Abstract
Chronic hepatitis B virus (HBV) infection, affecting approximately 240 million people worldwide, is a major public health problem that elevates the risk of developing liver cirrhosis and hepatocellular carcinoma. Given that current anti-HBV drugs are limited to interferon-based regimens and nucleos(t)ide analogs, the development of new anti-HBV agents is urgently needed. The viral entry process is generally an attractive target implicated in antiviral strategies. Using primary cells from humans and Tupaia belangeri, as well as HepaRG cells, important determinants of viral entry have been achieved. Recently, sodium taurocholate cotransporting polypeptide (NTCP) was identified as an HBV entry receptor and enabled the establishment of a susceptible cell line that can efficiently support HBV infection. This finding will allow a deeper understanding of the requirements for efficient HBV infection, including the elucidation of the molecular entry mechanism. In addition, pharmacological studies suggest that NTCP is able to serve as a therapeutic target. This article summarizes our current knowledge on the mechanisms of HBV entry and the role of NTCP in this process.
Keywords: HBV, infection, entry, replication, NTCP, SLC10A1, transporter, DMSO, myrcludex-B, cyclosporin
1. Introduction
Hepatitis B virus (HBV) infection constitutes a serious public health problem, affecting approximately 240 million carriers worldwide [1]. Chronic HBV infection significantly elevates the risk for developing liver cirrhosis and hepatocellular carcinoma. Currently, conventional interferon-α (IFNα) or PEGylated-IFNα and nucleos(t)ide analogs are available as anti-HBV agents [2,3]. However, IFN-based therapies, which cause significant side effects, yields long-term clinical benefits in less than 40% of treated patients [4]. Nucleos(t)ide analogs suppress an essential step in virus replication and thereby provide biochemical and histological improvement, but some of the early drugs give rise to drug-resistant viruses, which adversely affect long-term clinical outcome. Thus, in order to approach curative treatments, new anti-HBV agents targeting different molecules involved in HBV infection and propagation are needed. Nucleos(t)ide analogs suppress HBV replication mainly by inhibiting the reverse transcription process in the viral lifecycle (Figure 1) [3]. IFN functions as an immunomodulator and is also reported to directly interfere with HBV replication at multiple steps of the lifecycle [5]. Given that HBV encodes only one viral protein carrying enzymatic activity, polymerase, in its genome, strategies for inhibiting viral enzyme are limited. Although capsid or envelope protein assembly and the regulatory X-protein are possible future targets, it is critical for developing new classes of anti-HBV agents to identify cellular factors serving as possible drug targets.
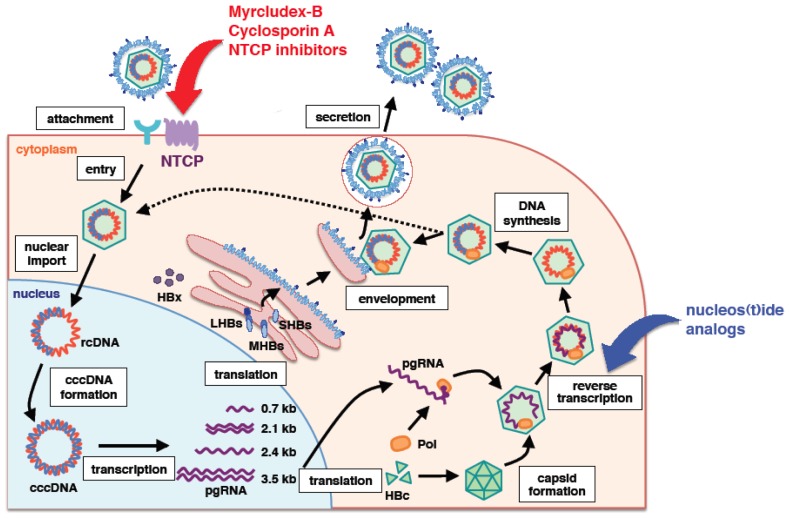
Figure 1.
Schematic representation of the hepatitis B virus (HBV) lifecycle. Nucleos(t)ide analogs inhibit reverse transcription. Myrcludex-B, cyclosporin A and some NTCP inhibitors can inhibit the viral entry process by targeting NTCP.
In general, the viral entry step is an attractive target for the development of antiviral agents [6–8]. The early HBV lifecycle, including the entry step, has gained significant attention very recently with regard to molecular mechanisms, triggered by the identification of sodium taurocholate cotransporting polypeptide (NTCP) as a cellular entry receptor [9]. This article summarizes the molecular evidence related to HBV entry, mainly focusing on recent findings, and its implications.
2. PreS1 Region of HBV Surface Protein Is Essential for Viral Entry
HBV infection into host hepatocytes follows a multiple step process: (1) initially, HBV reversibly attaches to host cell surface proteoglycans with a low affinity; (2) this is followed by the process involving more specific receptor(s) with high affinity to mediate the early entry step; and (3) after endocytosis-mediated internalization, the virus fuses with the cellular membrane compartment, probably in an endosomal compartment, although the mechanisms are not fully understood. Both initial attachment and, probably more importantly, specific receptor recognition contribute to host specificity and tissue tropism [10]. The initial attachment step is at least partly mediated by heparan sulfate proteoglycans [11–14]. The third internalization step is reported to involve caveolae-, clathrin- or macropinocytosis-dependent endocytosis, depending on the cell types and experimental systems [15–18]. However, cellular factors involved in the high-affinity binding and the early entry process remained to be elucidated until recently.
