S4B,C). Surprisingly, only U251 and LN18 cells show a significant increase in the cleaved caspase forms, suggesting a specific and prominent role of caspases in the apoptotic phenotype of these cell lines (Figure 5C). Additionally, these cell lines also exhibit a significant decrease of the anti-apoptotic protein Bcl-2 (Figure 5C). The Bax/Bcl-2 ratio was significantly increased in U251, LN18 and SF767 revealing an apoptotic mechanism (Figure 5C), and only a trend was observed for the U87 cell line. Since previous work has identified Bcl-XL to be regulated by eIF3e, we also assessed Bcl-XL expression in response to Int6/EIF3E silencing [18]. Interestingly, we did not observe any difference in Bcl-XL expression in our glioma cells when Int6/eIF3e was depleted (Figure 5C). To further understand the caspase involvement in our phenotypes, we assessed caspase-dependent cell death using the Z-VAD caspase inhibitor in glioma cells transfected or not with siInt6 (Figure 5D,E). Flow cytometry analyses of Annexin-V/PI staining demonstrate that Z-VAD treatment induced a partial, but significant reversal of glioma cell death for U251, LN18 and SF767 cells confirming a caspase role in glioma cell apoptosis when Int6 is inhibited (Figure 5D,E). Based on cleaved caspase expression and Z-VAD experiments, the U87 cell line seems to induce a caspase-independent apoptotic mechanism. These observations indicate that Int6 inhibition in human glioma cells induces apoptosis through caspase-dependent and caspase-independent pathways.
Figure 5.
Int6 gene silencing induces cell apoptosis. (A) Dot plot representing the percentage of Annexin V positive cells in two GBM cell lines (LN18 and U87) analyzed by flow cytometry and Annexin V/Propidium Iodide staining; (B) Quantification of Annexin V positive cells. Int6 inhibition (siInt6) results in a significant increase in apoptotic cells (** p < 0.01, *** p < 0.001 compared with the negative control, siScr, n = 5); (C) Western Blot analysis and quantifications of caspase 3, cleaved caspase 3, caspase 7, cleaved caspase 7, Bcl-2, Bcl-XL and Bax expression in LN18, SF767, U87 and U251 GBM cells transfected with siInt6 or siScr. Int6 silencing slightly increases Bax expression, reduces Bcl-2, caspase 3 and caspase 7 expression, and increases Bax/Bcl-2 ratio and cleaved caspase forms for U251 and LN18 cells (* p < 0.05, ** p < 0.01, n = 3); (D) Photographs representing glioma cells transfected with siScr or siInt6 and treated with 30 μM of Z-VAD caspase inhibitor (+Z-VAD) or DMSO (−Z-VAD). Z-VAD treatment partly rescues the deleterious effect of Int6 silencing (n = 3), scale bars: 20 μm; (E) Quantification of Annexin-V/PI staining analyzed by flow cytometry of glioma cells transfected with siScr or siInt6 and treated or not with Z-VAD caspase inhibitor. Z-VAD treatment partly reverses glioma cell death induced by Int6 inhibition (* p < 0.05, n = 3).
2.1.6. RNA Interference-Mediated Int6/EIF3E Silencing, Translation and HIFs
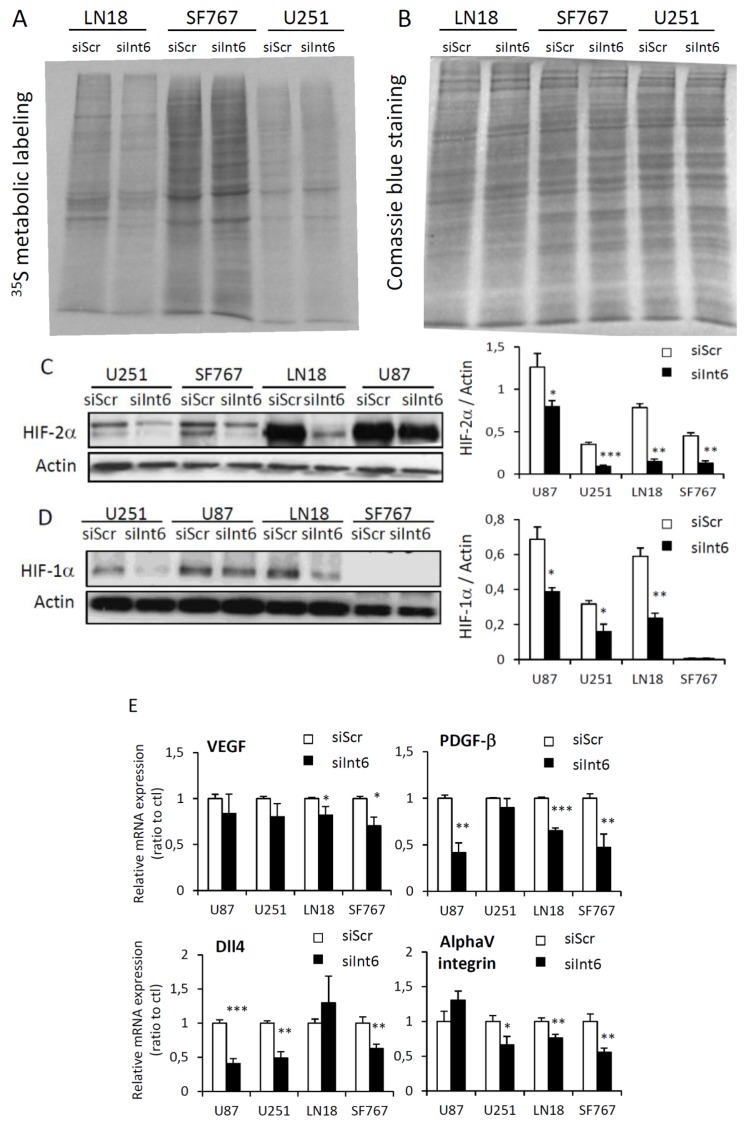
Int6/eIF3e is a component of the eukaryotic initiation factor 3 (eIF3), which is required for several steps in the initiation of protein synthesis. Int6 silencing in our glioma cell lines led to a strong inhibition of proliferation. We analyzed whether Int6/eIF3e inhibition was affecting global cell translation. Using 35S-Methionine metabolic labeling, we assessed de novo protein synthesis in LN18, SF767 and U251 cells (Figure 6A,B and Figure S5B,C). Interestingly, we did not observe any major difference in translation profiles between control cells (siScr) and cells where Int6 was silenced (siInt6) (Figure 6A,B). Furthermore, we did not observe any difference in protein expression profile on a coomassie blue stained gel loaded with proteins coming from the same number of control or cells transfected with siInt6 (Figure S5A). These results suggest that Int6/eIF3e depletion does not affect global protein synthesis in human GBM cells. Int6/eIF3e has also been identified as a potential regulator of the Hypoxia Inducible Factors, particularly HIF-α [6]. Based on the role of the HIFs