Abstract
Saframycin A (SafA) is a member of a class of natural products with potent antiproliferative effects in leukemia- and tumor-derived cells. This activity is frequently conjectured to derive from the ability of saframycins to covalently modify duplex DNA. We used a DNA-linked affinity purification technique to identify GAPDH as a protein target of DNA–small molecule adducts of several members of the saframycin class. Nuclear translocation of GAPDH occurs upon treatment of cancer cells with saframycins, and depletion of cellular GAPDH levels by small interfering RNA transfection confers drug resistance. Roeder and coworkers have recently suggested that GAPDH is a key transcriptional coactivator necessary for entry into S phase. Our data suggest that GAPDH is also capable of forming a ternary complex with saframycin-related compounds and DNA that induces a toxic response in cells. These studies implicate a previously unknown molecular mechanism of antiproliferative activity and, given that one member of the saframycin class has shown efficacy in cancer treatment, suggest that GAPDH may be a potential target for chemotherapeutic intervention.
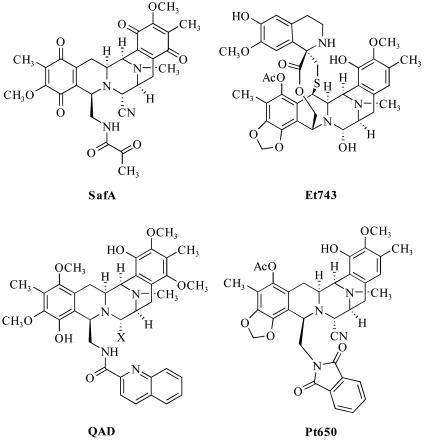
Many small-molecule agents for cancer therapy are known to covalently modify duplex DNA, but the pathway(s) that may link DNA alkylation with the inhibition of cancer cell growth are frequently not well understood. Examples are the structurally related natural products saframycin A (SafA) (Fig. 1), a bacterial fermentation product active against various tumor cell lines (1), and ecteinascidin (Et743) (2), a marine alkaloid currently in advanced clinical trials for the treatment of sarcomas of soft tissue and bone (3). Both natural products have been shown to bind reversibly in the minor groove of duplex DNA by covalent bond formation between a drug-derived iminium ion intermediate and the exocyclic amino group of guanosine residues (4–10). The bis-quinone alkaloid SafA requires prior reductive activation for iminium ion formation and DNA alkylation to occur (4), whereas the nonquinoid molecule Et743 does not (10). The development of efficient and versatile synthetic routes to each compound series has led to the discovery of synthetic molecules with enhanced antiproliferative activities and/or improved properties, such as the Et743 analog phthalascidin (Pt650) (11), and the nonquinoid, quinaldic acid derivatives (QAD) of SafA (Fig. 1) (12). Chemical and biological data suggest that the saframycin and Et743 compound series are related, but the basis for their activity is not known (13, 14). In one study, growth of a colon cancer cell line through several generations of exposure to Et743 led to the isolation of a strain with reduced sensitivity to drug (IC50 = 3.7 nM, 9-fold resistance); this resistance was traced to a mutation in a transcription-coupled nucleotide excision repair protein, leading to the proposal that this protein provides one determinant of sensitivity to Et743 (15). Other studies have shown that Et743 may activate more than one signaling pathway in cancer cells; these include a transcription-coupled process leading to cell-cycle arrest, and another transcription-independent pathway leading to apoptosis (16). In studies of the Et743 analog Pt650, drug exposure was shown to produce DNA–protein crosslinks, but the identities of bound proteins were not established (17). Here, we show that GAPDH is a common protein target of SafA–, QAD–, and Pt650–DNA adducts by using a DNA-linked affinity chromatography technique. We also provide several independent lines of experimental evidence to support the hypothesis that the interaction of these molecules with duplex DNA and GAPDH is an integral and common component of their antiproliferative effects.
Fig. 1.
Molecular structures of SafA, Et743, the quinaldic acid derivatives (QAD) of SafA (QADCN, X = CN; QADOH, X = OH), and phthalascidin (Pt650), an analog of Et743.
