Abstract
Chitinases (EC 3.2.2.14), expressed during the plant-pathogen interaction, are associated with plant defense against pathogens. In the present study, a positive correlation between chitinase activity and sugarcane smut resistance was found. ScChi (GenBank accession no. KF664180), a Class III chitinase gene, encoded a 31.37 kDa polypeptide, was cloned and identified. Subcellular localization revealed ScChi targeting to the nucleus, cytoplasm and the plasma membrane. Real-time quantitative PCR (RT-qPCR) results showed that ScChi was highly expressed in leaf and stem epidermal tissues. The ScChi transcript was both higher and maintained longer in the resistance cultivar during challenge with Sporisorium scitamineum. The ScChi also showed an obvious induction of transcription after treatment with SA (salicylic acid), H2O2, MeJA (methyl jasmonate), ABA (abscisic acid), NaCl, CuCl2, PEG (polyethylene glycol) and low temperature (4 °C). The expression levels of ScChi and six immunity associated marker genes were upregulated by the transient overexpression of ScChi. Besides, histochemical assay of Nicotiana benthamiana leaves overexpressing pCAMBIA 1301-ScChi exhibited deep DAB (3,3′-diaminobenzidinesolution) staining color and high conductivity, indicating the high level of H2O2 accumulation. These results suggest a close relationship between the expression of ScChi and plant immunity. In conclusion, the positive responses of ScChi to the biotic and abiotic stimuli reveal that this gene is a stress-related gene of sugarcane.
Keywords: chitinase, Saccharum officinarum, Sporisorium scitamineum, subcellular localization, gene expression, abiotic stress
1. Introduction
Plants are vulnerable to external environmental stimuli, as they lack an effective immune system. When under attack from pathogen infection, plants activate a number of defense responses to cope with infection and to protect themselves [1]. Chitinases (EC 3.2.2.14), which can catalyze poly chitin, are present in the cell walls of most fungi and homologues in plant typical pathogenesis-related (PR) proteins. Previous research has revealed that they play an important role in plant defense mechanisms [2,3]. Chitinases isolated from both monocot and dicot plants have been shown to inhibit the growth of chitin-containing fungi, both in vitro [4,5] and in vivo [6,7]. Plant chitinases also have resistance characteristics implicated in plant defense to different kinds of pathogens, including bacteria, viruses and other abiotic stresses [8]. Chitinase isozymes are a diverse group of enzymes that differ in enzymatic activities, primary sequence, isoelectric point (pI) and cellular localization [9]. Chitinase genes have been grouped into seven classes (Class I–VII) belonging to the glycoside hydrolase families 18 and 19 [2,10].
Sugarcane smut, caused by the fungus Sporisorium scitamineum (S. scitamineum), is a worldwide disease and causes serious losses in sugarcane yield and sucrose content [11–13]. Several methods, such as effective agronomic practices and chemical control, have been recommended for the control of smut infection. However, only the use of resistant cultivars has proven to be practical and economical [14]. With the development of biotechnological approaches, crop protection by engineering genetic disease resistance is becoming a promising tool to eliminate the disadvantages of traditional methods [10]. As reported, transgenic plants overexpressing PR proteins could enhance resistance against fungal diseases (e.g., the isolectin I purified from the transgenic Nicotiana tabacum, which transformed an isolectin I gene from Urtica dioica, showed in vitro antifungal activities on germinated spores of the fungi, Botrytis cinerea, Trichoderma viride and Colletotrichum lindemuthianum) [15]. However, isolating and characterizing the gene that encodes a broad-spectrum antifungal protein is a prerequisite for obtaining promising transgenic plants resistant to fungal pathogens.
Plant chitinases have been the most extensively studied and applied PR proteins. To date, a wide variety of plant chitinases have been identified from several plant species, including Arabidopsis thaliana, Oryza sativa, Nicotiana tabacum, Triticum aestivum and Zea mays [7,10,16]. Class I chitinase from Hordeum vulgare and Class VII chitinase from T. aestivum have been reported to demonstrate antifungal activities in recent studies [10,17]. Transgenic banana (Musa acuminata) integrating rice chitinase gene exhibited resistance to black leaf streak disease caused by the pathogenic fungus, Mycosphaerella fijiensis [16]. In sugarcane, only one full-length chitinase gene, ScChiB1 (EU914815.1), belonging to the Class IV of family 19 glycosyl hydrolases, has been amplified from the red rot resistant cultivar, Co93009 [18]. The relative expression of this chitinase gene in both the red rot resistant (Co 93009) and susceptible (CoC 671) cultivars by real-time quantitative PCR (RT-qPCR) showed a high level of expression only in the resistant cultivar during interaction with Colletotrichum falcatum. In addition, the chitinases from the leaves of the chewing cane (Saccharum officinarum) cultivar Fuan were upregulated after C. falcatum infection, and an increasing expression pattern of a partial chitinase gene has also been validated by RT-qPCR at 24, 48 and 72 h of interaction [19].
In the present study, an activity assay of chitinase enzymes was firstly conducted on sugarcane cultivars (resistant and susceptible genotypes) in response to S. scitamineum pathogen. Secondly, a full-length chitinase gene, ScChi (GenBank accession no. KF664180) from sugarcane challenged by S. scitamineum was isolated and characterized. Finally, its expression in Escherichia coli (E. coli) cells, probable localization, relative mRNA expression in sugarcane tissues and various expression profiles under different biotic/abiotic stresses, as well as transient expression in Nicotiana benthamiana were investigated. This study aims to obtain better knowledge of sugarcane chitinases and the function of their encoding genes.
2. Results and Discussion
2.1. Enzyme Activity of Chitinase
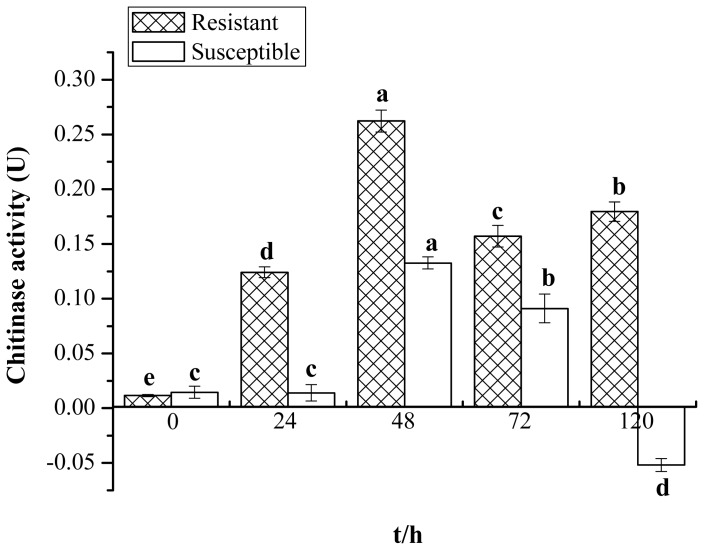
A biochemical method for the determination of enzyme activity was applied to analyze the change in chitinase activity in Yacheng05–179 (resistant) and Liucheng03–182 (susceptible) harvested at different time points after smut infection. Figure 1 shows that the elevated activity of chitinase in Yacheng05–179 can be observed at an early stage of 24 h compared to the control. Activity remains at a higher level at all later time points. The maximum level of activity was observed at 48 h; then, the accumulated level slightly decreases at 72 h before increasing to 0.1795 U (unit of chitinase activity (U)) at 120 h. The activity of chitinase in Liucheng03–182 remained unchanged at the first 24 h and reached the peak value of 0.1325 U at 48 h, then decreased at 72 h before being much reduced at 120 h. It is interesting that chitinase activity in the resistant cultivar is always higher than that of the mock and the susceptible cultivar challenged by smut pathogen, suggesting that a positive correlation may exist between sugarcane chitinase activity and smut resistance.
Figure 1.
The chitinase activity in smut-resistant (Yacheng05–179) and smut-susceptible (Liucheng03–182) sugarcane cultivars inoculated with Sporisorium scitamineum. All data points (with the deduction of their mocks) are the means ± SE (n = 3). Different lowercase letters indicate a significant difference, as determined by the least-significant difference test (p-value < 0.05). U, unit of chitinase activity.
2.2. Cloning and Sequence Analysis of the Chitinase Gene, ScChi
A sugarcane chitinase-like gene, ScChi (GenBank accession no. KF664180), was isolated from Yacheng05–179 post-48 h S. scitamineum inoculation by RT-PCR-based cloning. ScChi had a full length of 1001 bp, with an open reading frame (ORF) of 861 bp (encoding a polypeptide of 286 amino acids) (Figure 2). The predicted protein had a molecular weight of 31.37 kDa with a pI of 4.69. MotifScan software (SIB, Geneva, Switzerland) showed that ScChi contained a glycosyl hydrolase family 18 (Pfam) domain from position 32 to 266 aa, which suggested that sugarcane ScChi encoded a putative chitinase. The NetNGlyc online program analysis predicted three putative N-glycosylation sites (at position 65, 214 and 232 aa) with probabilities of 20.18%, 49.02% and 68.47%, respectively. There was no signal peptide and transmembrane helix domain in ScChi by SignalP (CBS, New York, NY, USA) and TMHMM Server v. 2.0 programs (CBS, New York, NY, USA), which suggested that ScChi was not a secretory protein. ScChi contained the active site of the catalytic residues, Asp137 (D137), Asp140 (D140), Asp142 (D142) and Glu146 (E146) (Figure 2).
Figure 2.