in promoting metabolism, migration, proliferation and cell growth in glioma, we decided to assess HIF status and their target genes in our human GBM cell lines by western blot and qRT-PCR. Int6 silencing has been shown to promote HIF-2α stability, however and very surprisingly, in human glioma cells, Int6 inhibition leads to a significant decrease of HIF-1α and HIF-2α expression with a more pronounced down-regulation of HIF-2α (Figure 6C,D and Figure S1), particularly in U251, LN18 and SF767 cells. We also assessed several HIF target genes by qRT-PCR. HIF reduced expression was correlated with diminished Vascular Endothelial Growth Factor (VEGF), Platelet-Derived Growth Factor β (PDGF-β), AlphaV integrin (αV integrin) and Delta Like Ligand 4 (Dll4) mRNAs (Figure 6E). Specifically, VEGF mRNA level is slightly decreased in all glioma cell lines when Int6 is silenced. With siInt6, PDGF-β mRNA is down-regulated in LN18, U87, and U251 cells, αV integrin mRNA is decreased in U251, LN18, and SF767 cells, and Dll4 mRNA is down-regulated in SF767, U87, and U251 cells. These results demonstrate that Int6 silencing, and consequently Int6/eIF3e inhibition, decreased HIF expression and their transcriptional activity and that Int6/eIF3e could be a potential positive HIF regulator in human GBM cells.
Figure 6.
Int6 gene silencing downregulates the Hypoxia Inducible Factors expression without affecting global cell translation. (A,B) De novo protein synthesis in GBM cell lines (72 h after siScr or siInt6 transfection) was assessed by 35S Methionine metabolic labeling. Proteins were resolved by SDS-polyacrylamide gel, autoradiographed (A) and stained with Coomassie (B); (C,D) Western blot analysis of HIF-2α (C) and HIF-1α (D) expression in human GBM cells transfected or not with siInt6 showing decreased HIF expression when Int6/EIF3E is knockdowned, n = 5; (E) Graphs representing HIF target gene (VEGF, PDGF-β, Dll4, αV integrin) expression in control or Int6 silenced GBM cells, assessed by qRT-PCR. Int6 inhibition significantly reduces HIF target gene expression. (* p < 0.05, ** p < 0.01, *** p < 0.001, n = 4).
2.2. Discussion
Glioblastoma (GBM) are very hypoxic, aggressive and resistant tumors and identification of new biological pathways is crucial to improve anti-cancer therapeutics [1,2]. Hypoxia Inducible Factors (HIFs), particularly HIF-2α, are important regulators involved in GBM resistance through the control of cell response and adaptation to low oxygen conditions. Targeting the HIFs remains challenging and several teams are currently involved in the development of specific inhibitors of HIF-1α and HIF-2α [5]. Recent research on translation and protein synthesis in malignant tumors also demonstrates the key role played by translational control in tumorigenic processes [13]. Our study identified a new biological pathway regulated by Int6/eIF3e, which modulates GBM cell growth and proliferation. Int6/eIF3e was recently identified as a novel regulator of HIF-2α [6,9] and is primarily known to form, with 12 other subunits, the Eukaryotic Initiation Factor 3 (eIF3) which facilitates the interaction between ribosome and mRNA [14,16]. The eIF3 consists of five subunits (eIF3a, eIF3b, eIF3c, eIF3g and eIF3i), which represent the essential core for the initiation of translation [15,16]. The remaining subunits have been described as modulators of its activity. Specifically, eIF3e, eIF3f and eIF3h are involved in the stabilization of the main core [16]. Interestingly, it has been shown that eIF3 subunits play a role in tumorigenesis. For instance, eIF3a, eIF3c or eIF3h are overexpressed in many tumors, leading to increased protein synthesis and supporting tumor development, while their inhibition can reverse the malignant phenotypes [20–22]. Recently, Liang et al. demonstrated that eIF3b was strongly expressed in human GBM and human GBM cell lines (U251, U373, U87 and A172) [23]. In this study, eIF3b depletion induces glioma cell apoptosis and cell cycle inhibition leading to decreased GBM cell proliferation, which is similar to what we observed with EIF3E silencing [23].