Materials and Methods
Cells and Reagents. HeLa-S3 and A549 cells (American Type Culture Collection) were cultured as monolayers in DMEM supplemented with 10% FBS at 37°C in a 5% CO2 atmosphere. Resins used were Toyopearl Oligo-Affinity Resin (Glen Research, Sterling, VA), and Toyopearl AF-carboxy-650M (Tosoh, Tokyo). Primary antibodies used were mouse monoclonal antibody to GAPDH (Research Diagnostics), goat polyclonal antibody to high-mobility group protein (HMG1), mouse monoclonal antibody to 14-3-3β, and mouse monoclonal antibody to Sp1 (Santa Cruz Biotechnology). Mitomycin C (MMC), cisplatin, and transplatin were purchased from Sigma. Pt650 was a gift from E. J. Corey (Harvard University). SafA, QADOH, and QADCN were synthesized by published procedures (12, 18). A GAPDH small interfering RNA (siRNA) transfection kit was purchased from Ambion (Austin, TX). A CelLytic nuclear extraction kit was purchased from Sigma. A CellTiter96AQ nonradioactive cell proliferation assay kit was purchased from Promega. Frozen bovine brain was purchased from Pel Freez Biologicals (Rogers, AR).
Construction of DNA–Small Molecule Affinity Reagents. The single-stranded (ss) 26-mer oligonucleotide 5′-CCTTGGCCCGAGCCCGGTTCCTATT-3′ was synthesized on ToyoPearl Oligo-Affinity Resin or on ToyoPearl Oligo-Affinity Resin functionalized with a photocleaveable spacer phosphoramidite. Hybridization to form a resin-bound duplex oligonucleotide was achieved by annealing the resin-based 26-mer ss oligonucleotide with the complementary 21-mer oligonucleotide 5′-GGAACCGGGCTCGGGCCAAGG-3′. Affinity resin displaying 1 nmol of double-stranded (ds) oligonucleotide was washed twice with buffer A (40 mM NaH2PO4, pH 5.8/5 mM MgCl2/0.1 mM Na2EDTA) and to the washed resin was added 10 nmol of drug (QAD, Pt650, and SafA) in buffer A. In the case of SafA, 1 μmol of DTT (1 M aqueous solution) was also added to the mixture of SafA and affinity resin to promote reductive activation. Reaction suspensions were mixed on an Eppendorf Mixer 5432 at 37°C for 15 h to complete formation of the DNA–small molecule affinity reagents. Literature procedures for the formation of DNA–small molecule complexes were followed to prepare MMC (19), cisplatin (20), and transplatin (20) affinity reagents with the modification that these reactions were mixed on an Eppendorf Mixer 5432 at 37°C for 15 h.
Affinity Chromatography Experiments. Bovine brain tissue (40 g) was homogenized in 40 ml of buffer B (20 mM Tris·HCl, pH 7.4/15 mM 2-mercaptoethanol/0.3% Triton X-100/0.015 mM pepstatin A/0.025 mM leupeptin/1 mM PMSF) at 4°C. The homogenate was centrifuged (100,000 × g) for 30 min at 4°C, and the supernatant obtained was clarified by filtration through a Bio-spin column (Bio-Rad). Lysate from an A549 tumor-cell line was prepared by incubating A549 cells (250-μl packed cell volume) with 1 ml of buffer C (50 mM Tris·HCl, pH 7.4/1% NP-40/0.25% sodium deoxycholate/150 mM NaCl/1 mM Na3VO3/1 mM NaF/1 mM EGTA) for 1 h at 4°C on an end-over-end rotator. To reduce nonspecific binding of proteins, protein pools from clarified bovine homogenate or A549 cell lysate were preincubated for 1 h at 4°C with a guard column prepared by conjugation of Toyopearl AF-carboxy-650M resin with ethanolamine in the presence of 1-ethyl-3-(3′-dimethyl-aminopropyl)carbodiimide hydrochloride. After this preincubation step, treated bovine brain homogenate or A549 cell lysate was filtered through a Bio-spin column to further reduce the level of nonspecific protein binding. Bovine brain lysate or A549 cell lysate pretreated in this fashion was then incubated with poly(dI-dC) (a nonspecific competitor) and a DNA–small molecule affinity reagent at 4°C for 15 h on an end-over-end rotator. After washing the resin three times with buffer D (20 mM Tris·HCl, pH 7.4/15 mM 2-mercaptoethanol/0.3% Triton X-100/0.5 M NaCl), bound proteins were released from the affinity resin by heat denaturation at 95°C for 5 min, the denatured proteins were resolved by SDS/PAGE (4–20% gradient gels), and protein bands were visualized by silver staining or identified by protein mass spectrometry sequencing and Western blotting.
