Abstract
The periplasmic aldehyde oxidoreductase PaoABC from Escherichia coli is a molybdenum enzyme involved in detoxification of aldehydes in the cell. It is an example of an αβγ heterotrimeric enzyme of the xanthine oxidase family of enzymes which does not dimerize via its molybdenum cofactor binding domain. In order to structurally characterize PaoABC, X-ray crystallography and small angle X-ray scattering (SAXS) have been carried out. The protein crystallizes in the presence of 20% (w/v) polyethylene glycol 3350 using the hanging-drop vapour diffusion method. Although crystals were initially twinned, several experiments were done to overcome twinning and lowering the crystallization temperature (293 K to 277 K) was the solution to the problem. The non-twinned crystals used to solve the structure diffract X-rays to beyond 1.80 Å and belong to the C2 space group, with cell parameters a = 109.42 Å, b = 78.08 Å, c = 151.77 Å, β = 99.77°, and one molecule in the asymmetric unit. A molecular replacement solution was found for each subunit separately, using several proteins as search models. SAXS data of PaoABC were also collected showing that, in solution, the protein is also an αβγ heterotrimer.
Keywords: periplasmic aldehyde oxidoreductase, X-ray crystallography, small angle X-ray scattering, crystal twinning
1. Introduction
Molybdenum is a transition metal that is incorporated, as a biologically active cofactor (molybdenum cofactor, Moco), in a class of widely distributed proteins collectively known as molybdoenzymes [1]. Moco is associated with a wide range of redox enzymes and is found in most organisms from bacteria to humans. The metal in Moco is coordinated to a pterin derivative called molybdopterin to form the molybdenum containing molybdopterin (Mo-MPT) cofactor [2]. A wide variety of transformations are catalyzed by these enzymes at carbon, sulfur and nitrogen atoms, which include the transfer of an oxo group or two electrons to or from the substrate. Depending on the ligands of the molybdenum center in Moco, molybdoenzymes are classified into three families: (i) the xanthine oxidase (XO) family, characterized by a cyanolyzable equatorial sulfur ligand coordinated to the molybdenum atom; (ii) the sulfite oxidase family, with two oxo ligands at the molybdenum center; and (iii) the dimethylsulfoxide (DMSO) reductase family, where one molybdenum atom is coordinated by two dithiolene groups [1,3,4]. While in eukaryotes Moco is present solely in its Mo-MPT form, in bacteria additional variability of the Moco is achieved by attachment of a second nucleotide, GMP or CMP to the phosphate group of MPT [5–7].
The XO family of molybdoenzymes comprises a number of different enzymes in prokaryotes and eukaryotes, all transferring oxygen derived from water to their substrate. Most enzymes of the XO family are well characterized as purine and/or aldehyde oxidizing enzymes with broad substrate specificities, but also several more specific enzymes like carbon monoxide dehydrogenase and nicotine dehydrogenase have been described [8,9]. Well-characterized enzymes with aldehyde-oxidizing activity are Desulfovibrio gigas aldehyde oxidoreductase and mammalian aldehyde oxidases [10]. Mammalian aldehyde oxidases are expressed at high levels in the liver and in the lung and have been implicated in the detoxification of environmental pollutants and xenobiotics [11]. Bacterial aldehyde oxidases and aldehyde dehydrogenases were identified in different bacteria, including Methylococcus sp., Pseudomonas sp., Streptomyces moderatus [12], Amycolatopsis methanolica [13] and Pseudomonas testosteroni [14]. In addition, xanthine dehydrogenases (XDH) capable of oxidizing various purine and aldehyde substrates were characterized in bacteria like Rhodobacter capsulatus [15], Pseudomonas putida 86 [16,17], and Veillonella atypica [18,19]. With the exception of R. capsulatus and Pseudomonas aeruginosa XDH [20,21] which binds Mo-MPT, all bacterial XDHs characterized to date bind the molybdopterin cytosine dinucleotide (MCD) form of Moco. In these XDHs, molecular masses range from 140 to 300 kDa and different subunit structures were observed, like α2 in Steptomyces cyanogenus [22], αβγ in V. atypica [19], α3 in P. putida [23], α2β2 in R. capsulatus [15], α2β2 in Comamonas acidovorans [24,25], and α4β4 in P. putida 86 [17]. However, in general, enzymes of the XO family possess the same overall architecture [26], with two distinct [2Fe–2S] clusters bound to the N-terminal domain or subunit, a flavin adenine dinucleotide (FAD) bound to a central domain or subunit (with the exception of D. gigas aldehyde oxidoreductase in which the FAD binding domain is absent [27]) and the Moco-binding domain at the C-terminus.
Among the members of the xanthine oxidase family in E. coli are the xanthine dehydrogenase XdhABC, the periplasmic aldehyde oxidoreductase PaoABC, and the so far uncharacterized xanthine dehydrogenase homologue XdhD [28]. The paoABCD operon encodes for a molybdenum-containing iron–sulfur flavoprotein that is located in the periplasm [29]. The 135 kDa enzyme comprises a noncovalent (αβγ) heterotrimer with a large (78.1 kDa) molybdenum cofactor (Moco)-containing PaoC subunit, a medium (33.9 kDa) FAD-containing PaoB subunit, and a small (21.0 kDa) 2× [2Fe2S]-containing PaoA subunit, which also contains the Tat-leader peptide for the localization to the periplasm. PaoD is not a subunit of the mature enzyme, and the protein is expected to be involved in Moco modification and insertion into PaoABC. Analysis of the form of Moco present in PaoABC revealed the presence of the MCD cofactor [29]. Kinetic characterization of the enzyme showed that PaoABC converts a broad spectrum of aldehydes, with a preference for aromatic aldehydes. The terminal electron acceptor of the enzyme has not been identified to date, however, NAD+ is not used as terminal electron acceptor and the reactivity with molecular oxygen is very slow. Complete growth inhibition of E. coli cells devoid of genes from the paoABC operon was observed by the addition of cinnamaldehyde to a low-pH medium. This finding showed that PaoABC might have a role in the detoxification of aromatic aldehydes for E. coli under certain growth conditions [29].