The HBV surface proteins are composed of three proteins, termed the large (LHBs), middle (MHBs) and small (SHBs) surface proteins, and include the preS1, preS2 and S regions: LHBs encompasses the preS1, preS2 and S regions; MHBs encompasses the preS2 and S; and SHBs comprise the S region [19,20]. The molecular requirement of HBV envelope proteins for HBV infection has been studied for more than a decade using primary hepatocytes from humans and Tupaia belangeri, as well as HepaRG cells [21]. A series of analyses using neutralizing antibodies and introduced point mutations suggested that the S and preS1, but not the preS2, regions play a significant role in HBV infection [22–27]. In a direct approach, the preS1 region in the LHBs has been shown to be essentially involved in the HBV infection process. This was demonstrated by the introduction of mutations in the viral context, infection competition with anti-preS1 antibodies and with peptides mimicking this region [28–34]. A myristoylated peptide encompassing amino acids 2–48 of the preS1 region turned out to be the most efficient in infection inhibition of HBV and also the envelope protein-related hepatitis D virus (HDV) [30,31]. Such a peptide has been used as a tool for characterizing the early infection step, including the identification of NTCP as an entry receptor [9] and as a lead substance (Myrcludex-B) presently in the clinical development (see below) [10,35,36].
3. Sodium Taurocholate Co-Transporting Polypeptide (NTCP) as a Bona Fide HBV Receptor
One of the recent milestones in the field in HBV molecular biology is the identification of NTCP as a host entry receptor, as reported by Yan and Zhong et al. in late 2012 [9]. By affinity purification and mass spectrometry analysis using an HBV preS1-derived lipopeptide as bait, they identified Tupaia belangeri NTCP (tsNTCP) as a cellular factor interacting with this lipopeptide. NTCP is a transporter residing in the basolateral membrane of hepatocytes and is involved in the hepatic uptake of mostly conjugated bile salts (see below). The lipopeptide was confirmed to specifically bind human NTCP (hNTCP), as well as tsNTCP, but surprisingly not crab-eating monkey NTCP (mkNTCP), which correlated with the species specificity of HBV infection: HBV is able to efficiently infect humans and Tupaia, but not crab-eating monkey [9]. Interestingly, this result also correlated with the in vitro binding activity of the peptide to the respective primary hepatocytes [10] and their in vivo hepatotropism [37]. The role of NTCP in the viral infection of HBV, its satellite virus, HDV, and a closely related primate hepadnavirus wooly monkey HBV was further examined by knockdown and overexpression analyses [9,38,39]. siRNA-mediated knockdown of NTCP in primary human hepatocytes (PHH), primary Tupaia hepatocytes and differentiated HepaRG cells reduced HBV and HDV infection, while ectopic expression of NTCP conferred HBV susceptibility in HepG2 cells, which originally did not support efficient infection [9]. This strongly argues that NTCP is an essential factor for HBV infection. The expression of NTCP in different cells was consistent with the HBV susceptibility, as it was significantly expressed in HBV-susceptible cells, PHH and differentiated HepaRG cells, but was weakly expressed or absent in HepG2, Huh-7, FLC4 and HeLa cells, which show little to no infection [40–42]. The introduction of NTCP into Huh-7 and undifferentiated HepaRG cells conferred HBV infection to these cells to some extent [38]. Although the total expressions in these transduced cells were comparable, hNTCP-expressing HepG2 cells showed much higher infection efficiency when compared with other human hepatocyte cell lines [38,43,44]. In the initial study, infection efficiency was ~10% in NTCP-overexpressing HepG2 cells cultured with medium containing 2% dimethyl sulfoxide (DMSO) [9]. Subsequent analysis showed that increasing the DMSO concentration to more than 2.5%~3% augmented infection efficiency to 50%~70%, as evaluated by immunofluorescence of HBV proteins, although the virus inoculum was different in these studies [38,43]. The speculations include that DMSO augmented the gene expression of NTCP, promoted the membrane localization of NTCP and changed the post-translational modification of NTCP, but the detailed molecular mechanisms for DMSO-mediated promotion of HBV infection is open for further studies. It remains unknown why not all of the cells were infected with HBV in these reports, but it is possible that the NTCP function for supporting HBV entry is reflected by post-translational modification, subcellular localization or other factors that are governed by cell conditions or by more general conditions, such as the cell cycle, cellular microenvironment or architecture. Another open question is on the high susceptibility for HDV, but not HBV, in Huh-7 cells overexpressing hNTCP [9,38]. Future analysis of this issue is necessary in order to establish a cell culture model that is 100% susceptible to HBV infection.
Crucial amino acid sequences in NTCP involved in HBV infection have been analyzed. By sequence comparison between hNTCP and mkNTCP, replacement of amino acids 157–165 of hNTCP with the respective sequence from mkNTCP abrogated the ability to support HBV preS1-binding and, subsequently, infection, while mkNTCP carrying a conversion to this region from hNTCP conferred HBV susceptibility. Thus, amino acids 157–165 of NTCP are crucial for NTCP-mediated HBV binding and infection [9,45]. It has also been shown that hNTCP bearing a substitution of the 84–87 aa from the mouse counterpart was able to bind preS1, but was not functional for HBV infection, while replacing these residues in mouse NTCP (mNTCP) with the human counterparts supported the infection [38,44]. These data indicate that the 84–87 aa residues are a determinant for NTCP function as an HBV entry receptor. It remains to be elucidated why mNTCP does not support HBV infection, but mNTCP was shown to support specific binding of the preS1-lipopeptide on the cell surface, although the binding capacity of mNTCP to the preS1 region appears to be weaker than that of hNTCP [44]. It is possible that the binding of HBV to NTCP is not sufficient and requires an additional molecule or mechanism to trigger the following early infection process.