Nucleotide acid sequences obtained by real-time (RT)-PCR and the deduced amino acid sequences of ScChi. The deduced amino acid sequences are shown as a one-letter code under the cDNA sequences. The underline indicates the catalytic domains (DXXDXDXXXE) of the glycosyl hydrolase family 18. The eight red squares of Tyr10 (Y10), Phe37 (F37), Asp140 (D140), Asp142 (D142), Phe144 (F144), Gln195 (Q195), Phe259 (F259) and Trp261 (W261) show putative carbohydrate binding sites.
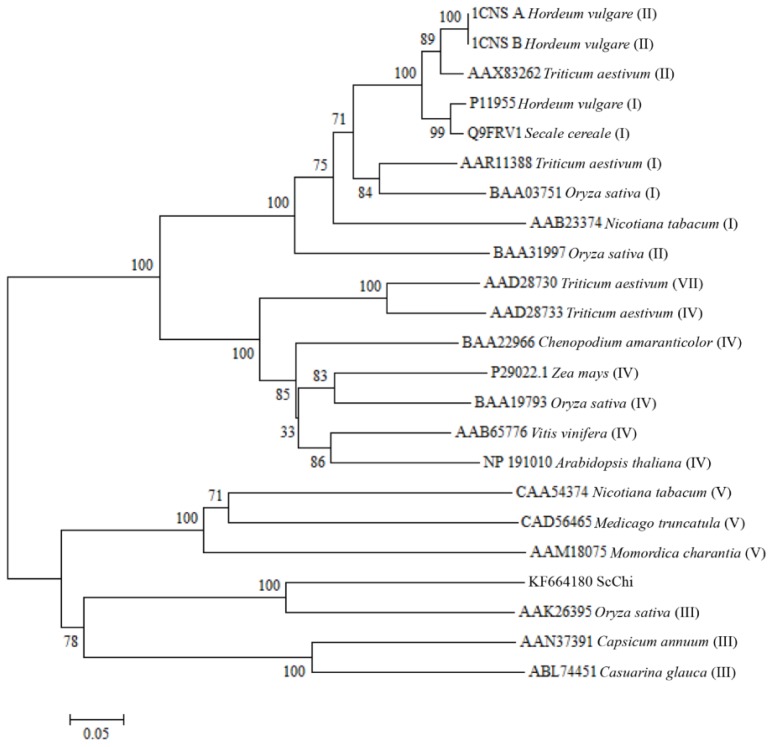
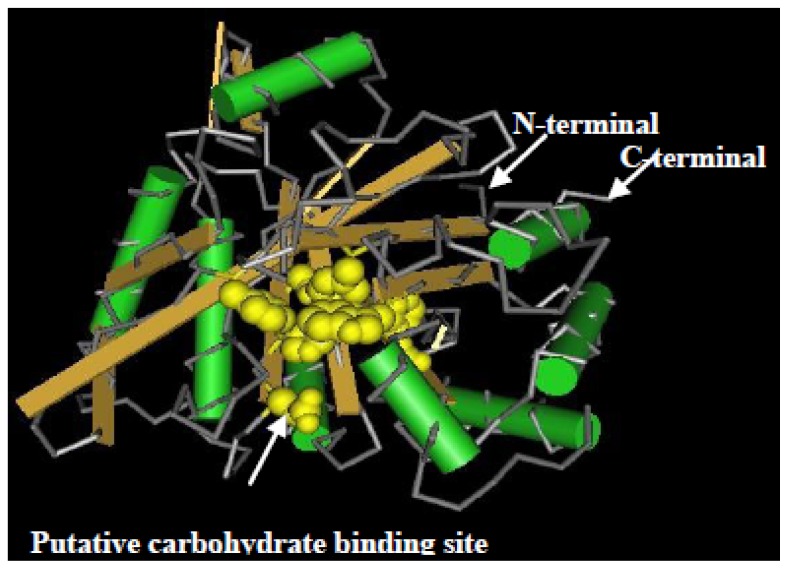
Basic Local Alignment Search Tool (BLAST) search of the amino acid sequences indicated that ScChi shared high sequence similarity with diverse plant chitinases in the GenBank database. The predicted amino acid sequence of ScChi showed 84.27% identity with that of Z. mays (AFW72831.1), 75.78% with that of Aegilops tauschii (EMT04102.1), 73.61% with that of O. sativa (NP_001064607.1) and 73.45% with that of Triticum urartu (EMS51632.1). The phylogenetic tree showed that ScChi was closely related to O. sativa chitinase (III) (AAK26395) (Figure 3). Three-dimensional structures (3D) of the GH18_narbonin (cd06544) similar to ScChi domain were found by Blastp in NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?RID=FSWDHR9T014&mode=all) (Figure 4). The structure of GH18_narbonin showed the presence of a putative carbohydrate binding site, which was highly homologous to ScChi (the residues were Tyr10 (Y10), Phe37 (F37), Asp140 (D140), Asp142 (D142), Phe144 (F144), Gln195 (Q195), Phe259 (F259) and Trp261 (W261)) (Figure 2). These combined data suggested that ScChi belonged to an acid Class III chitinase, the member of family 18 glycosyl hydrolase.
Figure 3.
Phylogenetic analysis of deduced amino acid sequence from ScChi (KF664180) and other plant chitinases. The neighbor-joining method was used. Plant chitinase classes are represented in parentheses.
Figure 4.
Three-dimensional structural analysis of the GH18_narbonin (cd06544) domain, which was similar to ScChi by Blastp. The presence of putative carbohydrate binding sites, which was highly homologous to ScChi, is shown in the 3D structure of GH18_narbonin by arrows.
2.3. Subcellular Localization of ScChi
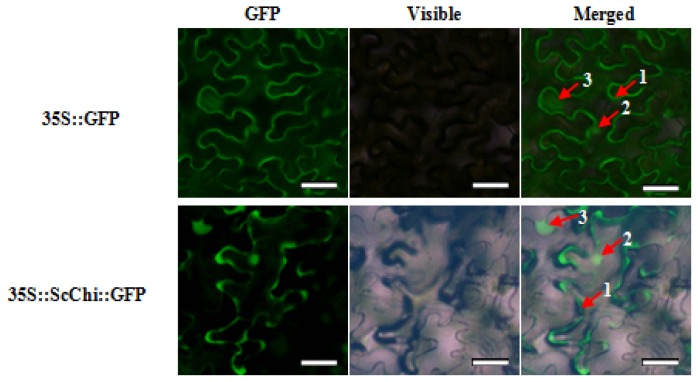
ScChi was cloned into the plant expression vector of pCAMBIA 2300 containing the CaMV 35S promoter and the GFP reporter gene. Agrobacterium-mediated transformation was then performed to identify the transient expression of the target gene and GFP in N. benthamiana. Infiltrated leaves incubated for 48 h at 24 °C were collected, and the green fluorescence was monitored under a fluorescence microscope. As illustrated in Figure 5, compared with the mock, ScChi::GFP fusion protein was observed in the nucleus, cytoplasm and potentially associated with the plasma membrane.
Figure 5.
The subcellular localization analysis of ScChi in Nicotiana benthamiana leaves 48 h after infiltration. The epidermal cells were used for taking images of green fluorescence, visible light and merged light. Read Arrows 1, 2 and 3 indicated plasma membrane, nucleus and cytoplasm, respectively. Bar = 50 μm.
2.4. Prokaryotic Expression of ScChi in E. coli Rosetta Cells
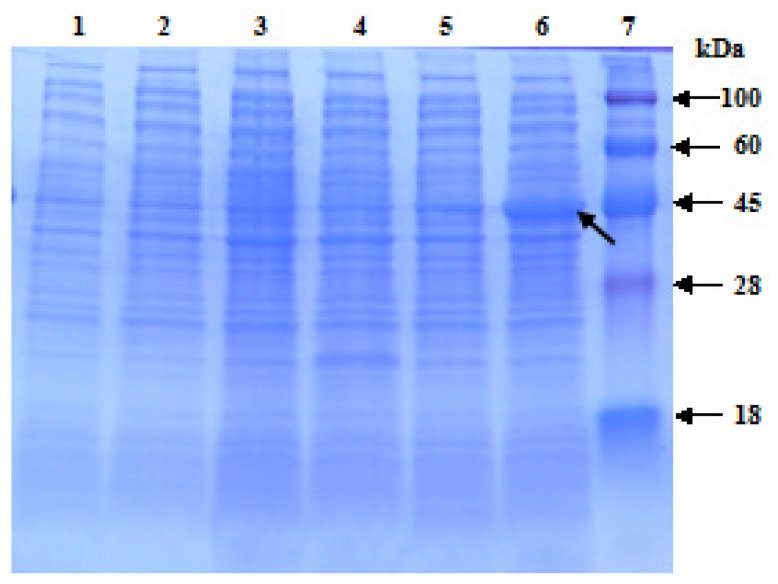
The ORF (861 bp) of ScChi was subcloned into expression vector pET 32a (+) under the control of an inducible promoter and transformed into E. coli Rosetta cells. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis showed the accumulation of the recombinant protein in LB medium in response to 1.0 mM isopropyl β-d-thiogalactoside (IPTG) induction at 28 °C (Figure 6). At the presence of 6× His-tag, the target protein showed higher molecular weight than the estimated one of 31.37 kDa.
Figure 6.
The prokaryotic expression of pET 32a-ScChi in Escherichia coli Rosetta cells. 1, blank (E. coli Rosetta cells) without induction; 2, blank induction for 1 h; 3, control (E. coli Rosetta cells containing pET 32a vector) without induction; 4, control induction for 1 h; 5, pET 32a-ScChi without induction; 6, pET 32a-ScChi induction for 1 h; 7, protein marker; the induced protein is shown by the arrow.