Here, we describe the crystallization experiments and preliminary X-ray diffraction data of the enzyme purified after homologous expression in E. coli. Additional SAXS experiments confirmed the αβγ heterotrimeric structure of the enzyme.
2. Results and Discussion
2.1. Crystallization and Data Processing
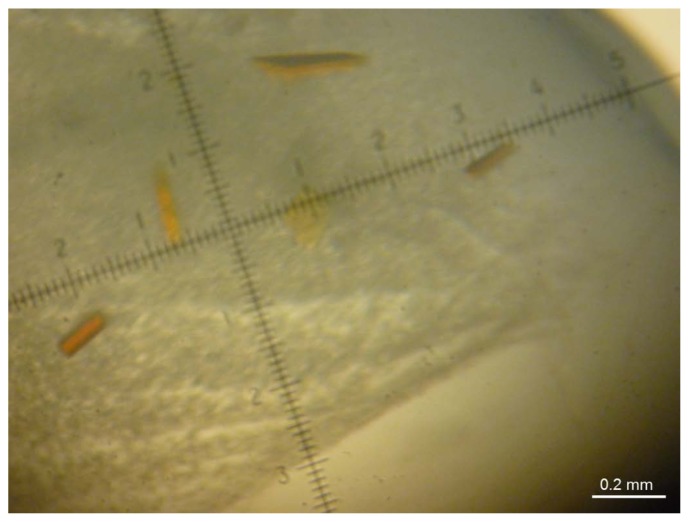
To crystallize the periplasmic aldehyde oxidoreductase (PaoABC) from E. coli several screens were tested. The protein crystallizes with 0.2 M ammonium iodide and 20% (w/v) polyethylene glycol 3350 (Figure 1). Two crystallization temperatures were used, 277 and 293 K (crystals_A and crystals_B, respectively), and several plate-like crystals appeared after four or two days, respectively. The datasets collected for both types of crystals diffracted to high resolution (1.67 and 1.8 Å, respectively) and presented similar processing statistics (Table 1). The crystals belong to the centred monoclinic space group C2 and the Matthews coefficient calculations suggest the presence of one heterotrimer (αβγ) per asymmetric unit, (Matthews coefficient of 2.7 Å3/Da) with a solvent content of 48%.
Figure 1.
Crystals of PaoABC grow in 0.2 M ammonium iodide and 22% (w/v) polyethylene glycol 3350.
Table 1.
Data-collection and processing statistics for two types of PaoABC crystals. Values in parentheses are for the highest resolution shell.
| Twinned crystals (A) (293 K) | Non-twinned crystals (B) (277 K) | |
|---|---|---|
| Data collection parameters | ||
|
| ||
| X-ray Source | ID14-1 beam line (ESRF, Grenoble) | X06DA—PXIII beam line (SLS, Villigen) |
| Detector | ADSC Q210 CCD | PILATUS 2 M |
| Wavelength (Å) | 0.934 | 0.976 |
|
| ||
| Processing statistics | ||
|
| ||
| Unit-cell parameters (Å, °) | a = 109.50 | a = 109.42 |
| b = 78.16 | b = 78.08 | |
| c = 151.84 | c = 151.77 | |
| β = 100.15 | β = 99.77 | |
| Space group | C2 | C2 |
| Molecules per AU | 1 | 1 |
| Matthews coefficient (Å3/Da) | 2.70 | 2.73 |
| Mosaicity (°) | 0.79 | 0.64 |
| Resolution range (Å) | 30.88–1.67 (1.76–1.67) | 41.23–1.80 (1.88–1.80) |
| <I/σI> | 12.8 (5.9) | 9.91 (2.41) |
| Rmerge (%) * | 6.1 (16.8) | 7.7 (46.2) |
| Rpim (%) + | 3.9 (11.1) | 5.2 (30.9) |
| Multiplicity | 3.0 (3.0) | 2.9 (2.8) |
| No. of observed reflections | 379108 (50315) | 324367 (40070) |
| No. of unique reflections | 124566 (16919) | 108448 (9450) |
| Completeness (%) | 85.2 (79.7) | 92.3 (80.7) |
Rmerge = ∑hkl∑i|Ii (hkl) − 〈I(hkl)〉|/∑hkl∑iIi(hkl);
.
Analysis using TRUNCATE [30] and XTRIAGE [31] showed normalized structure amplitudes <E> of 0.903 and 0.936, for the crystals grown at 277 K and 293K, respectively. Since the expected value for a non-twinned dataset is 0.886 and for a perfectly twinned data set is 0.94, the obtained values suggest the presence of twinning for the crystals prepared at highest temperature. As several problems may arise during phasing and refinement [32] for twinned data, the non-twinned dataset was used to solve the PaoABC structure.