Southwestern Blotting Experiments. Southwestern blotting experiments were performed according to a published procedure (21) by using purified human GAPDH or human HMG1 in the assay. A 32P-labeled 21-mer dsDNA corresponding in sequence to the duplex region of DNA–small molecule affinity reagents was used as a probe (specific activity, 0.1 μCi; 1 Ci = 37 GBq), alone or alkylated with one of the different small molecules of this study.
Nuclear Translocation Studies. Cytoplasmic and nuclear protein fractions (CelLytic nuclear extraction kit) derived from HeLa-S3 cells treated (0, 1, 2, or 3 days) with QADOH (17.5 nM) were separately resolved by SDS/PAGE (equivalent amounts of total protein from each fraction were loaded onto the gel), and the resolved protein bands were Western blotted for quantitative GAPDH detection. To ensure the integrity of the fractionation procedure, resolved protein bands from cytoplasmic and nuclear fractions were separately Western blotted using mouse monoclonal antibody to 14-3-3β (cytosolic protein) and mouse monoclonal antibody to Sp1 (nuclear protein).
Confocal microscopy experiments were performed by using control or drug-treated (17.5 nM of QADOH for 2 days) populations of HeLa-S3 cells. The cells were fixed, permeabilized, and immunolabeled according to a published procedure (22). The immunolabeled cells were visualized by using a Zeiss LSM510 confocal microscope.
GAPDH siRNA Transfection Studies. A549 cells were plated on a 24-well culture plate such that each well contained ≈50,000 cells in 1 ml of DMEM containing 10% FBS. Cell populations were transfected with control siRNA or GAPDH siRNA according to the manufacturer's recommended procedure. The cells were harvested and lysed after siRNA treatment for the indicated amount of time. Equal amounts of total protein from each supernatant solution were resolved by SDS/PAGE; resolved GAPDH was then quantified by Western blotting analysis.
Effects of QAD or Cisplatin Treatment on the Growth of A549 Cells Transfected with GAPDH siRNA. A549 cells transfected with GAPDH siRNA or a control siRNA (nontransfected cells were also examined as a further control) were cultured 6 h after transfection in 1 ml of DMEM containing 10% FBS and a defined concentration of QADCN or cisplatin in an atmosphere of 95% air/5% CO2 for 72 h at 37°C. Viable cells in each well were quantified by using the CellTiter96AQ nonradioactive cell proliferation assay kit following the manufacturer's recommended procedure. Percent cell survival versus log drug concentration was plotted in each case; GI50 values were calculated by interpolation using the cubic spline method.
Results and Discussion
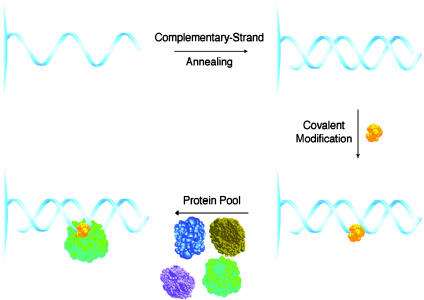
Development of an Affinity Technique for the Identification of Proteins Recognizing DNA–Small Molecule Adducts. To identify proteins that bound DNA–small molecule alkylation products, we developed a technique for the preparation of a series of DNA-linked affinity reagents that was initiated by the automated synthesis of a 26-mer ss oligonucleotide from a commercial solid support bearing chemically or photochemically cleavable linkers, followed by 21-mer complementary-strand hybridization (Scheme 1). The DNA duplex that was produced contained multiple known binding sites for the saframycin class of antiproliferative agents as well as cisplatin, transplatin, and MMC, used as controls, and was linked to the resin at the cleavable site by way of an intervening, unpaired sequence TATTT. Conditions for optimal binding of each drug to the duplex were then developed, guided by gel-shift characterization data for each DNA–small molecule complex after photochemical cleavage from the resin. Affinity columns prepared from the quinone-containing molecules SafA and MMC used DTT for reductive alkylation, whereas no such activation was required for QAD, Pt650, or platinum-based alkylating agents. Exposure of the duplex DNA–small molecule affinity columns to protein pools derived initially from bovine-brain lysate and, later, from human A549-cell lysate, followed by washing of nonbound proteins from the column, release of bound proteins by heat denaturation, and separation of the latter by SDS/PAGE, allowed for the identification of proteins that recognized each DNA–drug conjugate specifically. Preincubation of the protein pools with two guard columns in series, incorporation of poly(dI-dC) during protein binding to the affinity column, and the use of optimal salt and protein concentrations essentially abolished any nonspecific protein binding (identified by comparison with appropriate controls employing DNA duplexes that had not been exposed to drug). Specific protein-binding interactions were confirmed by competitive binding studies with solutions of free duplex DNA alkylated by the appropriate drug. The identities of bound proteins were established by mass spectrometric protein sequencing of bands isolated by SDS/PAGE, and were confirmed by Western blotting experiments.