HDV is a virusoid-like particle, which depends on HBV for assembly and propagation [46]. HDV shares the HBV envelope proteins, LHBs, MHBs and SHBs, and its attachment/early entry mechanism seems to be very similar to that for HBV. Due to its completely different replication strategy, it is very likely that it depends on different cellular factors and follows different pathways after membrane fusion. Intriguingly, HDV infection can be observed by complementing hNTCP in either mouse-derived Hepa1-6, MMHD3 and Hep56.1D cells, rat hepatocyte TC5123 cells or non-hepatocyte HeLa, CHO and Vero cells. This is in stark contrast to HBV, which cannot infect these cells [38,44]. This suggests that HBV requires additional host factors for infection or is restricted at a post-entry step prior to covalently closed circular DNA (cccDNA) formation. It is of particular interest to clarify the molecular mechanisms underlying the different cellular requirements between infection by HBV and HDV, especially when trying to establish a susceptible mouse model in the future.
4. Other Factors Essential for HBV Infection?
It is presently unclear whether there are additional cellular factors besides NTCP required for viral infection and determining the tissue and species tropism of HBV. These include factors essentially involved in the viral lifecycle during attachment, internalization, endocytosis, membrane fusion, uncoating, nuclear translocation and cccDNA formation and those affecting post-entry restriction. Overexpression of hNTCP in mouse hepatocyte cell lines, such as Hepa1-6 and MMHD3 cells, did not confer susceptibility to HBV infection, in contrast to the HBV infection observed after NTCP introduction into HepG2 cells [44]. hNTCP conferred efficient HBV infection in HepG2 cells, but only a low efficiency of infection was observed in Huh-7 and undifferentiated HepaRG cells and no detectable infection to mouse and rat hepatoma cells, including Hep56.1D, Hepa1-6 and TC5123 cells [38]. We also showed that different HepG2 clone isolates that similarly expressed high levels of ectopic NTCP, but were likely to have different cellular genetic backgrounds, had diverse efficiencies of HBV infection [43]. These observations favor the existence of additional host factors determining susceptibility to HBV infection. For hepatitis C virus (HCV) infection, multiple cellular factors are required for efficient viral entry, including low density lipoprotein receptor (LDLR), scavenger receptor class B type I (SR-BI), CD81, occludin (OCLN) and claudin-1 (CLDN-1) as viral entry receptors and Niemann-Pick C1-like 1 (NPC1L1), epidermal growth factor receptor (EGFR) and ephrin A2 (EphA2) as other factors involved in entry [21,47,48]. It has been reported that the complementation of both hCD81 and hOCLN are required for rendering high HCV susceptibility in mice [49]. Furthermore, in the case of duck hepatitis B virus (DHBV), multiple factors are suggested to be essential for efficient viral infection. Carboxypeptidase D was confirmed to bind the DHBV envelope and function in viral attachment and entry [50]. However, overexpression of this protein alone in Huh-7 cells did not support DHBV infection [51]. Carboxypeptidase D was able to bind to DHBV and heron HBV, which did not infect primary duck hepatocytes, and this protein is also expressed in non-liver tissues [52]. Thus, additional factors are likely to be required to explain DHBV susceptibility, one candidate of which can include duck NTCP [53]. These examples in viruses that utilize multiple receptors favor pursuing the identification of additional cellular factors crucial for HBV entry.
5. General Characteristic Features of NTCP
NTCP, also designated as solute carrier family 10A1 (SLC10A1), is a member of the SLC10 transporter gene family. The SLC10 family consists of seven members (SLC10A1–7). Among these, NTCP and apical sodium-dependent bile salt transporter (ASBT), also known as SLC10A2, are sodium-dependent transporters for bile acids [54]. NTCP is mainly distributed at the basolateral membrane of hepatocytes and plays a major role in the hepatic influx of conjugated bile salts from portal circulation [55,56]. NTCP on the plasma membranes in hepatocytes binds two sodium ions together with one molecule of preferentially conjugated bile salt for uptake. In addition to bile salts, NTCP, like other transporters, binds and/or transports other molecules, including steroid hormones, thyroid hormones, drug-conjugated bile salt and a variety of xenobiotics [57,58]. hNTCP is a 349 aa protein with an apparent mass of 56 kDa and includes a putative seven or nine transmembrane domains with a predicted topology of N-terminal extracellular and C-terminal intracellular ends [59–61]. While the structure of NTCP has not been resolved, the crystal structures of the ASBTs from Neisseria meningitis (ASBTNM) and Yersinia frederiksenii (ASBTYf) were recently reported [62,63]. ASBTNM shows a ten transmembrane domain and a hydrophobic inward-facing binding cavity. This structure is different from the model for hASBT currently favored based on bioinformatic prediction and experimental data, which carry seven to nine transmembrane helices with N-terminal extracellular and the C-terminal cytoplasmic domain [60,64,65]. A structural analysis of ASBTYf proposed two conformations, inward- and outward-open structures of bile salt transporters by rotating two core helices transmembrane (TM)-4 and TM9 [63]. Because ASBTNM and ASBTYf has only 26% and 22% homology, respectively, with hASBT and even lower homology with hNTCP [62,63], it is uncertain whether the structural features of ASBTNM or ASBTYf are useful for designing drugs targeting hASBT and hNTCP.