2.2. Structure Determination
To solve the structure of PaoABC, sequence alignment searches were performed separately for each subunit in order to find the best homologous models that could lead to good initial phases obtained by molecular replacement. For subunit A, 4-hydroxybenzoyl-CoA reductase from Thauera aromatica (TaHBRC, PDB code 1rm6) [33]), quinoline 2-oxidoreductase from Pseudomonas Putida 86 (PpQoR, PDB code 1t3q) [34] and carbon monoxide dehydrogenase from Hydrogenophaga pseudoflava (HpCODH, PDB code 1ffu) were used; for subunit B, TaHBCR, HpCODH (PDB code 1ffu and 1ffv) were used; and for subunit C, TaHBCR, Bos taurus xanthine oxidase complexed with xanthine (BtXO, PDB code 3eub) and reduced BtXO in complex with arsenite (PDB code 3sr6) [35] were used as models.
In order to optimize the structure solution, Chainsaw [36] was used, pruning non-conserved residues prior to PHASER [37], from the CCP4 package. Three steps of molecular replacement were necessary to solve the structure of the protein, finding the position of each subunit separately. In the final solution, positive electron density was found at the expected position of the cofactors MCD, 2× [2Fe-2S] clusters and FAD, that had been excluded from the search models, thus confirming the correctness of the solution. Anomalous maps have also been calculated in order to help positioning heavy atoms in the electron density. Even though molybdenum and iron have low anomalous contribution at 0.976 Å wavelength (distant from the adsorption peak at 0.62 Å and 1.74 Å, respectively), strong anomalous peaks could be found for the Mo atom at the enzyme active site and for iron at the electron transfer centres.
The data-collection and processing statistics are presented in Table 1 and the crystal structure is currently under refinement.
2.3. Quaternary Structure Analysis of PaoABC
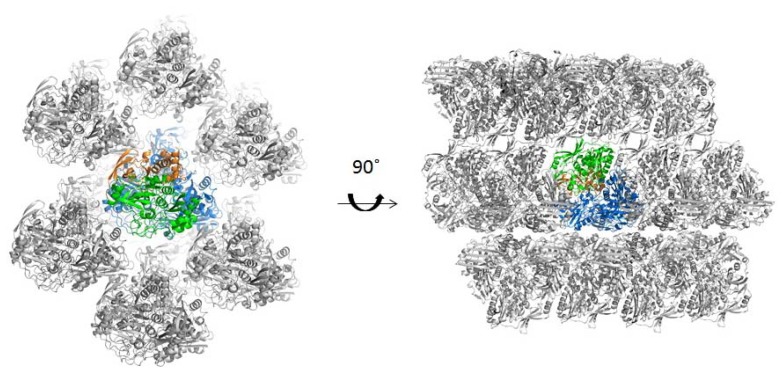
While all the enzymes of the XO family that have so far been analysed by crystallography exhibit dimeric structures (homodimers or dimers of heterotrimers (heterodimers) [38]), biochemical analysis suggested that PaoABC does not dimerize via its Moco-binding domain and remains an αβγ heterotrimer in solution. Although only one molecule is found in the asymmetric unit, we analysed the crystal packing from the position of the crystallographic dyads. This was first done by visual inspection and no tight packing could be identified (Figure 2). In addition, we used the PISA server (Protein Interfaces, Surfaces and Assemblies) [39] that predicted the ABC arrangement based on chemical thermodynamic calculations. The solvation free energy gain (ΔGint) of −163.1 kcal/mol and the free energy of assembly dissociation (ΔGdiss) of 22.8 kcal/mol shows that PaoABC is thermodynamically stable as αβγ heterotrimer [39].
Figure 2.
Crystal packing of PaoABC. Cartoon representation of several asymmetric units of PaoABC untwinned crystals with subunit A in orange, B in green and C in blue when viewed along the c axis and rotated 90°. Picture prepared using PyMOL 1.5.0.3 [40].
2.4. Small-Angle X-ray Scattering of PaoABC
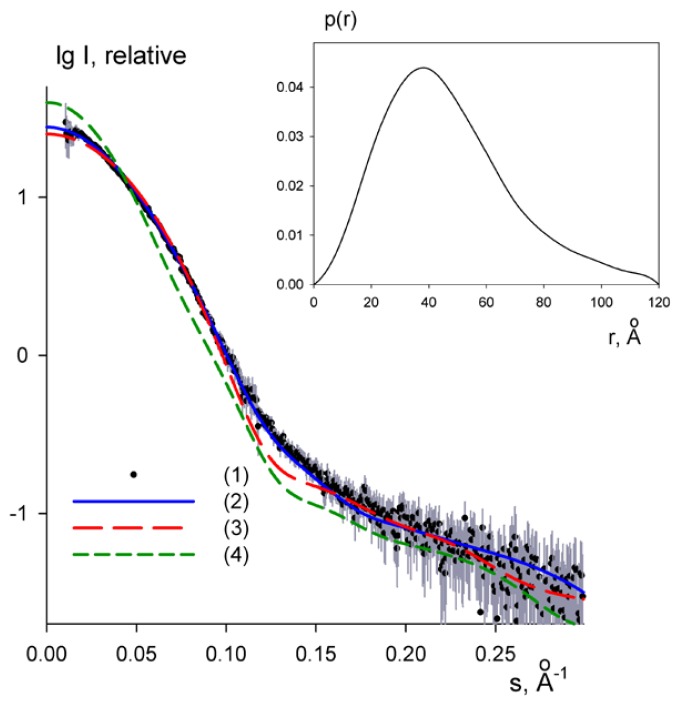
SAXS experiments have also been performed for PaoABC in order to further explore the structure and oligomerization state of the protein in solution. Monodisperse concentrated solutions of PaoABC were measured and the processed scattering profile of PaoABC are presented in Figure 3. The structural parameters including the radius of gyration (Rg), the maximum particle dimension (Dmax) and the excluded (Porod) volume of the hydrated particle computed from the experimental data are summarized in Table 2. The distance distribution function, P(r), obtained from the experimental data suggests that the protein is well folded and has a globular structure (Figure 3). The molecular mass (MM) of the solute estimated from the forward scattering I(0) and also from the Porod volume (Table 2) are both close to that of the monomeric PaoABC (136 kDa) indicating that the protein is a heterotrimer in solution.