The EIF3E/Int6 gene was originally identified as the site of frequent insertion of the mouse mammary tumor virus [24,25]. However, its role in carcinogenesis is still controversial in the literature and could depend on the tumor stage and type. On one hand, Int6/eIF3e has been described as a potential tumor suppressor with low expression associated with a loss of heterozygosity at the Int6 locus in breast and lung cancer [26,27]. Supporting these data, eIF3e inhibition, in vitro, in human mammary epithelial cells favored transformation [28] and induced epithelial-to-mesenchymal transition [19]. On the other hand, previous studies have reported that Int6 silencing inhibits the proliferation of various tumor cells suggesting a role for Int6 in tumor progression and development. Particularly, eIF3e depletion in ovarian (HeLa) and breast (MDA-MB-231) cancer cells or osteosarcoma cells (U2OS) induces apoptosis, and consequently leads to an inhibition of proliferation. Int6/eIF3e function in the complex mechanism underlying tumor formation and maintenance is still under debate and nothing is currently known in regards to the effect of Int6/eIF3e inhibition in human glioma cells.
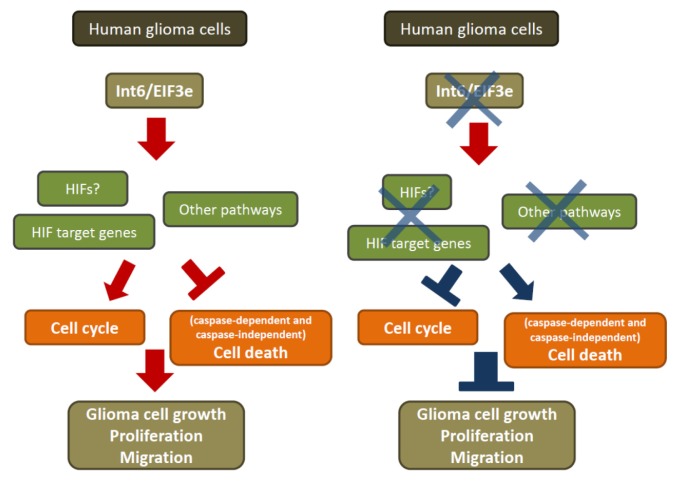
Therefore, the present study aimed to determine the role of Int6/eIF3e in glioma cells (Figure 7) and its molecular mechanism. RT-PCR and western blot analysis were performed on four different glioma cell lines (LN18, SF767, U87 and U251) revealing strong Int6 gene and protein expression. To establish the effect of Int6 on these GBM cells, we used small RNA interfering technology to silence Int6 expression. Previous studies have shown that Int6 silencing does not have any impact on MCF-10A cell proliferation [28]. However, Int6 inhibition in HeLa, U2OS and MDA-MB-231 cells results in decreased cell growth [17–19,28,29]. Following Int6 inhibition in human glioma cells, we observed that proliferation was markedly reduced, indicating that Int6/eIF3e is involved in the regulation of glioma cell proliferation.
Figure 7.
Int6/eIF3e is essential for human glioblastoma cell growth. Int6 inhibition leads to decreased glioma cell proliferation and migration, presumably through decreased HIF expression and activity and increased cell cycle arrest and apoptosis.
Flow cytometry analyses revealed that Int6 depletion arrested glioma cells in the G0/G1 phase. This observation indicates that Int6/eIF3e inhibition leads to a reduction in the number of glioma cells undergoing cell division and increases cell cycle arrest. We also demonstrated that apoptosis occurred in human GBM cells following Int6/EIF3E silencing confirming previous studies reporting that Int6 inhibition leads to apoptosis in HeLa cells [17,29]. Consistent with these results, we noticed that caspase pathways were activated, particularly in U251 and LN18 glioma cells, following Int6 inhibition. This activation of members of the caspase family is an important prerequisite for apoptosis and could partly explain our apoptotic phenotype. Furthermore, experiments with Z-VAD caspase inhibitor allowed us to confirm the partial caspase involvement in glioma cell death when Int6/EIF3E is silenced. However, for the SF767 and U87 cell lines, the apoptotic process seems to be caspase-independent. The different responses of glioma cells to Int6 inhibition requires clarifications but could potentially be explained by the diverse genetic/metabolic backgrounds of the cells, which were selected for their different mutations found in glioblastomas. Based on these various backgrounds, we did not expect all the cells to behave in the same manner following Int6 depletion as different cell death pathways could be involved. Our study focused primarily on typical cancer cell phenotypes, such as proliferation, cell cycle and apoptosis. However, glioblastoma cells are also known to be highly invasive. Thus, we also performed migration assays with glioma cells transfected with control or Int6/EIF3E specific siRNAs (Figure S6). Although Int6 inhibition in breast epithelial cells (MCF-10A) has been demonstrated to induce a constitutive migratory phenotype suggesting a role for Int6 in epithelial-mesenchymal transition [19], we observed that Int6/EIF3E silencing led to a decreased migration in our glioma cell lines. These results are currently under further investigation to explain this discrepancy.
It has been shown that Int6 regulates the MAPK pathway through control of MEK expression during zebrafish development [30]. This mechanism could explain the proliferation defect we observed following a knockdown of Int6. However, we did not observe any significant differences in activation of the MAPK pathway in our glioma cells, demonstrating that Int6 silencing inhibits cell proliferation independently of the MAPK pathway (Figure S7). Based on the Int6/eIF3e role within eIF3 and translation, we also assessed translation and protein synthesis in human glioma cells [13]. Impairment of protein synthesis could explain the strong cell growth inhibition. Very interestingly, we did not notice any difference in translation between control cells and cells where Int6 was inhibited. However, Int6/eIF3e depletion could affect specific mRNA translation without impacting global protein translation. Indeed, our results support the work from Grzmil et al. who reported that Int6 inhibition failed to modify the global translation in breast cancer and osteosarcoma cells [18]. In MDA-MB-231 cells, the analysis of polysome-bound mRNA revealed that Int6 positively and negatively regulates specific mRNAs, including PLAU, BCL2L1 (encoding BCL-XL) and MAD2L1 (MAD2 mitotic arrest deficient-like 1), respectively. Isolation of polysome-bound mRNA from control and siInt6 glioma cells would allow us to identify such mRNAs.