Scheme 1.
DNA-linked affinity reagents for the isolation of proteins that bind small molecule-DNA adducts. The resin-bound 26-mer ss-oligonucleotide (5′-CCTTGGCCCGAGCCCGGTTCCTATTT-3′) was prepared by automated synthesis on Toyopearl Oligo-Affinity Resin. Complementary-strand annealing with the 21-mer ss-oligonucleotide 5′-GGAACCGGGCTCGGGCCAAGG-3′ followed by covalent modification of the resulting DNA duplex with DNA-reactive small molecules (represented in yellow) provided affinity reagents used to identify specific binding proteins within a pool derived from tissue lysate.
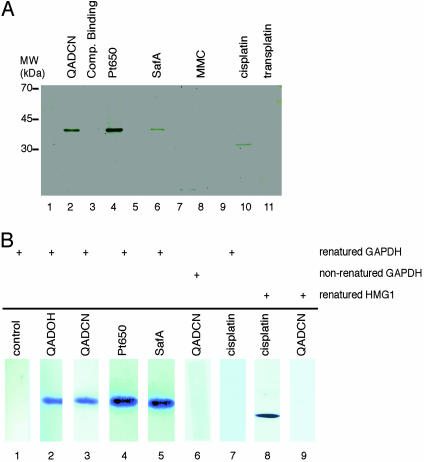
Results obtained by using affinity reagents prepared from cisplatin and transplatin and a protein pool derived from bovine brain established the potential of DNA-linked, small-molecule modified affinity reagents to identify specific binding proteins, in that the cisplatin-derived reagent provided a single major protein band, shown to be the 33-kDa (bovine) high-mobility group protein HMG1, whereas no such binding of HMG1 (nor any other protein) was observed to occur with the transplatin-derived reagent (Fig. 2A). HMG1 binding was not observed to occur with affinity reagents prepared from SafA, QAD, Pt650, nor MMC. HMG1 had previously been determined to be a potential protein target of cisplatin–DNA adducts by sequence analysis of cDNA isolated in an expression-library screening (23). Cisplatin–DNA adducts (but not transplatin–DNA adducts) were subsequently shown to bind HMG1 specifically (24); this binding has been proposed to form the chemical basis of cisplatin antitumor activity and, similarly, the lack of HMG1 binding to account for the poor clinical efficacy of transplatin in cancer therapy.
Fig. 2.
Identification of proteins recognizing duplex DNA-small molecule adducts by a DNA-linked affinity chromatography technique and confirmation of a direct binding interaction of the protein GAPDH by Southwestern blot analysis. (A) SDS/PAGE of proteins bound in affinity chromatography experiments using a protein pool derived from bovine-brain lysate. Lane 1, dsDNA resin with no drug modification: control matrix to lanes 2–4; lane 2, QADCN-alkylated dsDNA resin plus 40 equivalents (equiv) poly(dI-dC); lane 3, QADCN-alkylated dsDNA resin plus 40 equiv dsDNA-QADCN complex; lane 4, Pt650-alkylated dsDNA resin plus 40 equiv poly(dI-dC); lane 5, dsDNA resin with no drug modification, control matrix to lane 6; lane 6, SafA-alkylated dsDNA resin plus 40 equiv poly(dI-dC); lane 7, dsDNA resin with no drug modification, control matrix to lane 8; lane 8, MMC-alkylated dsDNA resin plus 40 equiv poly(dI-dC); lane 9, dsDNA resin with no drug modification, control matrix to lanes 10 and 11; lane 10, cisplatin-alkylated dsDNA resin plus 40 equiv poly(dI-dC); lane 11, transplatin-alkylated dsDNA resin plus 40 equiv poly(dI-dC). (B) Results of Southwestern blotting experiments. Purified human GAPDH or HMG1 was resolved by SDS/PAGE, electroblotted onto nitrocellulose, and then renatured (except for lane 6) on the membrane. The renatured blots were probed with a 32P-labeled 21-mer dsDNA (sequence: Scheme 1) unmodified (lane 1) or alkylated with the various compounds of study (as indicated, lanes 2–9).