Figure 3.
SAXS data from PaoABC in solution. The experimental data (1) are displayed as black dots with grey error bars, and the scattering computed from the models is shown as smooth lines: (2) is the scattering from the ab initio shape, (3) and (4) are scattering computed from the monomer and dimer of 1rm6, respectively. Logarithm of the scattering intensity is displayed as a function of the momentum transfer s. Insert, the characteristic function of PaoABC computed from the scattering data.
Table 2.
SAXS Data collection and derived parameters for PaoABC.
| Data collection parameters | |
|---|---|
| Instrument | EMBL X33 beam line (DORIS-III, DESY, Hamburg) |
| Beam geometry | 2.0 × 0.6 mm2 |
| Wavelength (Å) | 1.5 |
| q range (Å−1) a | 0.01–0.5 |
| Exposure time (s) | 8 × 15 |
| Concentration range (mg/mL) | 0.3–20 |
| Temperature (K) | 293 |
|
| |
| Structural parameters b | |
|
| |
| I(0) (relative) [from p(r)] | 120 ± 1 |
| Rg (Å) [from p(r)] | 36 ± 1 |
| I(0) (cm−1) (from Guinier) | 120 ± 1 |
| Rg (Å) (from Guinier) | 35 ± 1 |
| Dmax (Å) | 120 ± 10 |
| Porod volume estimate (Å3) | 200,000 ± 10,000 |
| Excluded volume estimate (Å3) | 240,000 ± 10,000 |
|
| |
| Molecular-mass determination | |
|
| |
| I(0) (cm−1) BSA (66,000 Da) | 60 ± 1 |
| Molecular mass Mr (Da) [from I(0)] | 130,000 ± 15,000 |
| Molecular mass Mr (Da) [from Porod volume (Vp/1.6)] | 125,000 ± 10,000 |
| Molecular mass Mr (Da) [from excluded volume (Vex/2)] | 120,000 ± 10,000 |
| Calculated monomeric Mr from sequence | 136,000 |
|
| |
| Software employed | |
|
| |
| Primary data reduction | Automated SAXS pipeline |
| Data processing | PRIMUS |
| Ab initio analysis | DAMMIN, DAMMIF |
| Validation and averaging | SUPCOMB, DAMAVER |
| Computation of model intensities | CRYSOL |
| 3D graphics representations | VMD |
Abbreviations: Mr: molecular mass; Rg: radius of gyration; Dmax: maximal particle dimension; Vp: Porod volume; Vex: Particle excluded volume.
Momentum transfer |q| = 4πsin(θ)/λ.
Values reported for the data merged data set (0.3 & 10 mg/mL).
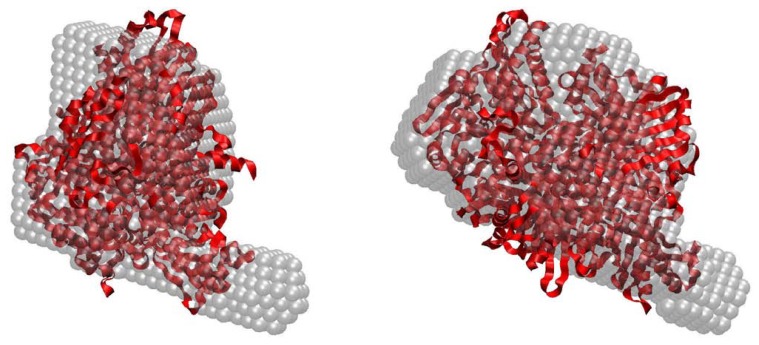
The overall shape of PaoABC was calculated ab initio from its scattering profile using programs DAMMIN [41] and DAMMIF [42]. The most typical model out of 20 reconstructions as analysed by DAMAVER [43] is overlapped in Figure 4 with the crystallographic structure of the monomer of TaHBRC (PDB code 1rm6, [33]). The reconstructed shape matches well with the overall appearance of the crystal structure. The experimental SAXS data were also fitted by the scattering profile calculated from the crystal structure of the monomeric 4-hydroxybenzoyl-Coa reductase using CRYSOL [44]. The crystal structure yielded Rg = 31 Å, somewhat smaller than the experimental value. The fit showed a discrepancy of χ = 1.3 (Figure 3) confirming that the overall shape of PaoABC is reasonably close to that of monomeric 1rm6, but also showing systematic deviations and indicating that PaoABC may be somewhat more extended in solution. In contrast, the scattering computed from the crystallographic dimer of 1rm6 provides an extremely poor fit to the SAXS data (Figure 3) with discrepancy χ = 3.1 and also the radius of gyration of the dimer (Rg = 43 Å) does not match the experimental data. Taken together, the SAXS data indicate that PaoABC is a heterotrimeric protein in solution with the overall structure similar but somewhat more extended than that of the monomeric 4-hydroxybenzoyl-Coa reductase.