2.5. Overexpression of ScChi in E. coli Enhances Cell Growth under Abiotic Stresses
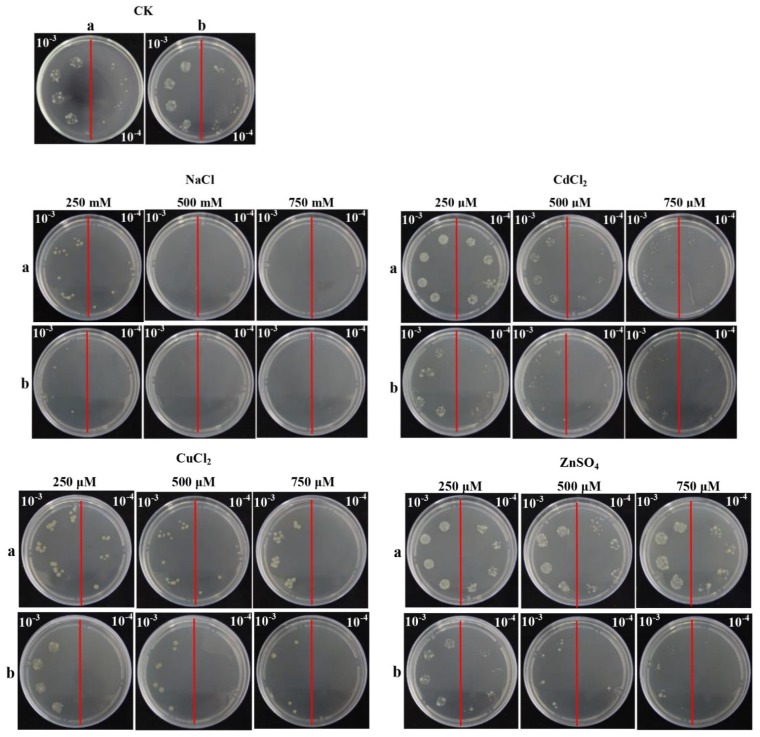
As reported, plant chitinase was a stress-related gene, which could be induced by different stresses, such as mechanical wounding, ABA (abscisic acid), SA (salicylic acid), ZnSO4 (zinc sulfate), MeJA (methyl jasmonate) and so on [20,21]. Spot assay was performed to ascertain the response of Rosetta + pET 32a-ScChi and control (Rosetta + pET 32a) to different supplements in vivo (Figure 7). After overnight culture, Rosetta + pET 32a-ScChi showed faster growth as compared to that of the control on LB plates containing NaCl (sodium chloride), CuCl2 (copper chloride), CdCl2 (cadmium chloride) and ZnSO4. These results demonstrated that the recombinant protein could enhance the growth of recombinant cells in high stress condition.
Figure 7.
Spot assays of Rosetta + pET 32a-ScChi (a) and Rosetta + pET 32a (control) (b) on Luria-Bertani (LB) plates with NaCl, CuCl2, CdCl2 and ZnSO4. Isopropyl β-d-thiogalactoside (IPTG) was added to the cultures of Rosetta + pET 32a-ScChi and Rosetta + pET 32a to induce the expression of recombinant protein. The cultures were adjusted to OD600 = 0.6. Ten microliters from 10−3 (left side of the red line on the plate) to 10−4 (right side of the red line on the plate) dilutions were spotted onto LB plates without any supplement (CK) or with NaCl (250, 500 and 750 mM), CuCl2 (250, 500 and 750 μM), CdCl2 (250, 500 and 750 μM) and ZnSO4 (250, 500 and 750 μM), respectively. NaCl, sodium chloride; CuCl2, copper chloride; CdCl2, cadmium chloride; ZnSO4, zinc sulfate.
2.6. Tissue-Specific Expression Analysis of ScChi in Different Sugarcane Tissues
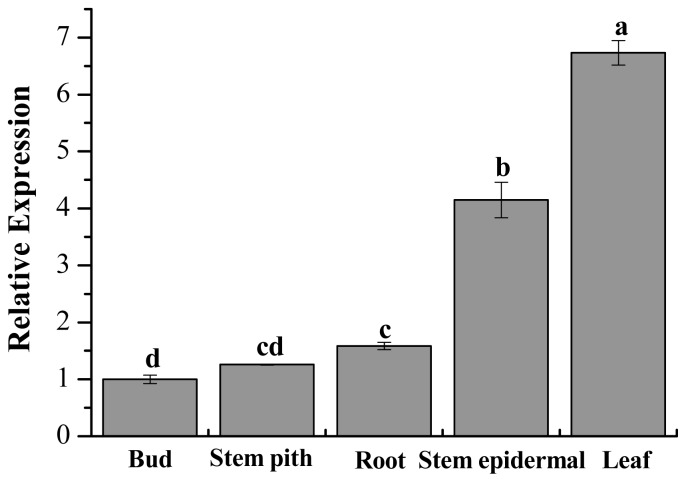
For tissue-specific expression analysis of ScChi, the sugarcane cultivar, Yacheng05–179, was used as the experimental material. The expression of ScChi in root, leaf, bud, stem epidermal and stem pith was detected, with the GAPDH gene being used as an internal control for RT-qPCR. The results suggested that ScChi was highly expressed in leaf and stem epidermal (Figure 8), while the root and stem pith exhibited a moderate mRNA expression level, and the bud showed a relatively low level in comparison.
Figure 8.
Tissue-specific expression analysis of the ScChi in different tissues of sugarcane cultivar Yacheng05–179. Data are normalized to the GAPDH expression level. All data points are the means ± SE (n = 3). Different lowercase letters indicate a significant difference, as determined by the least-significant difference test (p-value < 0.05).
2.7. Expression Profiles of ScChi under Different Stress Treatments
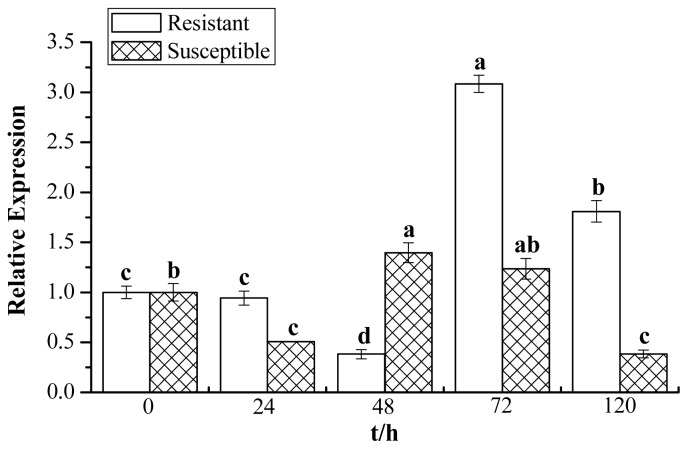
Expression profiles of ScChi were studied by RT-qPCR after sugarcane (Yacheng05–179 and Liucheng03–182) was challenged with smut pathogen (Figure 9). The ScChi transcript was calculated by the expression level of the inoculated sample minus the level of the mock at each corresponding time point, so as to eliminate any effect of wounding. In response to smut stress, the transcript level of ScChi in the resistant cultivar of Yacheng05–179 exhibited a little decrease at 48 h, but showed increasing accumulation at 72 and 120 h, which was higher than that of ScChi in susceptible cultivar Liucheng03–182. During the interaction of Liucheng03–182 and smut, ScChi transcript decreased at 24 h, then elevated at 48 and 72 h and, again, decreased rapidly at 120 h. We could observed that the expression profiles of ScChi in the Yacheng05–179 and smut interaction were higher than those of ScChi in the Liucheng03–182 and smut interaction, except at 48 h.
Figure 9.
RT-qPCR (quantitative PCR) analysis of the ScChi expression patterns during sugarcane-smut (Sporisorium scitamineum) interaction. Data were normalized to the GAPDH expression level. All data points (with the deduction of their mocks) are the means ± SE (n = 3). Different lowercase letters indicate a significant difference, as determined by the least-significant difference test (p-value < 0.05). Resistant: Yacheng05–179 cultivar; susceptible: Liucheng03–182 cultivar.