Hypoxia Inducible Factors are essential transcription factor for cellular response and adaption to low oxygen conditions [4]. Inhibition of these factors has been shown to prevent growth, proliferation, migration of human GBM cells in vitro and in vivo [31–34]. Recently, it has been shown in MCF-7 and endothelial cells that Int6/eIF3e, and particularly its overexpression, induces decreased HIF-2α expression. In contrast, its silencing promotes HIF-2α expression stability [9,11]. We decided to assess HIF-1α and HIF-2α expression in our glioma cells following Int6 silencing. Surprisingly, we observed decreased HIF-1α and HIF-2α expression. This was correlated with a down-regulation of some HIF target genes (VEGF, PDGF-β, AlphaV integrin and Dll4) involved in proliferation, migration and cell growth processes. These results indicate that Int6-dependent HIF-2α regulation is cell type specific and occurs in a different manner in glioblastoma cells. In human brain glioma cells, Int6 inhibition leads to decreased HIF-1α and HIF-2α expression. Despite a significant and reproducible decrease of HIF expression in the four tested glioma cell lines, HIF down-regulation following Int6 silencing occurs in different proportions depending on the cell lines. These differences could be due to the cellular genetic/metabolic backgrounds, which lead to various responses within each cell line when Int6 is inhibited. The extent to which the changes we observed in HIF and HIF target expression are a direct or indirect result of regulation by Int6/eIF3e is an interesting question. It has been reported that Int6 specifically regulates some mRNAs. Int6 suppression inhibits translation of mRNA encoding GADD45 or MAP3K14 in HeLa cells [35], affecting cell growth, and stimulates translation of mRNA encoding Snail1 or Zeb2 in MCF10A cells [19], inducing epithelial-mesenchymal transition. Decreased HIFs expression in GBM cells, may be due to a diminished HIF mRNA translation. These decreases in HIF expression could potentially explain the phenotypes we observe after Int6 silencing. Indeed, it is now clearly established in glioblastoma cells that HIF target inhibition induces decreased cell proliferation, migration or invasion. For instance, the Notch/Dll4 pathway inhibition has been shown to reduce the migratory and invasive properties of U87 and U251 glioma cells [36]. VEGF inhibition suppresses GBM stem-like cells tumorigenesis and angiogenesis through VEGF receptor 2 and decreases U251 and LN18 motility [37,38]. Glioblastoma cells express PDGF-β and its receptor, which both contribute to proliferation and angiogenic processes [39]. AlphaV integrin depletion in glioma cell lines impairs cell growth and invasion through induction of apoptosis [40,41]. Despite diminished HIF and HIF target expression, other mechanisms leading to decreased proliferation are likely involved in response to Int6 inhibition. However, to thoroughly characterize the degree to which the HIFs are directly involved in the defect of glioma cell proliferation after Int6 depletion, it will be essential to perform experiments under low oxygen conditions or using hypoxia mimetics, such as CoCl2. These investigations and the identification of new Int6/eIF3e targets are currently ongoing in our laboratory.
Finally, several studies have shown that the HIFs, particularly HIF-2α, are specifically expressed in glioma stem cells and regulate genes involved in the stemness of these cells such as Oct4, c-Myc or Sox2 [3,8,42,43]. Interestingly, Li et al. showed that HIF-2α inhibition decreases neurosphere formation and induces increased glioma stem cell apoptosis [8,42]. In the present study, we showed that Int6 silencing inhibits human glioma cell proliferation via induction of cell cycle arrest and apoptosis, potentially through HIF downregulation, supporting the idea that Int6/eIF3e could favor tumor growth as described in breast cancer and osteosarcoma cells. In consequence, Int6/eIF3e modulation in glioma stem cells could be of special interest for eradication of these treatment-resistant cells and needs to be studied further.
3. Experimental Section
3.1. Cell Culture
Four human glioblastoma cell lines, U87 (ATCC HTB-14), LN18 (ATCC CRL-2610), U251 and SF767 (obtained from M. Celeste Simon’s laboratory, University of Pennsylvania, Philadelphia, PA, USA), were used and routinely maintained in DMEM supplemented with 10% fetal calf serum at 37 °C in 5% CO2-humidified incubators and were subcultured once or twice a week.
3.2. siRNA Transfection
Human glioblastoma cells (2 × 105) were transfected with 20 nmol/L of siScramble (siScr, Qiagen, Venlo, Limburg, The Netherlands) or 20 nmol/L of siRNA against human Int6/EIF3E (siInt6, see below Table 1) combined with Lipofectamine, RNAimax reagent as recommended by the manufacturer (Invitrogen Life Technologies, Carlsbad, CA, USA).
Table 1.