Identification of GAPDH as a Protein Target of DNA Adducts of the Saframycin Antiproliferative Agents. Although affinity columns derived from SafA, QAD, and Pt650 did not bind HMG1, they were found to bind a different, 38-kDa bovine protein (Fig. 2 A) whose sequence was determined by mass spectrometry; comparison of the sequence with a protein database showed the 38-kDa binding protein to be GAPDH. This identification was confirmed by Western blotting. Binding of GAPDH was specific (binding did not occur in the absence of small-molecule modification of the DNA affinity reagent) and was subject to competitive displacement with the appropriate free dsDNA–drug complexes. GAPDH of human origin was also shown to bind selectively to affinity columns derived from QAD and Pt650 (but not cisplatin, transplatin, or mitomycin C) by using a protein pool derived from the lysate of a human cancer cell line (A549) for binding studies.
Confirmation That Direct Binding Occurs Between GAPDH and Saframycin–DNA Adducts by Southwestern Blotting Experiments. To establish that the binding of GAPDH to SafA–, QAD–, or Pt650–DNA affinity columns was the result of a direct interaction, and had not been mediated by another, relatively less abundant protein that was not detected by SDS/PAGE, we conducted Southwestern blotting experiments using human erythrocyte GAPDH (tetrameric form; molecular mass, 152 kDa) that was submitted to (denaturing) SDS/PAGE, blotted onto a nitrocellulose membrane, and renatured. Membranes were probed with the alkylation products of QAD, SafA, Pt650, or cisplatin and a 32P-labeled 21-mer dsDNA corresponding in sequence to the duplex region of our affinity columns, and were subsequently visualized by autoradiography. QAD-, SafA-, and Pt650-conjugated radiolabeled oligonucleotides were shown to bind membrane-bound, renatured GAPDH, whereas cisplatinconjugated and nonmodified oligonucleotides did not (Fig. 2B). Conversely, the cisplatin-conjugated oligonucleotide did bind membrane-bound human HMG1, but the QAD-conjugated DNA probe did not (Fig. 2B). Also, the QAD-conjugated DNA probe did not bind nonrenatured membrane-bound human GAPDH, suggesting that only folded GAPDH is recognized. Interestingly, purified human erythrocyte GAPDH was found not to bind saframycin DNA-linked affinity columns, but binding did occur after the tetrameric protein was subjected to a denaturation–renaturation protocol. These findings, in conjunction with results from Southwestern blotting experiments, lead us to conclude that a form of GAPDH other than the tetramer is recognized by saframycin–DNA adducts, perhaps the monomer.
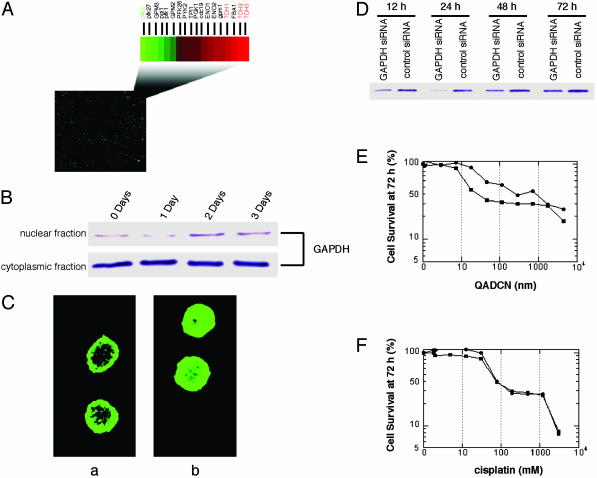
Saframycin–DNA Adducts Do Not Inhibit the Glycolytic Function of GAPDH. Human GAPDH is most commonly associated with glucose metabolism. In its native tetrameric form, GAPDH catalyzes the conversion of glyceraldehyde-3-phosphate and NAD+ into 3-bisphosphoglycerate and NADH. We found that duplex DNA–QAD adducts did not inhibit the ability of tetrameric human erythrocyte GAPDH to catalyze the conversion of glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate in an in vitro assay, consistent with the idea that a different function of GAPDH is targeted by saframycin–DNA adducts. Considering only its function in glycolysis, GAPDH may appear to be an unlikely target for an anticancer drug. However, GAPDH has a complex and evolving role in the nucleus, where it has been implicated to function as a monomer (25). GAPDH has been identified as a component of a multiprotein nuclear complex that recognizes oligonucleotides incorporating the antileukemic agent thioguanosine (26, 27), although GAPDH has not been shown to bind directly to thioguanosine-modified DNA. Monomeric GAPDH has also recently been found to be the principle component of a multiprotein nuclear complex involved in transcriptional coactivation of the histone 2B promoter, required for S phase progression in the cell cycle, whereas tetrameric GAPDH does not function in transcriptional coactivation (28). In light of these findings, we note that our earlier transcription profiling experiments using a SafA-sensitive strain of yeast exposed separately to SafA and QAD established in both cases that the greatest degree of overexpression in response to drug treatment occurred among transcripts for the three isoforms of GAPDH (1.7- to 3.8-fold), and that transcripts encoding each of the histones were substantially down-regulated (>2-fold, Fig. 3A) (14).