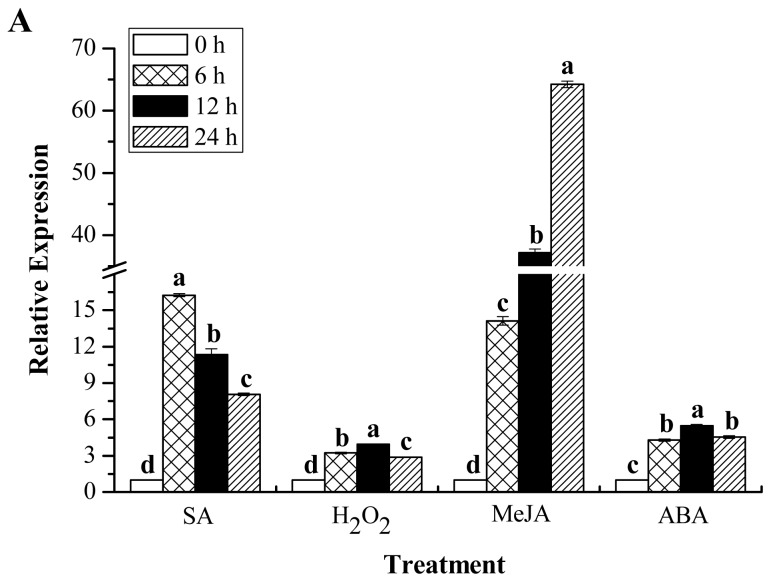
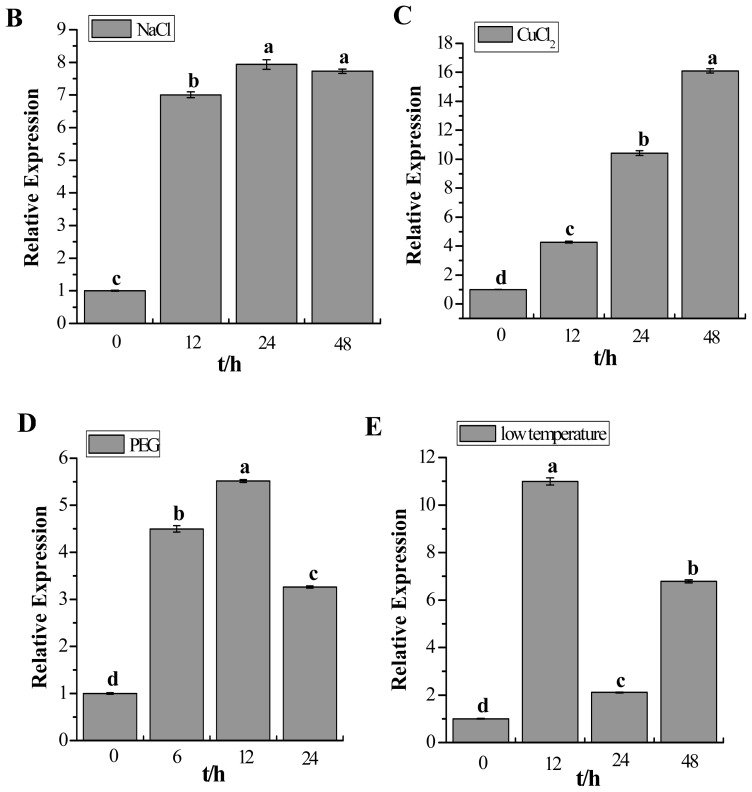
Furthermore, Yacheng05–179 plantlets under the stresses of 5 mM SA, 10 mM H2O2 (hydrogen peroxide), 25 μM MeJA, 100 μM ABA, 250 mM NaCl, 100 μM CuCl2, 25% PEG (polyethylene glycol) and 4 °C low temperature were carried out to examine the expression profiles of ScChi response to these stresses. As shown in Figure 10, the ScChi transcript was strongly induced by all of these exogenous stresses, including different plant hormones, oxidative stress, hyper-osmotic stress, metal stress, as well as temperature stress. It is interesting that under all the above abiotic treatments, ScChi showed a positive response in the early time of the stresses and maintained an increased transcript from 0 to 12 h or even to 24 h in the same cases of MeJA, NaCl and CuCl2 stresses.
Figure 10.
RT-qPCR analysis of the ScChi expression patterns in Yacheng05–179 plantlets under various abiotic stresses. Data were normalized to the GAPDH expression level. (A) The relative expression of ScChi under the stresses of 5 mM SA, 10 mM H2O2, 25 μM MeJA and 100 μM ABA; (B–E) the relative expression of ScChi under the stresses of 250 mM NaCl, 100 μM CuCl2, 25% PEG and 4 °C low temperature, respectively. All data points are the means ± SE (n = 3). Different lowercase letters indicate a significant difference, as determined by the least-significant difference test (p-value < 0.05). SA, salicylic acid; H2O2, hydrogen peroxide; MeJA, methyl jasmonate; ABA, abscisic acid; NaCl, sodium chloride; CuCl2, copper chloride; PEG, polyethylene glycol.
2.8. Transient Expression of ScChi Induces a Defense Response in N. benthamiana
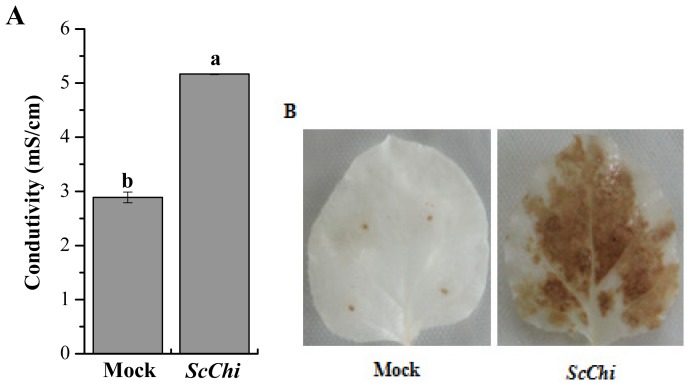
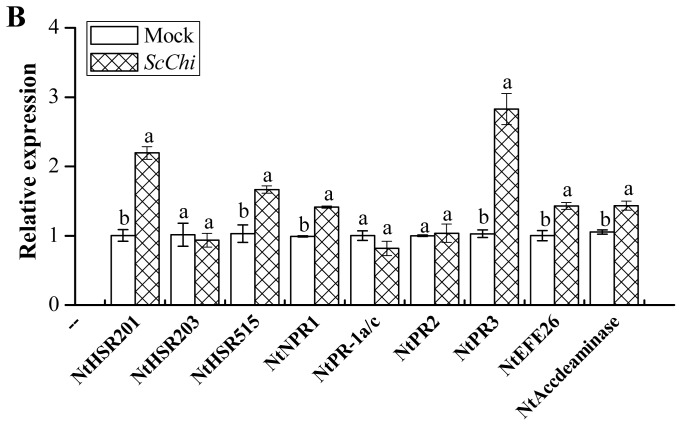
The overexpressed vector of pCAMBIA 1301-ScChi was constructed and validated by the enzyme digesting identification. An Agrobacterium-mediated transient expression method was carried out to identify the effect of ScChi expression on the induction of the defense response in N. benthamiana leaves (Figure 11A,B). After 48 h, a typical hyper-sensitive response (HR) symptom with enhanced conductivity and deeper DAB (3,3′-diaminobenzidinesolution) staining color was found in the tested leaves expressing ScChi. On the contrary, plants transiently expressing the mock of pCAMBIA 1301 did not trigger defense response in N. benthamiana leaves. Moreover, the expression levels of ScChi and six immunity associated marker genes, including the HR marker genes, NtHSR201 and NtHSR515, the SA-related gene, NtNPR1, the JA-associated gene, NtPR3, the ethylene-synthesis-depended genes NtEFE26, and NtAccdeaminase, were enhanced by the transient overexpression of ScChi (Figure 12).
Figure 11.
The transient expression of ScChi in Nicotiana benthamiana leaves. (A) Conductivity measurement of N. Benthamiana leaves infiltrated with the 35S::ScChi-containing Agrobacterium strain after 48 h. Mock: the Agrobacterium strain carrying 35S::00. All data points are the means ± SE (n = 3). Different lowercase letters indicate a significant difference, as determined by the least-significant difference test (p-value < 0.05); (B) DAB (3,3′-diaminobenzidinesolution) staining with N. benthamiana leaves 48 h after Agrobacterium strain infiltration.
Figure 12.
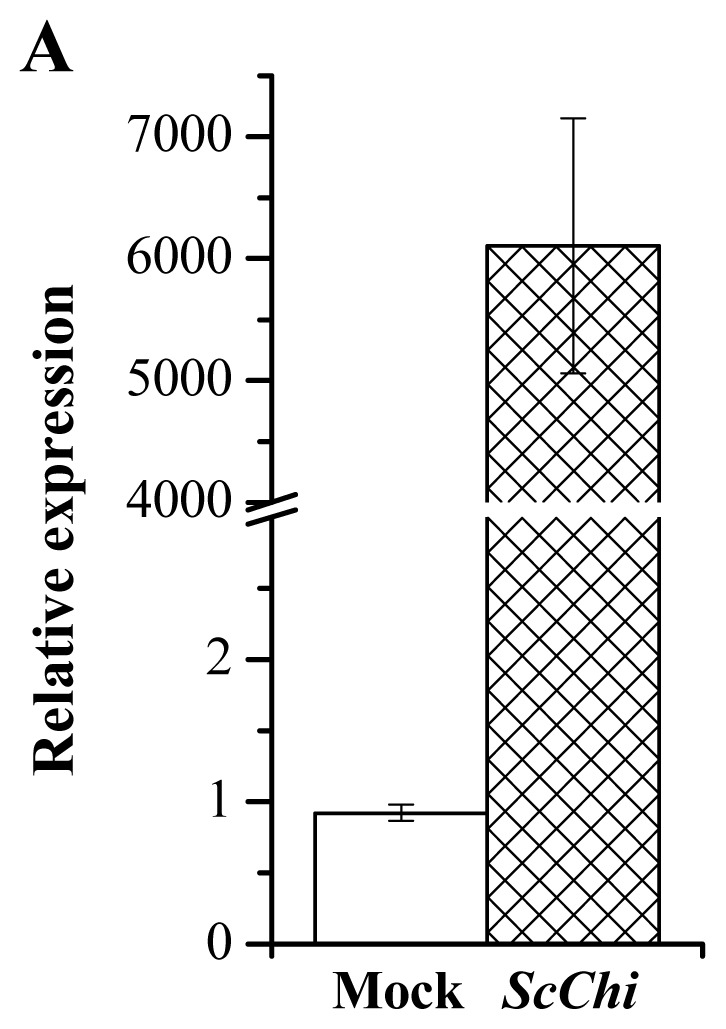
RT-qPCR analysis of ScChi and the immunity related marker genes in the Nicotiana benthamiana leaves 24 h after transiently expressing pCAMBIA 1301-ScChi and the empty vector. (A) The transcript analysis of ScChi; (B) the transcript analysis of the immunity related marker genes, including the hypersensitive response marker genes, NtHSR201, NtHSR203 and NtHSR515, the salicylic acid related gene, NtNPR1, the jasmonate associated genes, NtPR-1a/c, NtPR2 and NtPR3, and the ethylene synthesis depended genes, NtEFE26 and NtAccdeaminase. NtEF1-α was used to normalize the transcript levels. Mock: the Agrobacterium strain carrying 35S::00. All data points are the means ± SE (n = 3). Different lowercase letters indicate a significant difference, as determined by the least-significant difference test (p-value < 0.05).