Figure 4.
Superposition of the ab initio envelope of PaoABC (transparent beads, the most typical reconstruction from twenty DAMMIF runs) with the ribbon representation of the crystallographic monomer of 1rm6. Right panel is rotated 90 degrees counterclockwise along the vertical axis.
3. Experimental Section
3.1. Crystallization
PaoABC was expressed and purified using the procedure described previously [29]. The enzyme was concentrated to 20 mg/mL in 50 mM Tris-HCl pH 7.5, 250 mM NaCl, 1 mM EDTA with a Vivaspin 20 ultrafiltration device (Sartorius Stedim Biotech S.A., Goettingen, Germany). The final concentration was determined from the absorbance at 445 nm, using an extinction coefficient of 23,686 M−1·cm−1 for the native enzyme. The extinction coefficient was determined on the basis of FAD content after trichloroacetic acid precipitation [45].
The first crystallization screening experiments were performed at 293 K by hanging-drop vapour diffusion with 1 μL of protein to 1 μL of precipitant solution on 24 well crystallization plates (Molecular Dimensions, Suffolk, UK) using several commercial screenings, namely PEG/Ion HT (Hampton Research, Aliso Viejo, CA, USA), JBScreen Classic 1–10 (Jena Bioscience, Jena, Germany), and an 80 condition in-house screen (based on the screen of Jancarik et al. [46]).
PaoABC crystallized only in one condition of the commercial screen PEG/Ion HT that contains 0.2 M ammonium iodide and 20% (w/v) polyethylene glycol (PEG) 3350. Red, plate shape crystals appeared within two days. However, the first datasets (collected and processed) revealed that the crystals measured were twinned, with a twinning fraction of 50% [31]. To overcome this, several concentrations of ammonium iodide (between 0.1 M and 0.25 M), percentage of PEG 3350 (between 10% and 30%), proportions of drop and additives (Additive 1 and 2, Hampton Research) were tested but without success. When the crystallization temperature was changed from 293 K to 277 K, the crystals took four days to appear reaching maximum dimensions of 0.1 × 0.1 × 0.02 mm3 in the same crystallization condition. These crystals were untwinned and were used to solve the structure.
3.2. Data Collection and Processing
The crystals were flash-cooled directly in liquid nitrogen using as cryoprotectant a solution of 30% (v/v) glycerol, 0.2 M ammonium iodide and 22% (w/v) polyethylene glycol 3350 and maintained at 100 K under a stream of nitrogen gas during data collection.
For the crystals prepared at 293 K, several datasets were collected at beamline ID14-1 and BM14 at the European Synchrotron Radiation Facility (ESRF, Grenoble, France) (twinned crystals—A). For the crystals that were prepared at 277 K, the data was collected at X06DA—PXIII at Swiss Light Source (SLS, Villigen, Switzerland) (non-twinned crystals—B). Crystals A and B diffracted up to 1.67 Å at a wavelength of 0.934 Å and 1.80 Å at a wavelength of 0.976 Å, respectively. Both crystal types belong to C2 space group with similar unit-cell parameters: a = 109.50, b = 78.16, c = 151.84 Å, β = 100.15° for crystals A and a = 109.42, b = 78.08, c = 151.77 Å, β = 99.77° for crystals B. Matthews coefficient was calculated (ca 2.7 Å3/Da) [47] suggesting the presence of one heterotrimer per asymmetric unit, with a solvent content of 48%.
Data were processed with iMOSFLM v.1.0.7 [48] and SCALA [49] from the CCP4 program package v. 6.3.0 (Collaborative Computational Project, Number 4, 1994) [50].
3.3. Structure Solution
Structure determination was performed with PHASER [37] using as molecular models the 4-hydroxybenzoyl-CoA reductase from Thauera aromática (TaHBRC, PDB code 1rm6) [33], Quinoline 2-Oxidoreductase from Pseudomonas Putida 86 (PpQoR, PDB code 1t3q) [34] and Carbon monoxide dehydrogenase from Hydrogenophaga pseudoflava (HpCODH, PDB code 1ffu) [51] for subunit A; TaHBCR, hpCODH (PDB code 1ffu and 1ffv) for subunit B and, TaHBCR, Desulfo-Xanthine Oxidase with xanthine (PDB code 3eub) [52] and reduced Xanthine Oxidase in complex with arsenite (PDB code 3sr6) [35] from Bos taurus for subunit C. Density modification protocols [53] were applied giving initial phases with ca 0.7 mean figure of merit.
3.4. SAXS Assays
SAXS data were collected at the EMBL beamline X33 at DESY in Hamburg [54]. The measurements were performed at 293 K and different concentrations ranging from 0.3 to 30 mg/mL were used. Data were recorded using a Pilatus 1 M pixel detector at a sample-detector distance of 2.7 m and a wavelength of 1.5 Å, covering the range of momentum transfer 0.01 < q < 0.6 Å−1 (here, q = 4π sinθ/λ, where 2θ is the scattering angle). Sample solutions were circulated in a thermostated cuvette, positioned within a vacuum chamber. Eight frames of 15 s each were collected, normalized to the transmitted intensity, and subsequently averaged using AUTOSUB [55]. The data were processed with the ATSAS package [56] using standard procedures, corrected for buffer contribution, and extrapolated to infinite dilution using the program PRIMUS [57]. The forward scattering I(0) and the radii of gyration Rg were evaluated using the Guinier approximation assuming that at very small angles (q < 1.3/Rg) the intensity is represented as I(s) = I(0) exp(− ( qRg)2/3). These parameters were also computed from the entire scattering pattern using the indirect transform package GNOM [58] that also provides the maximum dimension of the particle Dmax and the distance distribution function P(r). The excluded volume of the hydrated particle (the Porod volume, Vp) was computed using the Porod invariant [59]. The program CRYSOL was used to compute the scattering from the known high-resolution models of TaHBRC (PDB code 1rm6) [44]. The ab initio modeling programs DAMMIN [41] and DAMMIF [42] were employed for low-resolution shape generation, and 20 models were calculated in the slow mode, using standard settings. The program DAMAVER [43] was utilized to superimpose individual structures, and to determine the averaged and the most probable reconstruction. The ab initio model was superimposed with the high-resolution structure of 4-hydroxybenzoyl-Coa reductase using SUPCOMB [60].