Int6/EIF3E siRNA sequences.
| Source | Sequence |
|---|---|
| Qiagen FlexiTube Gene Solution GS3646 for EIF3E, 1 nmol | SI02662499: 5′-CCCAAAGGUCGCGAUAAUAUU-3′ SI02661981: 5′-AAGCUGGCCUCUGAAAUCUUA-3′ SI02656094: 5′-AAGCUGAAAGGUGGAUUGUAA-3′ SI03153136: 5′-AUGGAAGACCUUACACGGUUA-3′ |
| Qiagen SI02662499 (FlexiTube EIF3E siRNA, 20 nmol) | 5′-CCCAAAGGUCGCGAUAAUAUU-3′ |
| Dharmacon (ON-TARGET plus SMART pool EIF3E siRNA 3646, 10 nmol) | #1: 5′-UGGCUUGUCUUGAGGAUUU-3′ #2: 5′-GGAUCGGCAUCUAGUCUUU-3′ #3: 5′-GGGUAACAAUGCAGUCUCA-3′ #4: 5′-AAAGGUCGCGAUAAUAUUA-3′ |
3.3. Proliferation Assay
Human glioblastoma cells (2.5 × 104) cells were transfected with siRNA (siScr or siInt6). Seventy-two hours after transfection, cells were collected daily for 5 days in 1 mL of trypsine. Twenty microlitres of cell suspension were mixed with 20 μL of Trypan Blue (Lonza, Bâle, Switzerland). Cells were counted using Malassez slide (Invitrogen Life Technologies, Carlsbad, CA, USA) and the number of cells per milliliter were determined by the following formula: (Number of cells/20 squares) × 2 × 100 × 1000. Pictures were taken with Nikon microscope (NIS Element, Nikon, Champigny sur Marne, France), 3 days after transfection with siRNA.
3.4. Western Blot Analysis
Cells, untransfected or transfected with siRNA, were lysed in 70 μL of lysis buffer (50 mM Tris HCl pH 7.5, 0.1% Triton, 5 mM EDTA complemented by Proteases and Phosphatases inhibitors (Chemicon Millipore, Billerica, MA, USA and Sigma Aldrich, Saint-Louis, MO, USA) at 1/100). Western blots were performed as previously described [44,45] using monoclonal rabbit antibodies, anti-Int6 (1/1000, Abcam, Paris, France), anti-HIF-2α (1/1000, Novus Biologicals, Cambridge, UK), anti-HIF-1α (1/1000, Cayman Chemical, Ann Harbor, MI, USA), anti-Caspase 3/7 (1/1000, Cell Signaling, Danvers, MA, USA), anti-cleaved caspase 3/7 (1/1000, Cell Signaling, Danvers, MA, USA) anti-PARP (1/1000, Cell Signaling, Danvers, MA, USA), anti-Bax (1/1000, Cell Signaling, Danvers, MA, USA), anti-Bcl-XL (1/1000, Cell Signaling, Danvers, MA, USA), anti-Bcl-2 (1/1000, Cell Signaling, Danvers, MA, USA), anti-MEK1 (1/1000, Cell Signaling, Danvers, MA, USA), anti-ERK (1/1000, Cell Signaling, Danvers, MA, USA), anti-phospho ERK (1/1000, Cell Signaling, Danvers, MA, USA), and were normalized using a mouse monoclonal antibody anti-actin (1/10,000, Chemicon Millipore, Billerica, MA, USA) or a rabbit polyclonal antibody anti-β-tubulin (1/1000, Cell Signaling, Danvers, MA, USA). Gel quantification was performed using ImageJ (Windows 1.47, Research Services Branch, NIH, Bethesda, MD, USA).
3.5. RT-qPCR
Total RNA was extracted using RNeasy Mini Kit (Qiagen, Venlo, Limburg, The Netherlands), treated with DNase I and subjected to reverse transcription (iScript™ cDNA Synthesis Kit, BioRad, Hercules, CA, USA). qPCR were performed with different primers (see below, Table 2) and normalized using β2-microglobuline and 18S genes, using a StepOnePlus (Applied Biosystems Life Technologies, Carlsbad, CA, USA).
Table 2.
qPCR primer sequences.
| Targets | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| β2m | ACCCCCACTGAAAAAGATGA | ATCTTCAAACCTCCATGATG |
| 18S | GTCTGTGATGCCCTTAGATG | CGTACAGGATGATGTCCGTATACCT |
| Int6 | TTCTTCAATCACCCCAAAGG | TAGAACCTGCCGACGTTTTC |
| HIF-2α | CCACCAGCTTCACTCTCTCC | TCAGAAAAAGGCCACTGCTT |
| Dll4 | TGGGTCAGAACTGGTTATTGGA | GTCATTGCGCTTCTTGCACAG |
| PDGFβ | GATCCCTCCTTTGATGATCTC | TCCAACTCGGCCCCATCT |
| VEGF | CTACCTCCACCATGCCAAGT | GCAGTAGCTGCGCTGATAGA |
| αV Integrin | GGAGCAATTCGACGAGCACT | TTCATCCCGCAGATACGCTA |
3.6. Flow Cytometry
Seventy-two hours after transfection with siRNA (siScr or siInt6) and treatment or not with 30 μM of Z-VAD-FMK caspase inhibitor (Sigma Aldrich, Saint-Louis, MO, USA), cells were collected, washed in PBS and incubated in 100 μL of Annexin-binding buffer 5× (10 mM HEPES pH7.4, 140 mM NaCl, 2,5 mM CaCl2), containing 5 μL of Annexin V-FITC antibody and 1 μL of Propidium Iodide (PI, Invitrogen Life Technologies, Carlsbad, CA, USA) solution at 100 μg/mL (FITC Annexin V/Dead Cell Apoptosis Kit with FITC annexin V and PI, Invitrogen Life Technologies, Carlsbad, CA, USA) during 15 min at room temperature (RT) in the dark. Four hundred microliters of Annexin-binding buffer 5× were then added after washes with PBS-BSA 1%. For cell cycle analysis, cells were fixed with cold ethanol 100% and then permeabilized with Triton ×100 at 0.25%. Cells were then labeled with Ki67 (Abcam, Paris, France) during 45 min at RT and treated with RNAse 1 μg/mL before labeling with PI during 2 hrs at RT. Labeled cells were preserved on ice and run on a flow cytometer (FACS Calibur, Becton-Dickinson, Franklin Lakes, NJ, USA).