Several single nucleotide polymorphisms (SNPs) that alter the transporter activity of NTCP have been reported [66,67]. As non-synonymous SNPs, I223T, a variant seen in 5.5% of allele frequencies in African Americans, decreased plasma membrane-localized NTCP and reduced its transporter activity. The S267F variant, seen in 7.5% of allele frequencies in Chinese Americans, exhibited almost complete loss of function for bile acid uptake, but possessed normal transport activity for the non-bile acid substrate, estrone sulfate. Another report showed that the A64T and S267F variants, carried by 1.0% and 3.1% of allele frequencies in Koreans, respectively, decreased the uptake of taurocholic acid. These polymorphisms are dependent on ethnicity. However, there have been no reports of serious diseases associated with defects in the NTCP gene. No reports describing NTCP knockout mice have been published to date. Thus, it is difficult to draw conclusions on whether the physiological roles of NTCP are complemented by other factors that share the redundant physiological function and whether NTCP inhibition is able to safely serve as an anti-HBV drug target.
Importantly, it was very recently reported that molecular determinants for the transporter function of NTCP overlapped with those for the ability to support HBV entry [68]. NTCP mutations in amino acids that were critical for bile salt binding (N262A, Q293A/L294A) abrogated both the binding to preS1 peptide and the infection of HBV. The S267F variant of NTCP could neither bind to the preS1 region nor support HBV infection in cell culture.
6. NTCP as a Target for Anti-HBV Agents
In general, the viral entry process is an attractive target for the development of antiviral agents. As noted above, the 2–48 aa region of preS1 in the LHBs protein is important for HBV infection [31]. Myrcludex-B, which is an optimized synthetic lipopeptide consisting of the myristoylated 2–48 aa region of preS1, is able to strongly inhibit HBV infection in both cell culture and an in vivo mouse model [36]. The IC50 in a cell culture model was reported to be approximately 100 pM [35]. Following the successful clinical development of enfuvirtide as the first peptidic HIV entry inhibitor mimicking the region derived from the viral gp41 envelope glycoprotein [69], Myrcludex-B is now under clinical development in phase Ib/IIa [70]. Mechanistically, Myrcludex-B binds hNTCP and inactivates its receptor function for HBV and HDV (Figure 1). Remarkably, IC50 to the transporter activity of NTCP was approximately 4 nM [38], showing that binding saturation is not required for receptor inactivation, thus allowing a therapeutic window for infection inhibition without a complete abrogation of bile salt transportation [38]. Thus, agents targeting NTCP are expected to be potent candidates that act as anti-HBV drugs.
Cyclosporin A (CsA) is the first line of such compounds revealed to inhibit HBV infection by targeting NTCP [42,45]. CsA is known to be an immunosuppressant classified as a calcineurin inhibitor and is clinically used for the suppression of the immunological failure of xenografts after tissue transplantation [71]. In cell culture analyses, CsA was also reported to suppress the replication of numerous viruses, including HIV, HCV, influenza virus, severe acute respiratory syndrome coronavirus, human papillomavirus, flaviviruses and HBV [72–79]. In most of these cases, cyclophilins (CyPs), cellular peptidyl prolyl cis-trans isomerases that catalyze conformational changes in proteins and are the primary cellular target for CsA, were critical for efficient viral replication, and CyP inhibition by CsA was responsible for antiviral activity. However, the anti-HBV entry activity of CsA was not mediated by the inhibition of CyP, but rather, via direct targeting of NTCP. CsA bound to NTCP on the plasma membrane and inhibited transporter activity (Figure 1) [42,45]. It also inhibited binding between LHBs and NTCP in vitro (Figure 1) [42]. This suggests that CsA interacted with NTCP, thus inhibiting the recruitment of LHBs of incoming HBV to NTCP on the plasma membrane and blocking HBV entry. The anti-HBV activity of CsA was pan-genotypic [42]. Moreover, our derivative analysis identified a series of CsA analogs having a stronger anti-HBV entry activity with a submicromolar IC50 [42]. Notably, non-immunosuppressive CsA analogs may be potent anti-HBV agents. Given that non-immunosuppressive CsA analogs, including alisporivir (Debio 025) and SCY-635, have significant activity in decreasing HCV viral load in clinical trials and are regarded as promising anti-HCV drug candidates [80,81], further derivative analysis of CsA may be a reasonable approach for drug development.
As other examples, compounds known to be NTCP inhibitors, including progesterone, propranolol and bosentan, have been shown to block HBV infection (Figure 1) [42]. NTCP substrates, such as taurocholate, tauroursodeoxycholate and bromosulfophthalein, also inhibited HBV infection [38,42,68]. An anticholesteremic drug, ezetimibe, has been shown to block HBV entry [82], and this drug was reported to inhibit the NTCP transporter [83]. These results indicate that compounds modulating NTCP function could substantially inhibit HBV infection. HepG2 cells engineered to overexpress NTCP are also useful for high-throughput screening to identify compounds targeting NTCP and inhibiting HBV infection. One example identified in such chemical screening is the oxysterols, which are oxidized derivatives of cholesterol or by-products of cholesterol biosynthesis [43].