3. Discussion
Plant diseases triggered by fungal pathogens are one of the major concerns in agriculture. Induced resistance to a plant pathogen is a complex mechanism, which involves the activation of various immune responses [1]. Many resistance genes, including PR proteins, have been isolated and used to enhance disease resistance in plants [22]. Initially, PR proteins were found commonly induced in resistant plants and present during HR to pathogens of bacteria, fungi and virus. However, it turned out that PR proteins can be triggered not only in resistant, but also in susceptible plants infected with pathogens, as well as in plants subjected to abiotic stresses [23,24]. Among these PR proteins, chitinases, which degrade the chitin walls of fungi, are widely distributed in nature and play a vital role in plant defense against pathogens. Hence, great effort has been made on the cloning and characterization of chitinase genes in plants [10]. Krishnaveni et al. have reported multiple antifungal chitinases, CH1, CH2 and CH3, from Sorghum bicolor [25]. Singh et al. found that a Class VII wheat chitinase exerted a broad-spectrum antifungal activity against Sarocladium oryzae, Alternaria sp., C. falcatum, Rhizoctonia solani, Pestalotia theae and Fusarium sp. [10].
Recently, Rahul et al. amplified several partial mRNA sequences of chitinase from both red rot-resistant and -susceptible sugarcane cultivars, and a full-length chitinase sequence, ScChiB1, was isolated from the resistant cultivar [18]. An upregulated (2.43-fold) expression protein of chitinase was identified in the leaves of chewing cane inoculated with C. falcatum, and the activity of chitinase was enhanced and differed significantly in the 10 chewing cane cultivars [19]. In addition, Viswanathan et al. performed a Western blotting study and found a higher and rapid accumulation of chitinase in a red rot-resistant cultivar [26]. The present study revealed that chitinase was also triggered during the interaction between sugarcane and S. scitamineum. What should also be stressed is that the chitinase enzyme activity in the resistant cultivar (Yacheng05–179) was almost always higher than those of the mock and the susceptible cultivar (Liucheng03–182) (Figure 1), suggesting that this chitinase may be involved in the induction of sugarcane resistance against S. scitamineum.
Chitinases have been reported to group into seven classes (Class I–VII) belonging to the glycoside hydrolase families 18 and 19, suggesting that chitinase isozymes were encoded by a multi-gene family [2,10]. In the present study, we isolated a chitinase gene that was induced upon infection of S. scitamineum at the time point of 48 h. Multiple sequence alignment showed 84.27% identity of ScChi (KF664180) with Z. mays chitinase (AFW72831.1), 75.78% with that of A. tauschii (EMT04102.1), 73.61% with that of O. sativa (NP_001064607.1) and 73.45% with that of T. urartu (EMS51632.1). The deduced amino acid sequences showed that there was an active site of the catalytic residues, Asp137 (D137), Asp140 (D140), Asp142 (D142) and Glu146 (E146) (DXXDXDXXXE), of glycosyl hydrolase family 18 in ScChi. Park et al. have reported that substitution at any of these sites (D or E) could result in the reduction or elimination of chitinase activity [27]. The obtained dendrogram in this study revealed that ScChi was closely related to O. sativa Class III chitinase (AAK26395) (Figure 3). Therefore, ScChi may be suggested for catalyzing the chitinase activity as an acid form of Class III (belonging to glycosyl hydrolase family 18). Using the Agrobacterium-mediated transformation in N. benthamiana leaves, the ScChi::GFP fusion protein was observed in the nucleus, cytoplasm and the plasma membrane (Figure 5). Results also revealed that ScChi, which contained no signal peptide and transmembrane helix domain, was unlikely to be a secretory protein. Previous studies even revealed extracellular localization of acidic Class III chitinases from chickpea (Cicer arietinum) and N. tabacum [3,28].
Chitinases are induced systematically on biotic, as well as abiotic treatments [1,29]. In the present study, the recombinant protein, ScChi, showed better growth in E. coli cells under all the four kinds of abiotic stress conditions, the NaCl, CuCl2, CdCl2 and ZnSO4 treatments. Previous studies have also demonstrated better growth of E. coli under stresses for recombinant protein production [30,31]. Chaurasia et al. found that phytochelatin synthase gene, PCS, was better expressed in E. coli by the addition of salt, pesticide, cadmium, copper, heat and UV [32]. The recombinant protein of sugarcane dirigent protein gene, ScDir, could tolerate high PEG and NaCl stresses [31]. Here, ScChi showed an upregulated transcript after challenge with smut pathogen. It also revealed that the expression level of ScChi in the incompatible interaction (Yacheng05–179 vs. smut pathogen) remained higher than that in a compatible interaction (Liucheng03–182 vs. smut pathogen) (Figure 9). Interestingly, the ScChi transcript was strongly induced by hormones stresses (SA, MeJA and ABA), especially by SA and MeJA, oxidative stress (H2O2), hyper-osmotic stresses (NaCl and PEG), metal stress (CuCl2), as well as low temperature (4 °C) stress (Figure 10). Li et al. found that the expression level of chitinase gene Mmchi1 in Mikania micrantha was significantly upregulated after two days post infection by Cuscuta campestris and also increased in response to the stresses of SA, ABA, ZnSO4 and wounding [20]. An acidic Class III chitinase, suggested to be a pathogenesis protein, was purified from N. tabacum leaves infected with Tobacco mosaic virus (TMV) [3]. Rahul et al. obtained a Class IV chitinase gene with a high level of expression observed in the incompatible interaction during challenge with C. falcatum causing red rot, although this gene was expressed in both resistant and susceptible sugarcane cultivars [18]. These phenomena suggest that the regulatory mechanism of chitinase genes might be similar in both eukaryotes and prokaryotes under biotic/abiotic stimuli. In the present study, enhanced chitinase activity and the expression profiles of the sugarcane chitinase gene, ScChi, during various environmental stresses suggest the positive responses to biotic and abiotic stresses.
It has been reported that many chitinase genes are developmentally regulated and may play a part in the specific physiological processes [33]. In our study, the transcript of ScChi accumulated in different sugarcane tissues (Figure 8). Constitutive expression of ScChi was detected, high in leaf and stem epidermal, moderate in root and stem pith and low in bud, indicating a specific role of ScChi in leaf and stem epidermal tissues. Infection with an avirulent pathogen strain or microbial triggering caused the rapid production of reactive oxygen intermediates (ROI) [34]. During the expression of plant disease resistance, H2O2 played an important role in the orchestration of a localized hypersensitive response [34]. In the present study, as indicated in Figure 11, pCAMBIA 1301-ScChi overexpressed in N. benthamiana leaves exhibited deep DAB staining color and high conductivity, indicating the high level of H2O2 accumulation. Additionally, the expression levels of ScChi and six immunity associated marker genes, NtHSR201, NtHSR515, NtNPR1, NtPR3, NtEFE26 and NtAccdeaminase, were upregulated (Figure 12). These results suggest a close relationship between the expression of ScChi and plant immunity, which was consistent with previous reports [34].
4. Experimental Section
4.1. Plant Materials and Treatments
Sugarcane cultivars of Yacheng05–179 (resistant) and Liucheng03–182 (susceptible) were cultivated in the Key Laboratory of Sugarcane Biology and Genetic Breeding, Ministry of Agriculture (Fuzhou, China). Smut whips were also collected from sugarcane cultivar “ROC”22, then sealed in a plastic bag and stored at 4 °C. The germination rate of the spores was checked before inoculation. A bath of two-bud sets of both sugarcane cultivars, Yacheng05–179 (smut resistant) and Liucheng03–182 (smut susceptible) (private bulletin), were inoculated with 0.5 μL suspension, which contained 5 × 106 spores/mL in 0.01% (v/v) Tween-20 [35]. As a control, a bath of two-bud sets were mock-inoculated with 0.01% (v/v) Tween-20 in sterile distilled water [36,37]. The treated samples were maintained at 28 °C in a photoperiod of 12 h light/12 h dark. Five buds were excised at 0, 24, 48, 72 and 120 h post-infection and stored at −80 °C, respectively.
To investigate gene expression in different tissues, six uniform 10-month-old plants of Yacheng05–179 were chosen for analysis. For each plant, the youngest fully expanded leaf (+1 leaf) with a visible dewlap (the collar between the leaf blade and sheath), the young root, the stem epidermal, stem pith and all the buds were collected and fixed in liquid nitrogen before RNA extraction.
For abiotic treatments, 4-month-old sugarcane tissue culture plantlets of Yacheng05–179 were subjected to the following eight different treatments in conical tubes at 28 °C with 16 h light/8 h dark. The plantlets were treated with 5 mM SA solution, 25 μM MeJA in 0.1% (v/v) ethanol and 0.05% (v/v) Tween-20, 100 μM ABA and 25% PEG, respectively. For the above four treatments, the whole plantlets were harvested at 0, 6, 12 and 24 h, respectively. In addition, the whole plantlets were separately treated with 250 mM NaCl, 100 μM CuCl2 and 4 °C low temperature for 0, 12, 24 and 48 h [37,38]. For H2O2 stress, leaves were sprayed with 10 mM H2O2, and the whole plantlets were sampled at 0, 6, 12 and 24 h [39]. All of the treatments were carried out in three replicates.