3.7. De Novo Protein Synthesis
Cells were starved with methionine/cysteine free medium +10% dialyzed serum for 1 h and then, incubated with 35S methionine (80 μCi/mL) for 30 min in order to assess de novo protein synthesis. Proteins were resolved by SDS-polyacrylamide gel, either dried and exposed to Biomax films (Kodak, Carestream Health, Rochester, NY, USA) or stained with Coomassie (Biorad, Hercules, CA, USA). The de novo synthesis of protein was detected by autoradiography.
3.8. Migration/in Vitro Wound-Closure Assay
For the wound-closure assay, human glioma cells transfected with siScr or siInt6 were seeded in 6 cm dishes and incubated overnight to generate confluent cultures. Of note, U87 cells do not form a monolayer of cells and we were therefore unable to perform the test for these cells. Cell layers were scraped with a plastic pipette tip and washed three times with serum-free media. The remaining cell culture was incubated 24 h to allow cells to migrate into the cleared space. To quantify cell migration, phase-contrast images of identical locations in each wound were taken at 0, 13 and 20 h after wounding. The rate of cell migration was then calculated as the average percentage of wound closure from at least three independent experiments using Nikon microscope and Image J software (Windows 1.47, Research Services Branch, NIH, Bethesda, MD, USA).
3.9. Statistical Analysis
Student’s test was done to compare the means of values from different experiments. Differences were considered statistically significant at p < 0.05. (* p < 0.05, ** p < 0.01, *** p < 0.001).
4. Conclusions
In conclusion, our study demonstrated that Int6, also known as eIF3e, plays an essential role in human glioblastoma cells controlling cell growth, cell cycle, and cell death. We highlighted that Int6/eIF3e inhibition through siRNA induces a diminution of glioblastoma cell proliferation through cell cycle arrest and an induction of caspase-dependent and caspase-independent cell death (Figure 7). With no modification in global cell translation following Int6 inhibition, the observed phenotypes could be, in part, explained by the decrease of HIF and HIF target expression in GBM cells. Although a deeper understanding of the molecular mechanisms involved in this Int6/eIF3e-HIFs pathway is necessary, Int6 could become a new therapeutic option for these aggressive tumors.
Supplementary Information
Acknowledgments
We thank the Cancer Research Center of Toulouse, and M. Celeste Simon’s laboratory for helpful discussions and comments. We sincerely thank Anne-Laure Iscache, and Pauline Jeanjean for technical support. This work was funded by INSERM, the Fondation ARC pour la Recherche sur le Cancer (Grant SF20121205349) and the Cancéropôle Grand Sud-Ouest (Grant RPS13005BBA).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Tanaka S., Louis D.N., Curry W.T., Batchelor T.T., Dietrich J. Diagnostic and therapeutic avenues for glioblastoma: No longer a dead end? Nat. Rev. Clin. Oncol. 2012;10:14–26. doi: 10.1038/nrclinonc.2012.204. [DOI] [PubMed] [Google Scholar]
- 2.Cloughesy T.F., Cavenee W.K., Mischel P.S. Glioblastoma: From molecular pathology to targeted treatment. Annu. Rev. Pathol. 2013 doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 3.Yang L., Lin C., Wang L., Guo H., Wang X. Hypoxia and hypoxia-inducible factors in glioblastoma multiforme progression and therapeutic implications. Exp. Cell Res. 2012;318:2417–2426. doi: 10.1016/j.yexcr.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Krock B.L., Skuli N., Simon M.C. Hypoxia-induced angiogenesis: Good and evil. Genes Cancer. 2011;2:1117–1133. doi: 10.1177/1947601911423654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keith B., Johnson R.S., Simon M.C. Hif1α and hif2α: Sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L., Endler A., Shibasaki F. Hypoxia and angiogenesis: Regulation of hypoxia-inducible factors via novel binding factors. Exp. Mol. Med. 2009;41:849–857. doi: 10.3858/emm.2009.41.12.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pietras A., Johnsson A.S., Pahlman S. The hif-2α-driven pseudo-hypoxic phenotype in tumor aggressiveness, differentiation, and vascularization. Curr. Top. Microbiol. Immunol. 2010;345:1–20. doi: 10.1007/82_2010_72. [DOI] [PubMed] [Google Scholar]
- 8.Li Z., Bao S., Wu Q., Wang H., Eyler C., Sathornsumetee S., Shi Q., Cao Y., Lathia J., McLendon R.E., et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L., Uchida K., Endler A., Shibasaki F. Mammalian tumor suppressor int6 specifically targets hypoxia inducible factor 2α for degradation by hypoxia- and pvhl-independent regulation. J. Biol. Chem. 2007;282:12707–12716. doi: 10.1074/jbc.M700423200. [DOI] [PubMed] [Google Scholar]