Host-targeting antivirals are generally expected to have significant advantages, including a much lower frequency drug resistance, universal antiviral effects beyond viral genotypes and complementary mechanisms of action that might act in a synergistic manner with currently available antiviral agents [48]. More importantly, they offer an additional therapeutic choice, given that only IFNs and nucleoside analogs are currently available as anti-HBV agents.
7. Conclusions
Identification of NTCP as an HBV entry receptor has accelerated the understanding of HBV molecular biology and offered useful experimental systems to analyze the HBV and HDV lifecycle, including the identification of host restriction and dependency factors. NTCP also represents a new therapeutic target in the development of new anti-HBV agents. Further analyses using a new cell culture system are necessary in order to clarify the molecular mechanisms underlying NTCP-mediated HBV infection and to establish an in vivo small animal model that fully supports HBV infection.
Acknowledgments
The authors are grateful to all of the members of the Department of Virology II, National Institute of Infectious Diseases, the Department of Infectious Diseases, Molecular Virology, University Hospital Heidelberg, and the Li lab at the National Institute of Biological Sciences, Beijing, for their research, technical support and discussions. Funding was provided by the Ministry of Health, Labor and Welfare, Japan, the Ministry of Education, Culture, Sports, Science and Technology, Japan, the Japan Society for the Promotion of Science, the Deutsche Zentrum für Infektionsforschung (DZIF), the Deutsche Krebshilfe, the Deutsche Forschungsgemeinschaft (DFG) UR72/7-1, FOR1202/UR72/5-1, the Ministry of Science and Technology, China (2010CB530101), and the National Science and Technology Major Project, China (2013ZX09509102).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Author Contributions
All of the authors wrote the paper.
References
- 1.Ott J.J., Stevens G.A., Groeger J., Wiersma S.T. Global epidemiology of hepatitis B virus infection: New estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212–2219. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 2.Zoulim F., Locarnini S. Optimal management of chronic hepatitis B patients with treatment failure and antiviral drug resistance. Liver Int. 2013;33:116–124. doi: 10.1111/liv.12069. [DOI] [PubMed] [Google Scholar]
- 3.Zoulim F., Perrillo R., Hepatitis B. Reflections on the current approach to antiviral therapy. J. Hepatol. 2008;48:S2–S19. doi: 10.1016/j.jhep.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 4.Marcellin P., Bonino F., Yurdaydin C., Hadziyannis S., Moucari R., Kapprell H.P., Rothe V., Popescu M., Brunetto M.R. Hepatitis B surface antigen levels: Association with 5-year response to peginterferon alfa-2a in hepatitis B e-antigen-negative patients. Hepatol. Int. 2013;7:88–97. doi: 10.1007/s12072-012-9343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belloni L., Allweiss L., Guerrieri F., Pediconi N., Volz T., Pollicino T., Petersen J., Raimondo G., Dandri M., Levrero M. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Invest. 2012;122:529–537. doi: 10.1172/JCI58847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haqqani A.A., Tilton J.C. Entry inhibitors and their use in the treatment of HIV-1 infection. Antiviral Res. 2013;98:158–170. doi: 10.1016/j.antiviral.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 7.Mercer J., Helenius A. Gulping rather than sipping: Macropinocytosis as a way of virus entry. Curr. Opin. Microbiol. 2012;15:490–499. doi: 10.1016/j.mib.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 8.Zeisel M.B., Fofana I., Fafi-Kremer S., Baumert T.F. Hepatitis C virus entry into hepatocytes: Molecular mechanisms and targets for antiviral therapies. J. Hepatol. 2011;54:566–576. doi: 10.1016/j.jhep.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z., Huang Y., Qi Y., Peng B., Wang H., et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meier A., Mehrle S., Weiss T.S., Mier W., Urban S. Myristoylated PreS1-domain of the hepatitis B virus L-protein mediates specific binding to differentiated hepatocytes. Hepatology. 2013;58:31–42. doi: 10.1002/hep.26181. [DOI] [PubMed] [Google Scholar]
- 11.Glebe D., Urban S. Viral and cellular determinants involved in hepadnaviral entry. World J. Gastroenterol. 2007;13:22–38. doi: 10.3748/wjg.v13.i1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamas Longarela O., Schmidt T.T., Schoneweis K., Romeo R., Wedemeyer H., Urban S., Schulze A. Proteoglycans act as cellular hepatitis delta virus attachment receptors. PLoS One. 2013;8:e58340. doi: 10.1371/journal.pone.0058340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leistner C.M., Gruen-Bernhard S., Glebe D. Role of glycosaminoglycans for binding and infection of hepatitis B virus. Cell. Microbiol. 2008;10:122–133. doi: 10.1111/j.1462-5822.2007.01023.x. [DOI] [PubMed] [Google Scholar]
- 14.Schulze A., Gripon P., Urban S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology. 2007;46:1759–1768. doi: 10.1002/hep.21896. [DOI] [PubMed] [Google Scholar]