4.2. Chitinase Activity Assay
Chitinase activity was tested by measuring the reducing end group N-acetamino-glucose produced from colloidal chitin [25,40]. Yacheng05–179 and Liucheng03–182 inoculated with smut pathogen, as well as mock controls (0, 24, 48, 72 and 120 h) were sampled as above. Buds of 0.5 g were homogenized with 3.0 mL of ice-cold sodium acetate buffer (pH 5.0), then centrifuged at 10,000× g for 10 min at 4 °C. Additionally, the supernatant was recentrifuged again in the same condition. The final supernatant was used as a crude enzyme solution. The reaction mixture (1.0 mL enzyme solution (for blank control, incubated in boiling water for 10 min), 2.0 mL of 0.1 M sodium acetate buffer (pH5.0) and 0.1 g colloidal chitin) was incubated at 37 °C overnight. Termination of the reaction was done by heating in boiling water for 10 min. The reaction was then cooled to room temperature and centrifuged at 4000 r/min for 5 min. The supernatant of 2.0 mL was mixed with 2.0 mL dinitrosalicylic acid reagent and subjected to boiling water for 10 min. After that, 6.0 mL distilled water was added, and spectrophotometric measurement by the Lambda 35 UV WinLab software (Perkin Elmer, New York, NY, USA) at 540 nm was conducted. One unit of chitinase activity (U) was defined as the amount of enzyme that liberates 1.0 μmol N-acetamino-glucose per gram of plant material. The activity of chitinase was calculated by the activity level of inoculation minus the level of the mock at each corresponding time point.
4.3. RNA Extraction and cDNA Synthesis
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad CA, USA), according to the manufacturer’s recommendations. RNA quality was determined by electrophoresis and spectrophotometer (NanoVue plus, GE, Schenectady, New York, NY, USA). DNase I (Promega, Madison, WI, USA) treated total RNA was used for first-strand cDNA synthesis by the Prime-Script™ RT Reagent Kit (Takala, Dalian, China).
4.4. Cloning, Sequencing and Bioinformatic Analysis
Twenty-two sugarcane expressed sequence tags (ESTs) were obtained from the sugarcane sequence database (cultivated sugarcanes (taxid: 286192); wild sugarcane (taxid: 62335); sugarcane (taxid: 128810); sugarcane (taxid: 4547)) in GenBank, which share high homology with the Z. mays chitinase mRNA sequence (GenBank accession no. NM_001154758.1). A putative cDNA sequence of sugarcane chitinase gene was assembled by the CAP3 sequence assembly program [41]. PCR was performed with primers ScChi-cDNAF and ScChi-cDNAR (Table 1) derived from the assembled sequence, using the Yacheng05–179 post-48 h S. scitamineum inoculation cDNA sample as a template. The PCR products were gel-purified, cloned into the pMD18-T vector (TaKaLa, Dalian, China) and sequenced (Shenggong, Shanghai, China).
Table 1.
Primers used in this study.
| Primer | Sequence (5′-3′) | Strategy |
|---|---|---|
| ScChi-cDNAF | ATACAGGTCCATTTTGTCAGC | RT-PCR |
| ScChi-cDNAR | AAACCAAGCAAGCCCTAA | RT-PCR |
| ScChi-SublocF | TGCTCTAGAATGACGAACGGCTACC | Subcellular localization vector construction |
| ScChi-SublocR | GGACTAGTGTGGTTGGCAACGATCT | Subcellular localization vector construction |
| ScChi-32aF | CGGGATCCATGACGAACGGCTACCT | prokaryotic expression vector construction |
| ScChi-32aR | CCAAGCTTTCAGTGGTTGGCAACGA | prokaryotic expression vector construction |
| ScChi-QF | ACGGCTACGGCGACAACA | RT-qPCR |
| ScChi-QR | GTCCGCTGACCAGATGAAGAG | RT-qPCR |
| GAPDH-QF | CACGGCCACTGGAAGCA | RT-qPCR |
| GAPDH-QR | TCCTCAGGGTTCCTGATGCC | RT-qPCR |
| ScChi-1301F | TGCTCTAGAATGACGAACGGCTACCTG | Over expression vector construction |
| ScChi-1301R | CGGGATCCTCAGTGGTTGGCAA | Over expression vector construction |
| NtHSR201 F | CAGCAGTCCTTTGGCGTTGTC | RT-qPCR |
| NtHSR201 R | GCTCAGTTTAGCCGCAGTTGTG | RT-qPCR |
| NtHSR203 F | TGGCTCAACGATTACGCA | RT-qPCR |
| NtHSR203 R | GCACGAAACCTGGATGG | RT-qPCR |
| NtHSR51 F | TTGGGCAGAATAGATGGGTA | RT-qPCR |
| NtHSR51 R | TTTGGTGAAAGTCTTGGCTC | RT-qPCR |
| NtNPR1 F | GGCGAGGAGTCCGTTCTTTAA | RT-qPCR |
| NtNPR1 R | TCAACCAGGAATGCCACAGC | RT-qPCR |
| NtPR-1a/c F | AACCTTTGACCTGGGACGAC | RT-qPCR |
| NtPR-1a/c R | GCACATCCAACACGAACCGA | RT-qPCR |
| NtPR2 F | TGATGCCCTTTTGGATTCTATG | RT-qPCR |
| NtPR2 R | AGTTCCTGCCCCGCTTT | RT-qPCR |
| NtPR3 F | CAGGAGGGTATTGCTTTGTTAGG | RT-qPCR |
| NtPR3 R | CGTGGGAAGATGGCTTGTTGTC | RT-qPCR |
| NtEFE26 F | CGGACGCTGGTGGCATAAT | RT-qPCR |
| NtEFE26 R | CAACAAGAGCTGGTGCTGGATA | RT-qPCR |
| NtAccdeaminase F | TCTGAGGTTACTGATTTGGATTGG | RT-qPCR |
| NtAccdeaminase R | TGGACATGGTGGATAGTTGCT | RT-qPCR |
| NtEF1-α F | TGCTGCTGTAACAAGATGGATGC | RT-qPCR |
| NtEF1-α R | GAGATGGGGACAAAGGGGATT | RT-qPCR |
The open reading frame (ORF) was analyzed using the ORF Finder online program [42]; ProtParam [43], SMART [44], NetNGlyc 1.0 Server [45], SignalP 4.0 Server [46], TargetP 1.1 server [47] and TMHMM Server v. 2.0 [48] were used to predicted the ScChi sequence. The ClustalW method available in the MEGA5.05 software [49] was employed for multiple sequence alignment of amino acid sequences of ScChi with other published plant chitinases in GenBank [10]. The phylogenetic tree was constructed by the neighbor-joining (NJ) method (1,000 bootstrap replicates) with the MEGA 5.05 program [50].
4.5. Subcellular Localization Assay
ScChi was PCR amplified from pMD18-T-ScChi using primers ScChi-SublocF and ScChi-SublocR (Table 1). Then, the fragment was fused with the pCAMBIA 2300-GFP constructor between the Xba I and Spe I sites to generate the subcellular location vector of 35S::ScChi::GFP. The recombinant vector was verified by PCR, double digestion and sequencing. A positive clone was transferred into Agrobacterium tumefaciens strain EHA105. N. benthamiana leaves at eight-leaf stage were chosen for the Agrobacterium-mediated transformation assay [51,52]. Agrobacterium cells containing the 35S::GFP or 35S::ScChi::GFP vector were grown overnight at 28 °C in LB liquid medium (containing 50 μg/mL kanamycin and 35 μg/mL rifampicin). The Agrobacterium solution (OD600 = 0.8) consisting of 200 μM acetosyringone was infiltrated into N. benthamiana leaves and cultured for 2 days (16 h light/8 h darkness) at 24 °C. The subcellular localization of the fusion protein was analyzed by fluorescence microscopy (Axio Scope A1, Heidenheim, Germany).
4.6. Expression in E. coli Rosetta
The ORF of ScChi was amplified by PCR from pMD18-T-ScChi, using the primers, ScChi-32aF and ScChi-32aR (Table 1), followed by initial denaturation for 4 min at 94 °C; 35 cycles of 30 s at 94 °C, 30 s at 58 °C and 1.5 min at 72 °C; and extension for 10 min at 72 °C. The PCR fragments were digested by the EcoRI and XhoI enzymes and ligated into the previously digested pET 32a (+) vector. The ligation mixture was used to transform E. coli Rosetta competent cells to construct the recombinants. The construct was verified by PCR amplification, double digestion and sequencing, and the positive clone was named pET 32a-ScChi. The prokaryotic expression of the recombinant protein was induced by the addition of 1.0 mM isopropyl β-d-thiogalactoside (IPTG) for 1 h at 28 °C. LB medium with blank E. coli Rosetta (blank) or Rosetta + pET 32a (control) was also induced in IPTG for 1 h. All the above prokaryotic expression samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
For the study of the stress responses of E. coli cells expressing the ScChi gene under different abiotic conditions, a spot assay in treatments of NaCl, CuCl2, CdCl2 or ZnSO4 was carried out. When OD600 of the LB medium (with 80 μg/mL ampicillin and 170 μg/mL chloramphenicol) with E. coli cells plus pET 32a-ScChi or pET 32a reached 0.6, 1.0 mM IPTG was added, and the cells were grown for 12 h at 37 °C. The cultures were firstly diluted to 0.6 at OD600 and, then, diluted to two levels (10−3 and 10−4). Ten microliters from each dilution were spotted on LB plates with 80 μg/mL ampicillin and 170 μg/mL chloramphenicol or containing NaCl (250, 500 and 750 mM), CuCl2 (250, 500 and 750 μM), CdCl2 (250, 500 and 750 μM) or ZnSO4 (250, 500 and 750 μM). The tested plates were cultured at 37 °C overnight and photographed.
4.7. Expression Patterns of ScChi in Different Tissues and Stress Treatments
Expression profiles of ScChi during Yacheng05–179-smut interaction and Liucheng03–182-smut interaction at 0, 24, 48, 72 and 120 h, as well as mock plants were investigated. The ScChi transcript under S. scitamineum stress was calculated by the expression level of the inoculated sample minus the level of the mock at each corresponding time point. Expression patterns of ScChi in different tissues (leaf, root, bud, stem epidermal and stem pith) and in different stress treatments (SA, H2O2, MeJA, ABA, NaCl, CuCl2, PEG and 4 °C low temperature) were performed by RT-qPCR.