4. Conclusions
In this work we reported the crystallization of a periplasmic aldehyde oxidoreductase from E. coli using polyethylene glycol as precipitating agent. The first diffraction experiments revealed the presence of twinning in the crystals that posed difficulties in solving the structure. We could however overcome the problem by changing the crystallization temperature, which was decreased from 293 to 277 K. Using non-twinned crystals (grown at 277 K) complete data could be collected that allowed the solution of the PaoABC crystal structure by molecular replacement using the structures of related molybdenum proteins as search models. The initial calculated maps showed good electron density for the Moco, the two [2Fe-2S] and FAD. The refinement of the crystallographic model of PaoABC is currently under way and will provide valuable information regarding the residues involved in the electron pathway between the existing centres as well as details of the active site.
SAXS data have also been collected and, combined with the X-ray crystallographic information, allowed to characterize PaoABC, the first example of an enzyme of the XO family to be structurally analysed which forms a heterotrimer in solution and does not dimerize via its Moco-binding domain.
Studies of PaoABC in solution and in the crystalline state will be pursued towards the understanding of its enzymatic mechanism as well as for the clarification of the interaction of this enzyme with its cognate chaperone, the dimeric protein PaoD.
Acknowledgments
This work was financially supported by Fundação para a Ciência e Tecnologia (FCT-MCTES) through project PTDC/BIA-PRO/118377/2010, grants SFRH/BD/85806/2012 (ARC), SFRH/BPD/64917/2009 (MC), DAAD-441.00 and Deutsche Forschungsgemeinschaft Grant Le1171/6-1 (to SL). The access to the EMBL facilities was supported by the EU FP7 infrastructure grants P-Cube (Project Number 227764) and BioStruct-X (Project Number 283570).
We thank the ID14-1 and BM14 staff of the European Synchrotron Radiation Facility (France) and X06DA-PXIII staff from the Swiss Light Source/Paul Scherrer Institut (Switzerland) for assistance during data collection.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hille R. The mononuclear molybdenum enzymes. Chem. Rev. 1996;96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- 2.Rajagopalan K.V., Johnson J.L. The pterin molybdenum cofactors. J. Biol. Chem. 1992;267:10199–10202. [PubMed] [Google Scholar]
- 3.Schindelin H., Kisker C., Hilton J., Rajagopalan K.V., Rees D.C. Crystal structure of DMSO reductase: Redox-linked changes in molybdopterin coordination. Science. 1996;272:1615–1621. doi: 10.1126/science.272.5268.1615. [DOI] [PubMed] [Google Scholar]
- 4.Kisker C., Schindelin H., Rees D.C. Molybdenum-cofactor-containing enzymes: Structure and mechanism. Annu. Rev. Biochem. 1997;66:233–267. doi: 10.1146/annurev.biochem.66.1.233. [DOI] [PubMed] [Google Scholar]
- 5.Neumann M., Mittelstädt G., Seduk F., Iobbi-Nivol C., Leimkühler S. MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in Escherichia coli. J. Biol. Chem. 2009;284:21891–21898. doi: 10.1074/jbc.M109.008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neumann M., Seduk F., Iobbi-Nivol C., Leimkühler S. Molybdopterin dinucleotide biosynthesis in Escherichia coli: Identification of amino acid residues of molybdopterin dinucleotide transferases that determine specificity for binding of guanine or cytosine nucleotides. J. Biol. Chem. 2011;286:1400–1408. doi: 10.1074/jbc.M110.155671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iobbi-Nivol C., Leimkühler S. Molybdenum enzymes, their maturation and molybdenum cofactor biosynthesis in Escherichia coli. Biochim. Biophys. Acta. 2013;1827:1086–1101. doi: 10.1016/j.bbabio.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Meyer O., Rajagopalan K.V. Molybdopterin in carbon monoxide oxidase from carboxydotrophic bacteria. J. Bacteriol. 1984;157:643–648. doi: 10.1128/jb.157.2.643-648.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freudenberg W., König K., Andreesen J.R. Nicotine dehydrogenase from Arthrobacter oxidans: Containing hydroxylase. FEMS Microbiol. Lett. 1988;52:13–17. [Google Scholar]
- 10.Romao M.J., Archer M., Moura I., Moura J.J., LeGall J., Engh R., Schneider M., Hof P., Huber R. Crystal structure of the xanthine oxidase-related aldehyde oxido-reductase from D. gigas. Science. 1995;270:1170–1176. doi: 10.1126/science.270.5239.1170. [DOI] [PubMed] [Google Scholar]
- 11.Calzi M.L., Raviolo C., Ghibaudi E., de Gioia L., Salmona M., Cazzaniga G., Kurosaki M., Terao M., Garattini E. Purification, cDNA cloning, and tissue distribution of bovine liver aldehyde oxidase. J. Biol. Chem. 1995;270:31037–31045. doi: 10.1074/jbc.270.52.31037. [DOI] [PubMed] [Google Scholar]