- 15.Cooper A., Shaul Y. Clathrin-mediated endocytosis and lysosomal cleavage of hepatitis B virus capsid-like core particles. J. Biol. Chem. 2006;281:16563–16569. doi: 10.1074/jbc.M601418200. [DOI] [PubMed] [Google Scholar]
- 16.Gao Z., Li M., He W., Li W. Hepatitis B virus may enter HepG2 cells complemented with human NTCP via macropinocytosis. Proceedings of the 2013 International Meeting on Molecular Biology of Hepatitis B Viruses; Shanghai, China. 20–23 October 2013; p. O-2. [Google Scholar]
- 17.Huang H.C., Chen C.C., Chang W.C., Tao M.H., Huang C. Entry of hepatitis B virus into immortalized human primary hepatocytes by clathrin-dependent endocytosis. J. Virol. 2012;86:9443–9453. doi: 10.1128/JVI.00873-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macovei A., Radulescu C., Lazar C., Petrescu S., Durantel D., Dwek R.A., Zitzmann N., Nichita N.B. Hepatitis B virus requires intact caveolin-1 function for productive infection in HepaRG cells. J. Virol. 2010;84:243–253. doi: 10.1128/JVI.01207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stibbe W., Gerlich W.H. Structural relationships between minor and major proteins of hepatitis B surface antigen. J. Virol. 1983;46:626–628. doi: 10.1128/jvi.46.2.626-628.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heermann K.H., Goldmann U., Schwartz W., Seyffarth T., Baumgarten H., Gerlich W.H. Large surface proteins of hepatitis B virus containing the pre-S sequence. J. Virol. 1984;52:396–402. doi: 10.1128/jvi.52.2.396-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baumert T.F., Meredith L., Ni Y., Felmlee D.J., McKeating J.A., Urban S. Entry of hepatitis B and C viruses—Recent progress and future impact. Curr. Opin. Virol. 2014;4C:58–65. doi: 10.1016/j.coviro.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Abou-Jaoude G., Sureau C. Entry of hepatitis delta virus requires the conserved cysteine residues of the hepatitis B virus envelope protein antigenic loop and is blocked by inhibitors of thiol-disulfide exchange. J. Virol. 2007;81:13057–13066. doi: 10.1128/JVI.01495-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bremer C.M., Sominskaya I., Skrastina D., Pumpens P., El Wahed A.A., Beutling U., Frank R., Fritz H.J., Hunsmann G., Gerlich W.H., et al. N-terminal myristoylation-dependent masking of neutralizing epitopes in the preS1 attachment site of hepatitis B virus. J. Hepatol. 2011;55:29–37. doi: 10.1016/j.jhep.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 24.Iwarson S., Tabor E., Thomas H.C., Goodall A., Waters J., Snoy P., Shih J.W., Gerety R.J. Neutralization of hepatitis B virus infectivity by a murine monoclonal antibody: An experimental study in the chimpanzee. J. Med. Virol. 1985;16:89–96. doi: 10.1002/jmv.1890160112. [DOI] [PubMed] [Google Scholar]
- 25.Ni Y., Sonnabend J., Seitz S., Urban S. The pre-s2 domain of the hepatitis B virus is dispensable for infectivity but serves a spacer function for l-protein-connected virus assembly. J. Virol. 2010;84:3879–3888. doi: 10.1128/JVI.02528-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salisse J., Sureau C. A function essential to viral entry underlies the hepatitis B virus “a” determinant. J. Virol. 2009;83:9321–9328. doi: 10.1128/JVI.00678-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shearer M.H., Sureau C., Dunbar B., Kennedy R.C. Structural characterization of viral neutralizing monoclonal antibodies to hepatitis B surface antigen. Mol. Immunol. 1998;35:1149–1160. doi: 10.1016/s0161-5890(98)00110-2. [DOI] [PubMed] [Google Scholar]
- 28.Barrera A., Guerra B., Notvall L., Lanford R.E. Mapping of the hepatitis B virus pre-S1 domain involved in receptor recognition. J. Virol. 2005;79:9786–9798. doi: 10.1128/JVI.79.15.9786-9798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engelke M., Mills K., Seitz S., Simon P., Gripon P., Schnolzer M., Urban S. Characterization of a hepatitis B and hepatitis delta virus receptor binding site. Hepatology. 2006;43:750–760. doi: 10.1002/hep.21112. [DOI] [PubMed] [Google Scholar]
- 30.Glebe D., Urban S., Knoop E.V., Cag N., Krass P., Grun S., Bulavaite A., Sasnauskas K., Gerlich W.H. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology. 2005;129:234–245. doi: 10.1053/j.gastro.2005.03.090. [DOI] [PubMed] [Google Scholar]
- 31.Gripon P., Cannie I., Urban S. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J. Virol. 2005;79:1613–1622. doi: 10.1128/JVI.79.3.1613-1622.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong H.J., Ryu C.J., Hur H., Kim S., Oh H.K., Oh M.S., Park S.Y. In vivo neutralization of hepatitis B virus infection by an anti-preS1 humanized antibody in chimpanzees. Virology. 2004;318:134–141. doi: 10.1016/j.virol.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Le Seyec J., Chouteau P., Cannie I., Guguen-Guillouzo C., Gripon P. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J. Virol. 1999;73:2052–2057. doi: 10.1128/jvi.73.3.2052-2057.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeng C.Y., Ryu C.J., Gripon P., Guguen-Guillouzo C., Hong H.J. Fine mapping of virus-neutralizing epitopes on hepatitis B virus PreS1. Virology. 2000;270:9–16. doi: 10.1006/viro.2000.0250. [DOI] [PubMed] [Google Scholar]