RT-qPCR was carried out with the SYBR Green Master (ROX) kit (Roche, Shanghai, China) on a 7500 RT-qPCR system (Applied Biosystems, Foster, CA, USA). The primer sequences for ScChi (ScChi-QF/ScChi-QR) and GAPDH (GAPDH-QF/GAPDH-QR) [53] were shown in Table 1. The RT-qPCR was performed in a final volume of 20 μL, which contained 0.5 μM of each primer, 10 μL FastStart Universal SYBR Green PCR Master (ROX) (Roche, Shanghai, China) and 2.0 μL template (10× diluted cDNA). PCR with distilled water as the template was performed as the blank control. The RT-qPCR cycling conditions were as follows: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s and 60 °C for 1 min; and the melting curve was established at the end of the amplification. Each RT-qPCR was conducted in triplicate. The 2−Δ ΔCt method was used to calculate the relative gene expression [54].
4.8. Transient Expression of ScChi in N. benthamiana
The primers of ScChi-1301F/ScChi-1301R in Table 1 were used to construct the overexpression vector pCAMBIA 1301-ScChi to analyze its defense response. Agrobacterium strain EHA105 carrying the recombinant vector was grown overnight in LB liquid medium containing 50 μg/mL kanamycin and 35 μg/mL rifampicin at 28 °C. Culture cells were collected and resuspended in MS liquid medium containing 200 μM acetosyringone at OD600 = 0.8. Then, cells were infiltrated into eight-leaf stage-old N. benthamiana leaves [51,52]. For comparison, the Agrobacterium strain containing the pCAMBIA 1301 vector alone was also transiently expressed in N. benthamiana leaves. One of the materials were incubated at 24 °C for 24 h (16 h light/8 h darkness) and used for the RT-qPCR analysis of the expression of ScChi (ScChi-QF/ScChi-QR) and several immunity associated marker genes, including the hypersensitive response (HR) marker genes, NtHSR201, NtHSR203 and NtHSR515, the SA-related gene, NtNPR1, the JA-associated genes, NtPR-1a/c, NtPR2 and NtPR3, and the ethylene synthesis-dependent genes, NtEFE26 and NtAccdeaminase (Table 1), in N. benthamiana. NtEF1-α (Table 1) was used to normalize the transcript levels. Other materials were incubated at 24 °C for 2 days and applied to the following tests of the histochemical assay and ion conductivity determination. All of the treatments were carried out in three replicates.
4.9. Histochemical Assay
DAB (3,3′-diaminobenzidinesolution) was used to stain H2O2 produced in N. benthamiana leaves. The leaves were incubated in 1.0 mg/mL DAB-HCl solution in the dark overnight and destained by boiling in 95% ethanol for 5 min. The bronzing color of the leaves for H2O2 detection was photographed [52,55].
4.10. Measurement of Ion Conductivity
Ion conductivity determination was performed as in the research of Hwang and Hwang [51]. Six leaf discs (11 mm in diameter) per leaf were washed and incubated in 20 mL of distilled water and then shaken gently for 60 min at room temperature. The ion conductivity was recorded using a conductivity meter (SevenEasy, METTLER TOLEDO, Zurich, Switzerland).
5. Conclusions
In summary, this is a report of the activity and cloning of sugarcane chitinases involved in the defense response to S. scitamineum. The pathogenesis-related induction of a 31.37 kDa chitinase, ScChi, was clearly identified at the transcript and protein levels. ScChi showed a positive response to biotic and abiotic stresses. Although the defensive role of chitinase in sugarcane against fungi is far from being fully understood, our study is indicative of the involvement of chitinase in the resistance response of sugarcane to biotic and various abiotic stimuli. Whether chitinase induction plays a direct role in the resistance of sugarcane to S. scitamineum needs further study. The results obtained in this study can be used for smut management in sugarcane and the development of marker-assisted breeding if the positive response to smut challenge can be found in sugarcane from different genetic backgrounds.
Acknowledgments
We especially thank Andrew C. Allan in The New Zealand Institute for Plant and Food Research Ltd, (Plant and Food Research), Mt Albert Research Centre, Private Bag 92169, Auckland, New Zealand, for his critical revision and valuable comments on this manuscript. This work was funded by the earmarked fund for Modern Agriculture Technology of China (CARS-20), the National Natural Science Foundation of China (31340060), the 948 Program on the Introduction of International Advanced Agricultural Science and Techniques of the Department of Agriculture (2014-S18), the Research Funds for Distinguished Young Scientists in the Fujian Provincial Department of Education (K80MKT04A) and the Research Funds for Distinguished Young Scientists in Fujian Agriculture and Forestry University (xjq201202).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Author Contributions
Conceived and designed the experiments: Yachun Su, Liping Xu and Youxiong Que. Performed the experiments: Yachun Su, Zhiwei Fu, Yuting Yang, Jinlong Guo, and Shanshan Wang. Analyzed the data: Yachun Su, Liping Xu and Youxiong Que. Contributed reagents/materials/analysis tools: Yachun Su, Liping Xu and Youxiong Que. Wrote the paper: Yachun Su, Liping Xu and Youxiong Que. Revised and approved the final version of the paper: Liping Xu and Youxiong Que.
References
- 1.Walters D., Walsh D., Newton A., Lyon G. Induced resistance for plant disease control: Maximizing the efficacy of resistance elicitors. Phytopathology. 2005;95:1368–1373. doi: 10.1094/PHYTO-95-1368. [DOI] [PubMed] [Google Scholar]
- 2.Neuhaus J.M., Fritig B., Linthorst H.J.M., Meins F., Mikkelsen J.D., Ryals J. A revised nomenclature for chitinase genes. Plant Mol. Biol. Rep. 1996;14:102–104. [Google Scholar]
- 3.Lawton K., Ward E., Payne G., Moyer M., Ryals J. Acidic and basic class III chitinase mRNA accumulation in response to TMV infection of tobacco. Plant Mol. Biol. 1992;19:735–743. doi: 10.1007/BF00027070. [DOI] [PubMed] [Google Scholar]
- 4.Schlumbaum A., Mauch F., Vogeli U., Boller T. Plant chitinases are potent inhibitors of fungal growth. Nature. 1986;324:365–367. [Google Scholar]
- 5.Mauch F., Mauch-Mani B., Boller T. Antifungal hydrolases in pea tissue II. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol. 1988;88:936–942. doi: 10.1104/pp.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maximova S.N., Marelli J.P., Young A., Pishak S., Verica J.A., Guiltinan M.J. Over-expression of a cacao class I chitinase gene in Theobroma cacao L. enhances resistance against the pathogen Colletotrichum gloeosporioides. Planta. 2006;224:740–749. doi: 10.1007/s00425-005-0188-6. [DOI] [PubMed] [Google Scholar]
- 7.Xiao Y.H., Li X.B., Yang X.Y., Luo M., Hou L., Guo S.H., Luo X.Y., Pei Y. Cloning and characterization of a balsam pear class I chitinase gene (Mcchil1) and its ectopic expression enhances fungal resistance in transgenic plants. Biosci. Biotechnol. Biochem. 2007;71:1211–1219. doi: 10.1271/bbb.60658. [DOI] [PubMed] [Google Scholar]
- 8.Liu J.J., Ekramoddoullah A.K., Zamani A. A class IV chitinase is up-regulated by fungal infection and abiotic stresses and associated with slow-canker-growth resistance to Cronartium ribicola in western white pine (Pinus monticola) Phytopathology. 2005;95:284–291. doi: 10.1094/PHYTO-95-0284. [DOI] [PubMed] [Google Scholar]
- 9.Meins F., Fritig B., Linthorst H.J., Mikkelsen J.D., Neuhaus J.M., Ryals J. Plant chitinase genes. Plant Mol. Biol. Rep. 1994;12:S22–S28. [Google Scholar]
- 10.Singh A., Isaac-Kirubakaran S., Sakthivel N. Heterologous expression of new antifungal chitinase from wheat. Protein Expres. Purif. 2007;56:100–109. doi: 10.1016/j.pep.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Hoy J.W., Hollier C.A., Fontenot D.B., Grelen L.B. Incidence of sugarcane smut in Louisiana and its effects on yield. Plant Dis. 1986;70:59–60. [Google Scholar]
- 12.Padmanaban P., Alexander K.C., Shanmugan N. Effect of smut on growth and yield parameters of sugarcane. Indian Phytopath. 1998;41:367–369. [Google Scholar]
- 13.Que Y.X., Xu L.P., Lin J.W., Chen R.K., Grisham M.P. Molecular variation of Sporisorium scitamineum in Mainland China revealed by RAPD and SRAP markers. Plant Dis. 2012;96:1519–1525. doi: 10.1094/PDIS-08-11-0663-RE. [DOI] [PubMed] [Google Scholar]
- 14.Xu L.P., Chen R.K., Chen P.H. Analysis on infection index of smut caused by Ustilago scitaminea in sugarcane segregated population. Chin. J. Trop Crop. 2004;25:33–36. [Google Scholar]