- 12.Yasuhara A., Akiba-Goto M., Fujishiro K., Uchida H., Uwajima T., Aisaka K. Production of aldehyde oxidases by microorganisms and their enzymatic properties. J. Biosci. Bioeng. 2002;94:124–129. doi: 10.1263/jbb.94.124. [DOI] [PubMed] [Google Scholar]
- 13.Kim S.W., Luykx D.M., de Vries S., Duine J.A. A second molybdoprotein aldehyde dehydrogenase from Amycolatopsis methanolica NCIB 11946. Arch. Biochem. Biophys. 1996;325:1–7. doi: 10.1006/abbi.1996.0001. [DOI] [PubMed] [Google Scholar]
- 14.Poels P.A., Groen B.W., Duine J.A. NAD(P)+-independent aldehyde dehydrogenase from Pseudomonas testosteroni: A novel type of molybdenum-containing hydroxylase. Eur. J. Biochem. 1987;166:575–579. doi: 10.1111/j.1432-1033.1987.tb13552.x. [DOI] [PubMed] [Google Scholar]
- 15.Leimkühler S., Kern M., Solomon P.S., McEwan A.G., Schwarz G., Mendel R.R., Klipp W. Xanthine dehydrogenase from the phototrophic purple bacterium Rhodobacter capsulatus is more similar to its eukaryotic counterparts than to prokaryotic molybdenum enzymes. Mol. Microbiol. 1998;27:853–869. doi: 10.1046/j.1365-2958.1998.00733.x. [DOI] [PubMed] [Google Scholar]
- 16.Parschat K., Canne C., Huttermann J., Kappl R., Fetzner S. Xanthine dehydrogenase from Pseudomonas putida 86: Specificity, oxidation-reduction potentials of its redox-active centers, and first EPR characterization. Biochim. Biophys. Acta. 2001;1544:151–165. doi: 10.1016/s0167-4838(00)00214-4. [DOI] [PubMed] [Google Scholar]
- 17.Hettrich D., Lingens F. Microbial metabolism of quinoline and related compounds. VIII. Xanthine dehydrogenase from a quinoline utilizing Pseudomonas putida strain. Biol. Chem. Hoppe Seyler. 1991;372:203–211. doi: 10.1515/bchm3.1991.372.1.203. [DOI] [PubMed] [Google Scholar]
- 18.Smith S.T., Rajagopalan K.V., Handler P. Purification and properties of xanthine dehydroganase from Micrococcus lactilyticus. J. Biol. Chem. 1967;242:4108–4117. [PubMed] [Google Scholar]
- 19.Gremer L., Meyer O. Characterization of xanthine dehydrogenase from the anaerobic bacterium Veillonella atypica and identification of a molybdopterin-cytosine-dinucleotide-containing molybdenum cofactor. Eur. J. Biochem. 1996;238:862–866. doi: 10.1111/j.1432-1033.1996.0862w.x. [DOI] [PubMed] [Google Scholar]
- 20.Noriega C., Hassett D.J., Rowe J.J. The mobA gene is required for assimilatory and respiratory nitrate reduction but not xanthine dehydrogenase activity in Pseudomonas aeruginosa. Curr. Microbiol. 2005;51:419–424. doi: 10.1007/s00284-005-0125-8. [DOI] [PubMed] [Google Scholar]
- 21.Joshi M.S., Rajagopalan K.V. Specific incorporation of molybdopterin in xanthine dehydrogenase of Pseudomonas aeruginosa. Arch. Biochem. Biophys. 1994;308:331–334. doi: 10.1006/abbi.1994.1047. [DOI] [PubMed] [Google Scholar]
- 22.Ohe T., Watanabe Y. Purification and properties of xanthine dehydrogenase from Streptomyces cyanogenus. J. Biochem. 1979;86:45–53. [PubMed] [Google Scholar]
- 23.Woolfolk C.A. Purification and properties of a novel ferricyanide-linked xanthine dehydrogenase from Pseudomonas putida 40. J. Bacteriol. 1985;163:600–609. doi: 10.1128/jb.163.2.600-609.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sin I.L. Purification and properties of xanthine dehydrogenase from Pseudomonas acidovorans. Biochim. Biophys. Acta. 1975;410:12–20. doi: 10.1016/0005-2744(75)90203-x. [DOI] [PubMed] [Google Scholar]
- 25.Xiang Q., Edmondson D.E. Purification and characterization of a prokaryotic xanthine dehydrogenase from Comamonas acidovorans. Biochemistry. 1996;35:5441–5450. doi: 10.1021/bi952880d. [DOI] [PubMed] [Google Scholar]
- 26.Hille R. Molybdenum-containing hydroxylases. Arch. Biochem. Biophys. 2005;433:107–116. doi: 10.1016/j.abb.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Turner N., Barata B., Bray R.C., Deistung J., Le Gall J., Moura J.J. The molybdenum iron-sulphur protein from Desulfovibrio gigas as a form of aldehyde oxidase. Biochem. J. 1987;243:755–761. doi: 10.1042/bj2430755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann M., Leimkühler S. The role of system-specific molecular chaperones in the maturation of molybdoenzymes in bacteria. Biochem. Res. Int. 2011 doi: 10.1155/2011/850924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann M., Mittelstädt G., Iobbi-Nivol C., Saggu M., Lendzian F., Hildebrandt P., Leimkühler S. A periplasmic aldehyde oxidoreductase represents the first molybdopterin cytosine dinucleotide cofactor containing molybdo-flavoenzyme from Escherichia coli. FEBS J. 2009;276:2762–2774. doi: 10.1111/j.1742-4658.2009.07000.x. [DOI] [PubMed] [Google Scholar]
- 30.French S., Wilson K. On the treatment of negative intensity observations. Acta Cryst. A. 1978;34:517–525. [Google Scholar]