- 35.Schulze A., Schieck A., Ni Y., Mier W., Urban S. Fine mapping of pre-S sequence requirements for hepatitis B virus large envelope protein-mediated receptor interaction. J. Virol. 2010;84:1989–2000. doi: 10.1128/JVI.01902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen J., Dandri M., Mier W., Lutgehetmann M., Volz T., von Weizsacker F., Haberkorn U., Fischer L., Pollok J.M., Erbes B., et al. Prevention of hepatitis B virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat. Biotechnol. 2008;26:335–341. doi: 10.1038/nbt1389. [DOI] [PubMed] [Google Scholar]
- 37.Schieck A., Schulze A., Gahler C., Muller T., Haberkorn U., Alexandrov A., Urban S., Mier W. Hepatitis B virus hepatotropism is mediated by specific receptor recognition in the liver and not restricted to susceptible hosts. Hepatology. 2013;58:43–53. doi: 10.1002/hep.26211. [DOI] [PubMed] [Google Scholar]
- 38.Ni Y., Lempp F.A., Mehrle S., Nkongolo S., Kaufman C., Falth M., Stindt J., Koniger C., Nassal M., Kubitz R., et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014 doi: 10.1053/j.gastro.2013.12.024. in press. [DOI] [PubMed] [Google Scholar]
- 39.Zhong G., Yan H., Wang H., He W., Jing Z., Qi Y., Fu L., et al. Sodium taurocholate cotransporting polypeptide mediates woolly monkey hepatitis B virus infection of Tupaia hepatocytes. J. Virol. 2013;87:7176–7184. doi: 10.1128/JVI.03533-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotani N., Maeda K., Debori Y., Camus S., Li R., Chesne C., Sugiyama Y. Expression and transport function of drug uptake transporters in differentiated HepaRG cells. Mol. Pharm. 2012;9:3434–3441. doi: 10.1021/mp300171p. [DOI] [PubMed] [Google Scholar]
- 41.Kullak-Ublick G.A., Beuers U., Paumgartner G. Molecular and functional characterization of bile acid transport in human hepatoblastoma HepG2 cells. Hepatology. 1996;23:1053–1060. doi: 10.1002/hep.510230518. [DOI] [PubMed] [Google Scholar]
- 42.Watashi K., Sluder A., Daito T., Matsunaga S., Ryo A., Nagamori S., Iwamoto M., Nakajima S., Tsukuda S., Borroto-Esoda K., et al. Cyclosporin A and its analogs inhibit hepatitis B virus entry into cultured hepatocytes through targeting a membrane transporter NTCP. Hepatology. 2014 doi: 10.1002/hep.26982. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwamoto M., Watashi K., Tsukuda S., Aly H.H., Fukasawa M., Fujimoto A., Suzuki R., Aizaki H., Ito T., Koiwai O., et al. Evaluation and identification of hepatitis B virus entry inhibitors using HepG2 cells overexpressing a membrane transporter NTCP. Biochem. Biophys. Res. Commun. 2014;443:808–813. doi: 10.1016/j.bbrc.2013.12.052. [DOI] [PubMed] [Google Scholar]
- 44.Yan H., Peng B., He W., Zhong G., Qi Y., Ren B., Gao Z., Jing Z., Song M., Xu G., et al. Molecular determinants of hepatitis B and D virus entry restriction in mouse sodium taurocholate cotransporting polypeptide. J. Virol. 2013;87:7977–7991. doi: 10.1128/JVI.03540-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nkongolo S., Ni Y., Lempp F.A., Kaufman C., Lindner T., Esser-Nobis K., Lohmann V., Mier W., Mehrle S., Urban S. Cyclosporin A inhibits Hepatitis B and Hepatitis D Virus entry by Cyclophilin-independent interference with the NTCP receptor. J. Hepatol. 2014 doi: 10.1016/j.jhep.2013.11.022. in press. [DOI] [PubMed] [Google Scholar]
- 46.Taylor J.M. Hepatitis delta virus. Virology. 2006;344:71–76. doi: 10.1016/j.virol.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 47.Lindenbach B.D., Rice C.M. The ins and outs of hepatitis C virus entry and assembly. Nat. Rev. Microbiol. 2013;11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeisel M.B., Lupberger J., Fofana I., Baumert T.F. Host-targeting agents for prevention and treatment of chronic hepatitis C—Perspectives and challenges. J. Hepatol. 2013;58:375–384. doi: 10.1016/j.jhep.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 49.Dorner M., Horwitz J.A., Robbins J.B., Barry W.T., Feng Q., Mu K., Jones C.T., Schoggins J.W., Catanese M.T., Burton D.R., et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuroki K., Eng F., Ishikawa T., Turck C., Harada F., Ganem D. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J. Biol. Chem. 1995;270:15022–15028. doi: 10.1074/jbc.270.25.15022. [DOI] [PubMed] [Google Scholar]
- 51.Breiner K.M., Urban S., Schaller H., Carboxypeptidase D. (gp180), a Golgi-resident protein, functions in the attachment and entry of avian hepatitis B viruses. J. Virol. 1998;72:8098–8104. doi: 10.1128/jvi.72.10.8098-8104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Urban S., Breiner K.M., Fehler F., Klingmuller U., Schaller H. Avian hepatitis B virus infection is initiated by the interaction of a distinct pre-S subdomain with the cellular receptor gp180. J. Virol. 1998;72:8089–8097. doi: 10.1128/jvi.72.10.8089-8097.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor J.M. Virus entry mediated by hepatitis B virus envelope proteins. World J. Gastroenterol. 2013;19:6730–6734. doi: 10.3748/wjg.v19.i40.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Claro da Silva T., Polli J.E., Swaan P.W. The solute carrier family 10 (SLC10): Beyond bile acid transport. Mol. Aspects Med. 2013;34:252–269. doi: 10.1016/j.mam.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eloranta J.J., Jung D., Kullak-Ublick G.A. The human Na+-taurocholate cotransporting polypeptide gene is activated by glucocorticoid receptor and peroxisome proliferator-activated receptor-gamma coactivator-1alpha, and suppressed by bile acids via a small heterodimer partner-dependent mechanism. Mol. Endocrinol. 2006;20:65–79. doi: 10.1210/me.2005-0159. [DOI] [PubMed] [Google Scholar]