- 10.Miyashita R., Chen L., Oshiro H., Uchino H., Shibasaki F. Int6 silencing causes induction of angiogenic factors in neuronal cells via accumulation of hypoxia-inducible factor 2α and decreases brain damage in rats. Neurosci. Lett. 2006;528:83–88. doi: 10.1016/j.neulet.2012.08.033. [DOI] [PubMed] [Google Scholar]
- 11.Chen L., Endler A., Uchida K., Horiguchi S., Morizane Y., Iijima O., Toi M., Shibasaki F. Int6/eif3e silencing promotes functional blood vessel outgrowth and enhances wound healing by upregulating hypoxia-induced factor 2α expression. Circulation. 2010;122:910–919. doi: 10.1161/CIRCULATIONAHA.109.931931. [DOI] [PubMed] [Google Scholar]
- 12.Endler A., Chen L., Li Q., Uchida K., Hashimoto T., Lu L., Xu G.T., Shibasaki F. Int6/eif3e silenced hif2α stabilization enhances migration and tube formation of huvecs via IL-6 and IL-8 signaling. Cytokine. 2013;62:115–122. doi: 10.1016/j.cyto.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 13.Silvera D., Formenti S.C., Schneider R.J. Translational control in cancer. Nat. Rev. Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 14.Hershey J.W. Regulation of protein synthesis and the role of eif3 in cancer. Br. J. Med. Biol. Res. 2010;43:920–930. doi: 10.1590/s0100-879x2010007500098. [DOI] [PubMed] [Google Scholar]
- 15.Damoc E., Fraser C.S., Zhou M., Videler H., Mayeur G.L., Hershey J.W., Doudna J.A., Robinson C.V., Leary J.A. Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol. Cell. Proteomics. 2007;6:1135–1146. doi: 10.1074/mcp.M600399-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Marchione R., Leibovitch S.A., Lenormand J.L. The translational factor eif3f: The ambivalent eif3 subunit. Cell. Mol. Life Sci. 2013;70:3603–3616. doi: 10.1007/s00018-013-1263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris C., Tomimatsu N., Richard D.J., Cluet D., Burma S., Khanna K.K., Jalinot P. Int6/eif3e interacts with atm and is required for proper execution of the DNA damage response in human cells. Cancer Res. 2012;72:2006–2016. doi: 10.1158/0008-5472.CAN-11-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grzmil M., Rzymski T., Milani M., Harris A.L., Capper R.G., Saunders N.J., Salhan A., Ragoussis J., Norbury C.J. An oncogenic role of eif3e/int6 in human breast cancer. Oncogene. 2010;29:4080–4089. doi: 10.1038/onc.2010.152. [DOI] [PubMed] [Google Scholar]
- 19.Gillis L.D., Lewis S.M. Decreased eif3e/int6 expression causes epithelial-to-mesenchymal transition in breast epithelial cells. Oncogene. 2012;32:3598–3605. doi: 10.1038/onc.2012.371. [DOI] [PubMed] [Google Scholar]
- 20.Cappuzzo F., Varella-Garcia M., Rossi E., Gajapathy S., Valente M., Drabkin H., Gemmill R. Myc and eif3h coamplification significantly improve response and survival of non-small cell lung cancer patients (nsclc) treated with gefitinib. J. Thorac. Oncol. 2009;4:472–478. doi: 10.1097/JTO.0b013e31819a5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nupponen N.N., Porkka K., Kakkola L., Tanner M., Persson K., Borg A., Isola J., Visakorpi T. Amplification and overexpression of p40 subunit of eukaryotic translation initiation factor 3 in breast and prostate cancer. Am. J. Pathol. 1999;154:1777–1783. doi: 10.1016/S0002-9440(10)65433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scoles D.R., Yong W.H., Qin Y., Wawrowsky K., Pulst S.M. Schwannomin inhibits tumorigenesis through direct interaction with the eukaryotic initiation factor subunit c (eif3c) Hum. Mol. Genet. 2006;15:1059–1070. doi: 10.1093/hmg/ddl021. [DOI] [PubMed] [Google Scholar]
- 23.Liang H., Ding X., Zhou C., Zhang Y., Xu M., Zhang C., Xu L. Knockdown of eukaryotic translation initiation factors 3b (eif3b) inhibits proliferation and promotes apoptosis in glioblastoma cells. Neurol. Sci. 2012;33:1057–1062. doi: 10.1007/s10072-011-0894-8. [DOI] [PubMed] [Google Scholar]
- 24.Miyazaki S., Kozak C.A., Marchetti A., Buttitta F., Gallahan D., Callahan R. The chromosomal location of the mouse mammary tumor gene int6 and related pseudogenes in the mouse genome. Genomics. 1995;27:420–424. doi: 10.1006/geno.1995.1072. [DOI] [PubMed] [Google Scholar]
- 25.Miyazaki S., Imatani A., Ballard L., Marchetti A., Buttitta F., Albertsen H., Nevanlinna H.A., Gallahan D., Callahan R. The chromosome location of the human homolog of the mouse mammary tumor-associated gene int6 and its status in human breast carcinomas. Genomics. 1997;46:155–158. doi: 10.1006/geno.1997.4996. [DOI] [PubMed] [Google Scholar]
- 26.Buttitta F., Martella C., Barassi F., Felicioni L., Salvatore S., Rosini S., D’Antuono T., Chella A., Mucilli F., Sacco R., et al. Int6 expression can predict survival in early-stage non-small cell lung cancer patients. Clin. Cancer Res. 2005;11:3198–3204. doi: 10.1158/1078-0432.CCR-04-2308. [DOI] [PubMed] [Google Scholar]