Fig. 3.
The role of GAPDH in the response of eukaryotic cells to treatment with saframycin analogs (QAD). (A) Transcription-profiling studies performed in a SafA-sensitive yeast strain show that all three isoforms of GAPDH (TDH1, TDH2, and TDH3) are substantially up-regulated after treatment with QADCN or SafA, whereas the gene transcript for phosphfructokinase, pfk2, catalyzing the principle, rate-limiting step of glycolysis is down-regulated >2-fold. Transcripts for each of the histones were down-regulated >2-fold (14). (B) Western blots for the determination of GAPDH levels in the nuclear and cytoplasmic fractions of HeLa-S3 cells treated with QADOH for the indicated times. (C) Confocal microscopy of HeLa-S3 cells treated (b) or untreated (a) with QADOH (17.5 nM, 48 h) and visualized by using a standard FITC-conjugated antibody immunostaining protocol. Cell nuclei were visualized separately by Hoechst dye 33340 staining (not shown). (D) GAPDH levels in A549 cells that had been transfected with GAPDH siRNA or control siRNA, respectively, as determined by immunoblotting. (E) Percentage of viable cells transfected with GAPDH siRNA (filled circle) or control siRNA (filled square) after 72-h treatment with varying concentrations of QADCN. (F)Asin E, but treated with cisplatin in lieu of QADCN.
Exposure of Cancer Cells to Saframycins Leads to Translocation of GAPDH to the Nucleus. To explore the potential role of nuclear GAPDH in the antiproliferative activity of saframycin-like molecules, we sought to determine whether exposure of cancer cells to these agents would lead to translocation of GAPDH to the nucleus. We conducted both confocal microscopy and cellular fractionation experiments in a human cancer cell line (HeLa-S3) exposed to the analog QADOH. Both types of experiments showed that nuclear GAPDH levels were elevated relative to controls in which QADOH was not present (Fig. 3). Cellular fractionation studies showed that nuclear translocation of GAPDH was maximal after ≈48 h of drug exposure (2-fold increase, Fig. 3B). Despite the increased translocation of GAPDH into the nucleus, it should be noted that by far the larger proportion of cellular GAPDH remains in the cytosol even after drug treatment (Fig. 3B).† Cellular microscopy studies further confirmed that translocation of GAPDH to the nucleus occurred upon drug treatment (Fig. 3C). Visually, the cells exhibited morphological signs of apoptosis after drug treatment (membrane blebbing). Additional evidence for an apoptotic pathway was obtained by the treatment of A549 cells with QADOH, which produced a characteristic ladder of cleaved chromosomal DNA.