- 15.Does M.P., Houterman P.M., Dekker H.L., Cornelissen B.J. Processing, targeting, and antifungal activity of stinging nettle agglutinin in transgenic tobacco. Plant physiol. 1999;120:421–432. doi: 10.1104/pp.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovacs G., Sagi L., Jacon G., Arinaitwe G., Busogoro J.P., Thiry E., Strosse H., Swennen R., Remy S. Expression of a rice chitinase gene in transgenic banana (“Gros Michel” AAA genome group) confers resistance to black leaf streak disease. Transgen. Res. 2013;22:117–130. doi: 10.1007/s11248-012-9631-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirubakaran S.I., Sakthivel N. Cloning and overexpression of antifungal barley chitinase gene in Escherichia coli. Protein Expres. Purif. 2007;52:159–166. doi: 10.1016/j.pep.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Rahul P.R., Kumar V.G., Sathyabhama M., Viswanathan R., Sundar A.R., Malathi P. Characterization and 3D structure prediction of chitinase induced in sugarcane during pathogenesis of Colletotrichum falcatum. J. Plant Biochem. Biot. 2013:1–8. [Google Scholar]
- 19.Lin S., Zhou Y., Chen G., Zhang Y., Zhang Y., Ning W., Pan D. Molecular responses to the fungal pathogen Gibberella fujikuroi in the leaves of chewing cane (Saccharum officinarum L.) Sugar Tech. 2010;12:36–46. [Google Scholar]
- 20.Li D.M., Staehelin C., Wang W.T., Peng S.L. Molecular cloning and characterization of a chitinase-homologous gene from Mikania micrantha infected by Cuscuta campestris. Plant Mol. Biol. Rep. 2010;28:90–101. [Google Scholar]
- 21.Meuriot F., Noquet C., Avice J.C., Volenec J.J., Cunningham S.M., Sors T.G., Caillot S., Ourry A. Methyl jasmonate alters N partitioning, N reserves accumulation and induces gene expression of a 32-kDa vegetative storage protein that possesses chitinase activity in Medicago sativa taproots. Physiol. Plantarum. 2004;120:113–123. doi: 10.1111/j.0031-9317.2004.0210.x. [DOI] [PubMed] [Google Scholar]
- 22.Takken F.L., Joosten M.H. Plant resistance genes: Their structure, function and evolution. Eur. J. Plant Pathol. 2000;106:699–713. [Google Scholar]
- 23.Van-Loon L.C. Pathogenesis-related proteins. Plant Mol. Biol. 1985;4:111–116. doi: 10.1007/BF02418757. [DOI] [PubMed] [Google Scholar]
- 24.Brogue K., Chet I., Holliday M., Cressman R., Biddle P., Knowlton S., Mauvais C.J., Broglie R. Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science. 1991;254:1194–1197. doi: 10.1126/science.254.5035.1194. [DOI] [PubMed] [Google Scholar]
- 25.Krishnaveni S., Liang G.H., Muthukrishnan S., Manickam A. Purification and partial characterization of chitinases from sorghum seeds. Plant Sci. 1999;144:1–7. [Google Scholar]
- 26.Viswanathan R., Malathi P., Sundar A.R., Aarthi S., Premkumari S.M., Padmanaban P. Differential induction of chitinases and thaumatin-like proteins in sugarcane in response to infection by Colletotrichum falcatum causing red rot disease. J. Plant Dis. Protect. 2005;112:417–425. [Google Scholar]
- 27.Park S.K., Kim C.W., Kim H., Jung J.S., Harman G.E. Cloning and high-level production of a chitinase from Chromobacterium sp. and the role of conserved or nonconserved residues on its catalytic activity. Appl. Microbiol. Biot. 2007;74:791–804. doi: 10.1007/s00253-006-0614-0. [DOI] [PubMed] [Google Scholar]
- 28.Mackenbrock U., Vogelsang R., Barz W. Isoflavone and pterocarpan malonylglucosides and β-1,3-glucan-and chitin-hydrolases are vacuolar constituents in chickpea (Cicer arietinum L.) Z. Naturforsch. C. 1992;47:815–822. [Google Scholar]
- 29.Viswanathan R., Samiyappan R. Antifungal activity of chitinases produced by some fluorescent pseudomonads against Colletotrichum falcatum Went causing red rot disease in sugarcane. Microbiol. Res. 2001;155:309–314. doi: 10.1016/S0944-5013(01)80009-4. [DOI] [PubMed] [Google Scholar]
- 30.Gupta K., Agarwal P.K., Reddy M.K., Jha B. SbDREB2A, an A-2 type DREB transcription factor from extreme halophyte Salicornia brachiata confers abiotic stress tolerance in Escherichia coli. Plant Cell Rep. 2010;29:1131–1137. doi: 10.1007/s00299-010-0896-7. [DOI] [PubMed] [Google Scholar]
- 31.Guo J.L., Xu L.P., Fang J.P., Su Y.C., Fu H.Y., Que Y.X., Xu L.P. A novel dirigent protein gene with highly stem-specific expression from sugarcane, response to drought, salt and oxidative stresses. Plant Cell Rep. 2012;31:1801–1812. doi: 10.1007/s00299-012-1293-1. [DOI] [PubMed] [Google Scholar]
- 32.Chaurasia N., Mishra Y., Ai L.C. Cloning expression and analysis of phytochelatin synthase (pcs) gene from Anabaena sp. PCC 7120 offering multiple stress tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 2008;376:225–230. doi: 10.1016/j.bbrc.2008.08.129. [DOI] [PubMed] [Google Scholar]
- 33.Porat R., Vinokur V., Holland D., Gregory-McCollum T., Droby S. Isolation of a citrus chitinase cDNA and characterization of its expression in response to elicitation of fruit pathogen resistance. J. Plant Physiol. 2001;158:1585–1590. [Google Scholar]
- 34.Levine A., Tenhaken R., Dixon R., Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 35.Que Y.X., Lin J.W., Song X.X., Xu L.P., Chen R.K. Differential gene expression in sugarcane in response to challenge by fungal pathogen Ustilago scitaminea revealed by cDNA-AFLP. J. Biomed. Biotechnol. 2011;2011 doi: 10.1155/2011/160934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moosawi-Jorf S.A., Mahin B.I. In vitro detection of yeast-like and mycelial colonies of Ustilago scitaminea in tissue-cultured plantlets of sugarcane using polymerase chain reaction. J. Appl. Sci. 2007;7:3768–3773. [Google Scholar]
- 37.Su Y.C., Xu L.P., Xue B.T., Wu Q.B., Guo J.L., Wu L.G., Que Y.X. Molecular cloning and characterization of two pathogenesis-related β-1,3-glucanase genes ScGluA1 and ScGluD1 from sugarcane infected by Sporisorium scitamineum. Plant Cell Rep. 2013;32:1503–1519. doi: 10.1007/s00299-013-1463-9. [DOI] [PubMed] [Google Scholar]
- 38.Guo J.L., Xu L.P., Su Y.C., Wang H.B., Gao S.W., Xu J.S., Que Y.X. ScMT2-1-3, a metallothionein gene of sugarcane, plays an important role in the regulation of heavy metal tolerance/accumulation. BioMed Res. 2013;2013 doi: 10.1155/2013/904769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Que Y.X., Yang Z.X., Xu L.P., Chen R.K. Isolation and identification of differentially expressed genes in sugarcane infected by Ustilago scitaminea. Acta Agron. Sin. 2009;35:452–458. [Google Scholar]
- 40.Li S.G., Gu J.G., Jiang R.B., Niu Y.C. The research on biocontrol Trichoderma spp. strains producing chitinase. Chin. Biotech. Bull. 2009;30:135–138. [Google Scholar]
- 41.CAP3 sequence assembly program. [(accessed on 1 January 2014)]. Available online: http://pbil.univ-lyon1.fr/cap3.php.
- 42.ORF Finder online program. [(accessed on 2 December 2013)]. Available online: http://www.ncbi.nlm.nih.gov/gorf/gorf.html.
- 43.ProtParam. [(accessed on 1 January 2005)]. Available online: http://web.expasy.org/protparam/
- 44.SMART. [(accessed on 10 October 2011)]. Available online: http://smart.embl-heidelberg.de/
- 45.NetNGlyc 1.0 Server. [(accessed on 5 June 2013)]. Available online: http://www.cbs.dtu.dk/services/NetNGlyc/
- 46.SignalP 4.0 Server. [(accessed on 4 June 2013)]. Available online: http://www.cbs.dtu.dk/services/SignalP/
- 47.TargetP 1.1 server. [(accessed on 4 October 2013)]. Available online: http://www.cbs.dtu.dk/services/TargetP/
- 48.TMHMM Server v. 2.0. [(accessed on 12 June 2013)]. Available online: http://www.cbs.dtu.dk/services/TMHMM-2.0/
- 49.MEGA5.05 software. [(accessed on 4 May 2011)]. Available online: http://www.megasoftware.net/
- 50.Kumar S., Tamura K., Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 51.Hwang I.S., Hwang B.K. Requirement of the cytosolic interaction between pathogenesis-related protein10 and leucine-rich repeat protein1 for cell death and defense signaling in pepper. Plant Cell. 2012;24:1675–1690. doi: 10.1105/tpc.112.095869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su Y.C., Guo J.L., Ling H., Chen S.S., Wang S.S., Xu L.P., Allan A.C., Que Y.X. Isolation of a novel peroxisomal catalase gene from sugarcane, which is responsive to biotic and abiotic stresses. PLos One. 2014;9:e84426. doi: 10.1371/journal.pone.0084426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Que Y.X., Xu L.P., Xu J.S., Zhang J.S., Zhang M.Q., Chen R.K. Selection of control genes in real-time qPCR analysis of gene expression in sugarcane. Chin. J. Trop Crop. 2009;30:274–278. [Google Scholar]
- 54.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−Δ ΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 55.Huckelhoven R., Fodor J., Trujillo M., Kogel K.H. Barley Mla and Rar mutants compromised in the hypersensitive cell death response against Blumeria graminis f. sp. hordei are modified in their ability to accumulate reactive oxygen intermediates at sites of fungal invasion. Planta. 2000;212:16–24. doi: 10.1007/s004250000385. [DOI] [PubMed] [Google Scholar]