- 27.Umar A., Kang H., Timmermans A.M., Look M.P., Meijer-van Gelder M.E., den Bakker M.A., Jaitly N., Martens J.W., Luider T.M., Foekens J.A., et al. Identification of a putative protein profile associated with tamoxifen therapy resistance in breast cancer. Mol. Cell. Proteomics. 2009;8:1278–1294. doi: 10.1074/mcp.M800493-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suo J., Snider S.J., Mills G.B., Creighton C.J., Chen A.C., Schiff R., Lloyd R.E., Chang E.C. Int6 regulates both proteasomal degradation and translation initiation and is critical for proper formation of acini by human mammary epithelium. Oncogene. 2010;30:724–736. doi: 10.1038/onc.2010.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris C., Jalinot P. Silencing of human int-6 impairs mitosis progression and inhibits cyclin b-cdk1 activation. Oncogene. 2005;24:1203–1211. doi: 10.1038/sj.onc.1208268. [DOI] [PubMed] [Google Scholar]
- 30.Grzmil M., Whiting D., Maule J., Anastasaki C., Amatruda J.F., Kelsh R.N., Norbury C.J., Patton E.E. The int6 cancer gene and mek signaling pathways converge during zebrafish development. PLoS One. 2007;2:e959. doi: 10.1371/journal.pone.0000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kessler J., Hahnel A., Wichmann H., Rot S., Kappler M., Bache M., Vordermark D. Hif-1α inhibition by sirna or chetomin in human malignant glioma cells: Effects on hypoxic radioresistance and monitoring via ca9 expression. BMC Cancer. 2010;10:605. doi: 10.1186/1471-2407-10-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen R.L., Ragel B.T., Whang K., Gillespie D. Inhibition of hypoxia inducible factor-1α (hif-1α) decreases vascular endothelial growth factor (vegf) secretion and tumor growth in malignant gliomas. J. Neurooncol. 2006;78:233–247. doi: 10.1007/s11060-005-9103-z. [DOI] [PubMed] [Google Scholar]
- 33.Gillespie D.L., Whang K., Ragel B.T., Flynn J.R., Kelly D.A., Jensen R.L. Silencing of hypoxia inducible factor-1α by rna interference attenuates human glioma cell growth in vivo. Clin. Cancer Res. 2007;13:2441–2448. doi: 10.1158/1078-0432.CCR-06-2692. [DOI] [PubMed] [Google Scholar]
- 34.Franovic A., Holterman C.E., Payette J., Lee S. Human cancers converge at the hif-2α oncogenic axis. Proc. Natl. Acad. Sci. USA. 2009;106:21306–21311. doi: 10.1073/pnas.0906432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris C., Wittmann J., Jack H.M., Jalinot P. Human int6/eif3e is required for nonsense-mediated mrna decay. EMBO Rep. 2007;8:596–602. doi: 10.1038/sj.embor.7400955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X., Chen T., Zhang J., Mao Q., Li S., Xiong W., Qiu Y., Xie Q., Ge J. Notch1 promotes glioma cell migration and invasion by stimulating β-catenin and NF-κB signaling via akt activation. Cancer Sci. 2012;103:181–190. doi: 10.1111/j.1349-7006.2011.02154.x. [DOI] [PubMed] [Google Scholar]
- 37.Kil W.J., Tofilon P.J., Camphausen K. Post-radiation increase in vegf enhances glioma cell motility in vitro. Radiat. Oncol. 2012;7:25. doi: 10.1186/1748-717X-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu C., Wu X., Zhu J. Vegf promotes proliferation of human glioblastoma multiforme stem-like cells through vegf receptor 2. Sci. World J. 2013;2013:417413. doi: 10.1155/2013/417413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nazarenko I., Hede S.M., He X., Hedren A., Thompson J., Lindstrom M.S., Nister M. Pdgf and pdgf receptors in glioma. Ups. J. Med. Sci. 2012;117:99–112. doi: 10.3109/03009734.2012.665097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taga T., Suzuki A., Gonzalez-Gomez I., Gilles F.H., Stins M., Shimada H., Barsky L., Weinberg K.I., Laug W.E. αV-integrin antagonist emd 121974 induces apoptosis in brain tumor cells growing on vitronectin and tenascin. Int. J. Cancer. 2002;98:690–697. doi: 10.1002/ijc.10265. [DOI] [PubMed] [Google Scholar]
- 41.MacDonald T.J., Ladisch S. Antisense to integrin alpha v inhibits growth and induces apoptosis in medulloblastoma cells. Anticancer Res. 2001;21:3785–3791. [PubMed] [Google Scholar]
- 42.Heddleston J.M., Li Z., McLendon R.E., Hjelmeland A.B., Rich J.N. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8:3274–3284. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heddleston J.M., Li Z., Lathia J.D., Bao S., Hjelmeland A.B., Rich J.N. Hypoxia inducible factors in cancer stem cells. Br. J. Cancer. 2010;102:789–795. doi: 10.1038/sj.bjc.6605551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skuli N., Monferran S., Delmas C., Lajoie-Mazenc I., Favre G., Toulas C., Cohen-Jonathan-Moyal E. Activation of rhob by hypoxia controls hypoxia-inducible factor-1α stabilization through glycogen synthase kinase-3 in u87 glioblastoma cells. Cancer Res. 2006;66:482–489. doi: 10.1158/0008-5472.CAN-05-2299. [DOI] [PubMed] [Google Scholar]
- 45.Skuli N., Majmundar A.J., Krock B.L., Mesquita R.C., Mathew L.K., Quinn Z.L., Runge A., Liu L., Kim M.N., Liang J., et al. Endothelial hif-2α regulates murine pathological angiogenesis and revascularization processes. J. Clin. Investig. 2012;122:1427–1443. doi: 10.1172/JCI57322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.