Depletion of Cellular GAPDH by RNA Interference Confers Resistance to Saframycins. We sought to determine whether altering the levels of GAPDH in human cancer cells would influence their sensitivity to saframycin compounds and so modulated GAPDH levels by the reverse-genetic method of RNA interference (siRNA). Treatment of human non-small cell lung cancer cells (A549) with a commercial siRNA transfection vector targeting GAPDH led to time-dependent reduction of cellular GAPDH levels relative to a negative control cell population (treated with siRNA of a similar base composition, but with no homology to any known coding sequence). Maximal diminution of GAPDH levels (95% depletion versus control) was observed at 24 h; thereafter, GAPDH levels partially recovered (50% versus control at 72 h, Fig. 3D). GAPDH-depleted cells were found to be 8-fold resistant to growth inhibition by QADCN relative to controls with normal GAPDH levels (measured as percent cell survival at 72 h) (Fig. 3E). Furthermore, experiments using cisplatin in lieu of QADCN showed that there was no difference in sensitivity to growth inhibition by cisplatin in GAPDH-depleted and control cell populations (Fig. 3F). If GAPDH functioned as a general mediator of apoptosis, it might have been anticipated that the growth of cells treated with cisplatin would have been affected as well, although this need not necessarily have been the case, given the complexities of apoptotic signaling pathways.
Discussion. There is increasing evidence to suggest that GAPDH functions in one or more critical nuclear processes (25, 28). The demonstration here that a specific binding interaction occurs between GAPDH, duplex DNA, and several known members of the saframycin class of antiproliferative agents both implicates a previously unknown mechanism of action for the compound class and identifies GAPDH as a potential target for chemotherapeutic intervention. That the mechanism of action of the saframycins involves a direct interaction with GAPDH and DNA was not expected, but is supported herein by affinity experiments and Southwestern binding studies, by the fact that drug exposure leads to an increased concentration of GAPDH in the nucleus, and by the fact that depletion of cellular GAPDH levels confers drug resistance. The latter finding may be of significance, in that many tumor cells have been shown to overexpress GAPDH (30), providing a potential basis for tumor-cell specificity in cell killing. Exactly how a ternary complex of GAPDH, dsDNA, and a small-molecule agent of the saframycin class is toxic to the cell is not evident. That it is becomes all of the more intriguing in light of recent evidence implicating a role for GAPDH in transcriptional coactivation necessary for S-phase progression of the cell (28), but it is unclear at present whether or how these observations are related.
Acknowledgments
We thank Professor E. J. Corey for a generous gift of Pt650 and the Harvard Microchemistry Facility for protein sequence analysis. We gratefully acknowledge the National Institutes of Health for financial support, the Howard Hughes Medical Institute for a Predoctoral fellowship (to J.R.L.), and the American Cancer Society for a graduate research fellowship (to J.K.B.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SafA, saframycin A; Et743, ecteinascidin; QAD, quinaldic acid derivative; HMG, high-mobility group; MMC, mitomycin C; siRNA, small interfering RNA; ss, single-stranded; ds, double-stranded; Pt650, phthalascidin.
Footnotes
Cytosolic concentrations of GAPDH are typically >1,000-fold higher than the concentrations of saframycins used in these studies (GI50 values of saframycins in most cancer cell lines range from 1 to 100 nM; a typical measure of GAPDH in the cell is ≈70 μM) (29). These calculations, plus the fact that saframycin–DNA adducts do not inhibit the glycolytic function of GAPDH, point toward a mechanism of GAPDH-mediated cytotoxicity that is not related to an interference with glycolytic metabolism.
References
- 1.Arai, T. & Kubo, A. (1983) in The Alkaloids, ed. Brossi, A. (Academic, New York), pp. 56–98.