- 31.Zwart P.H., Grosse-Kunstleve R.W., Adams P.D. Xtriage and Fest: Automatic assessment of X-ray data and substructure structure factor estimation. CCP4 Newsl. 2005;43:27–35. [Google Scholar]
- 32.Chandra N., Acharya K.R., Moody P.C. Analysis and characterization of data from twinned crystals. Acta Cryst. D. 1999;55:1750–1758. doi: 10.1107/s0907444999009968. [DOI] [PubMed] [Google Scholar]
- 33.Unciuleac M., Warkentin E., Page C.C., Boll M., Ermler U. Structure of a xanthine oxidase-related 4-hydroxybenzoyl-CoA reductase with an additional [4Fe-4S] cluster and an inverted electron flow. Structure. 2004;12:2249–2256. doi: 10.1016/j.str.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 34.Bonin I., Martins B.M., Purvanov V., Fetzner S., Huber R., Dobbek H. Active site geometry and substrate recognition of the molybdenum hydroxylase quinoline 2-oxidoreductase. Structure. 2004;12:1425–1435. doi: 10.1016/j.str.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Cao H., Hall J., Hille R. X-ray crystal structure of arsenite-inhibited xanthine oxidase: μ-sulfifo, μ-oxo double bridge between molybdenum and arsenic in the active site. J. Am. Chem. Soc. 2011;133:12414–12417. doi: 10.1021/ja2050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stein N. CHAINSAW: A program for mutating pdb files used as templates in molecular replacement. J. Appl. Cryst. 2008;41:641–643. [Google Scholar]
- 37.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallography software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romão M.J. Molybdenum and tungsten enzymes: A crystallographic and mechanistic overview. Dalton Trans. 2009;21:4053–4068. doi: 10.1039/b821108f. [DOI] [PubMed] [Google Scholar]
- 39.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 40.DeLano W. The PyMOL Molecular Graphics System. CA DeLano Sci; San Carlos, CA, USA: 2002. [Google Scholar]
- 41.Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franke D., Svergun D.I. DAMMIF a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volkov V., Svergun D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svergun D.I., Barberato C., Koch M. CRYSOL—A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 45.Rajagopalan K.V., Fridovich I., Handler P. Hepatic aldehyde oxidase. J. Biol. Chem. 1962;237:922–928. [PubMed] [Google Scholar]
- 46.Jancarik J., Kim S.H. Sparse matrix sampling: A screening method for crystallization of proteins. J. Appl. Cryst. 1991;24:409–411. [Google Scholar]
- 47.Matthews B.W. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 48.Leslie A.G.W. Jt. CCP4 + ESF-EAMCB Newsl. Protein Cryst. CCP4; Oxon, UK: 1992. Recent Changes to the MOSFLM Package for Processing Film and Image Plate Data. [Google Scholar]
- 49.Kabsch W. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J. Appl. Cryst. 1988;21:916–924. [Google Scholar]
- 50.Number 4, C.C.P. The CCP4 suite: Programs for protein crystallography. Acta Cryst. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 51.Hänzelmann P., Dobbek H., Gremer L., Huber R., Meyer O. The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase. J. Mol. Biol. 2000;301:1221–1235. doi: 10.1006/jmbi.2000.4023. [DOI] [PubMed] [Google Scholar]
- 52.Pauff J.M., Cao H., Hille R. Substrate orientation and catalysis at molybdenum site in xanthine oxidase: Crystal structure in complex with xanthine and lumazine. J. Mol. Biol. 2009;284:8760–8767. doi: 10.1074/jbc.M804517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cowtan K.D., Main P. Phase combination and cross validation in iterated density-modification calculations. Acta Cryst. D. 1996;52:43–48. doi: 10.1107/S090744499500761X. [DOI] [PubMed] [Google Scholar]
- 54.Blanchet C.E., Zozulya A.V., Kikhney A.G., Franke D., Konarev P.V., Shang W., Klaering R., Robrahn B., Hermes C., Cipriani F., et al. Instrumental setup for high-throughput small- and wide-angle solution scattering at the X33 beamline of EMBL Hamburg. J. Appl. Cryst. 2012;45:489–495. [Google Scholar]
- 55.Franke D., Kikhney A.G., Svergun D.I. Automated acquisition and analysis of small angle X-ray scattering data. Nucl. Instrum. Methods. Phys. Res. Sect. A. 2012;689:52–59. [Google Scholar]
- 56.Petoukhov M.V., Konarev P.V., Kikhney A.G., Svergun D.I. ATSAS 2.1—Towards automated and web-supported small-angle scattering data analysis. J. Appl. Cryst. 2007;40:223–228. [Google Scholar]
- 57.Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 58.Mertens H.D., Svergun D.I. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J. Struct. Biol. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 59.Porod G. In Small Angle X-ray Scattering. Academic Press; New York, NY, USA: 1982. [Google Scholar]
- 60.Kozin M.B., Svergun D.I. Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 2001;34:33–41. [Google Scholar]