- 2.Rinehart, K. L., Holt, T. G., Fregeau, N. L., Keifer, P. A., Wilson, G. R., Perun, T. J., Jr., Sakai, R., Thompson, A. G., Stroh, J. G., Shield, L. S., et al. (1990) J. Nat. Prod. 53, 771–792. [DOI] [PubMed] [Google Scholar]
- 3.Aune, G., Furuta, T. & Pommier, Y. (2002) Anti-Cancer Drugs 13, 545–555. [DOI] [PubMed] [Google Scholar]
- 4.Ishiguro, K., Sakiyama, S., Takahashi, K. & Arai, T. (1978) Biochemistry 17, 2545–2550. [DOI] [PubMed] [Google Scholar]
- 5.Ishiguro, K., Takahashi, T., Yazawa, K., Sakiyama, S. & Arai, T. (1981) J. Biol. Chem. 256, 2162–2167. [PubMed] [Google Scholar]
- 6.Lown, J. W., Joshua, A. V. & Lee, J. (1982) Biochemistry 21, 428–436. [DOI] [PubMed] [Google Scholar]
- 7.Rao, K. E. & Lown, J. W. (1990) Chem. Res. Toxicol. 3, 262–267. [DOI] [PubMed] [Google Scholar]
- 8.Rao, K. E. & Lown, J. W. (1992) Biochemistry 31, 12076–12082. [DOI] [PubMed] [Google Scholar]
- 9.Hill, G. C. & Remers, W. A. (1991) J. Med. Chem. 34, 1990–1998. [DOI] [PubMed] [Google Scholar]
- 10.Pommier, Y., Kohlhagen, G., Bailly, C., Waring, M., Mazumder, A. & Kohn, K. W. (1996) Biochemistry 35, 13303–13309. [DOI] [PubMed] [Google Scholar]
- 11.Martinez, E. J. & Corey, E. J. (1999) Org. Lett. 1, 75–77. [DOI] [PubMed] [Google Scholar]
- 12.Plowright, A. T. & Myers, A. G. (2001) J. Am. Chem. Soc. 123, 5114–5115. [DOI] [PubMed] [Google Scholar]
- 13.Martinez, E. J., Corey, E. J. & Owa, T. (2001) Chem. Biol. 8, 1151–1160. [DOI] [PubMed] [Google Scholar]
- 14.Plowright, A. T., Schaus, S. E. & Myers, A. G. (2002) Chem. Biol. 9, 607–618. [DOI] [PubMed] [Google Scholar]
- 15.Takebayashi, Y., Pourquier, P., Zimonjic, D. B., Nakayama, K., Emmert, S., Ueda, T., Urasaki, Y., Kanzaki, A., Akiyama, S., Popescu, N., et al. (2001) Nat. Med. 7, 961–966. [DOI] [PubMed] [Google Scholar]
- 16.Gajate, C., An, F. & Mollinedo, F. (2002) J. Biol. Chem. 277, 41580–41589. [DOI] [PubMed] [Google Scholar]
- 17.Martinez, E. J., Owa, T., Schreiber, S. L. & Corey, E. J. (1999) Proc. Natl. Acad. Sci. USA 96, 3496–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers, A. G. & Kung, D. W. (1999) J. Am. Chem. Soc. 122, 10828–10829. [Google Scholar]
- 19.Tomasz, M., Chawla, A. K. & Lipman, R. (1988) Biochemistry 27, 3182–3187. [DOI] [PubMed] [Google Scholar]
- 20.Mansy, S., Rosenberg, B. & Thomson, A. J. (1973) J. Am. Chem. Soc. 95, 1633–1640. [DOI] [PubMed] [Google Scholar]
- 21.Philippe, J. (2000) in The Nucleic Acid Protocols Handbook, ed. Rapley, R. (Humana, Totowa, NJ), pp. 773–782.
- 22.Allan, V. J. (2000) in Protein Localization by Fluorescence Microscopy, ed. Allan, V. J. (Oxford Univ. Press, Oxford), pp. 1–26.
- 23.Toney, J. H., Donahue, B. A., Kellett, P. J., Bruhn, S. L., Essigmann, J. M. & Lippard, S. J. (1989) Proc. Natl. Acad. Sci. USA 86, 8328–8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pil, P. M. & Lippard, S. J. (1992) Science 256, 234–237. [DOI] [PubMed] [Google Scholar]
- 25.Sirover, M. A. (1999) Biochim. Biophys. Acta 1432, 159–184. [DOI] [PubMed] [Google Scholar]
- 26.Krynetski, E. Y., Krynetskaia, N. F., Gallo, A. E., Murti, K. G. & Evans, W. E. (2001) Mol. Pharmacol. 59, 367–374. [DOI] [PubMed] [Google Scholar]
- 27.Krynetski, E. Y., Krynetskaia, N. F., Bianchi, M. E. & Evans, W. E. (2003) Cancer Res. 63, 100–106. [PubMed] [Google Scholar]
- 28.Zheng, L., Roeder, R. G. & Luo, Y. (2003) Cell 114, 255–266. [DOI] [PubMed] [Google Scholar]
- 29.Furfine, C. S. & Velick, S. F. (1965) J. Biol. Chem. 240, 844–855. [PubMed] [Google Scholar]
- 30.Vila, M. R., Nicolas, A., Morote, J., de Torres, I. & Meseguer, A. (2000) Cancer 89, 152–164. [PubMed] [Google Scholar]