- 56.Ananthanarayanan M., Ng O.C., Boyer J.L., Suchy F.J. Characterization of cloned rat liver Na(+)-bile acid cotransporter using peptide and fusion protein antibodies. Am. J. Physiol. 1994;267:G637–G643. doi: 10.1152/ajpgi.1994.267.4.G637. [DOI] [PubMed] [Google Scholar]
- 57.Doring B., Lutteke T., Geyer J., Petzinger E. The SLC10 carrier family: Transport functions and molecular structure. Curr. Top. Membr. 2012;70:105–168. doi: 10.1016/B978-0-12-394316-3.00004-1. [DOI] [PubMed] [Google Scholar]
- 58.Kosters A., Karpen S.J. Bile acid transporters in health and disease. Xenobiotica. 2008;38:1043–1071. doi: 10.1080/00498250802040584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hagenbuch B., Meier P.J. Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter. J. Clin. Invest. 1994;93:1326–1331. doi: 10.1172/JCI117091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hallen S., Mareninova O., Branden M., Sachs G. Organization of the membrane domain of the human liver sodium/bile acid cotransporter. Biochemistry. 2002;41:7253–7266. doi: 10.1021/bi012152s. [DOI] [PubMed] [Google Scholar]
- 61.Mareninova O., Shin J.M., Vagin O., Turdikulova S., Hallen S., Sachs G. Topography of the membrane domain of the liver Na+-dependent bile acid transporter. Biochemistry. 2005;44:13702–13712. doi: 10.1021/bi051291x. [DOI] [PubMed] [Google Scholar]
- 62.Hu N.J., Iwata S., Cameron A.D., Drew D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature. 2011;478:408–411. doi: 10.1038/nature10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou X., Levin E.J., Pan Y., McCoy J.G., Sharma R., Kloss B., Bruni R., Quick M., Zhou M. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature. 2013;505:569–573. doi: 10.1038/nature12811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hallen S., Branden M., Dawson P.A., Sachs G. Membrane insertion scanning of the human ileal sodium/bile acid co-transporter. Biochemistry. 1999;38:11379–11388. doi: 10.1021/bi990554i. [DOI] [PubMed] [Google Scholar]
- 65.Zhang E.Y., Phelps M.A., Banerjee A., Khantwal C.M., Chang C., Helsper F., Swaan P.W. Topology scanning and putative three-dimensional structure of the extracellular binding domains of the apical sodium-dependent bile acid transporter (SLC10A2) Biochemistry. 2004;43:11380–11392. doi: 10.1021/bi049270a. [DOI] [PubMed] [Google Scholar]
- 66.Ho R.H., Leake B.F., Roberts R.L., Lee W., Kim R.B. Ethnicity-dependent polymorphism in Na+-taurocholate cotransporting polypeptide (SLC10A1) reveals a domain critical for bile acid substrate recognition. J. Biol. Chem. 2004;279:7213–7222. doi: 10.1074/jbc.M305782200. [DOI] [PubMed] [Google Scholar]
- 67.Pan W., Song I.S., Shin H.J., Kim M.H., Choi Y.L., Lim S.J., Kim W.Y., Lee S.S., Shin J.G. Genetic polymorphisms in Na+-taurocholate co-transporting polypeptide (NTCP) and ileal apical sodium-dependent bile acid transporter (ASBT) and ethnic comparisons of functional variants of NTCP among Asian populations. Xenobiotica. 2011;41:501–510. doi: 10.3109/00498254.2011.555567. [DOI] [PubMed] [Google Scholar]
- 68.Yan H., Peng B., Liu Y., Xu G., He W., Ren B., Jing Z., Sui J., Li W. Viral entry of Hepatitis B and D viruses and bile salts transportation share common molecular determinants on sodium taurocholate cotransporting polypeptide. J. Virol. 2014 doi: 10.1128/JVI.03478-13. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kilby J.M., Hopkins S., Venetta T.M., DiMassimo B., Cloud G.A., Lee J.Y., Alldredge L., Hunter E., Lambert D., Bolognesi D., et al. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 70.Warner N., Locarnini S. The new front-line in hepatitis B/D research: Identification and blocking of a functional receptor. Hepatology. 2013;58:9–12. doi: 10.1002/hep.26292. [DOI] [PubMed] [Google Scholar]
- 71.Watashi K., Shimotohno K. Cyclophilin and viruses: Cyclophilin as a cofactor for viral infection and possible anti-viral target. Drug Target Insights. 2007;2:9–18. [PMC free article] [PubMed] [Google Scholar]
- 72.Watashi K., Hijikata M., Hosaka M., Yamaji M., Shimotohno K. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology. 2003;38:1282–1288. doi: 10.1053/jhep.2003.50449. [DOI] [PubMed] [Google Scholar]
- 73.Bienkowska-Haba M., Patel H.D., Sapp M. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog. 2009;5:e1000524. doi: 10.1371/journal.ppat.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu X., Zhao Z., Li Z., Xu C., Sun L., Chen J., Liu W. Cyclosporin A inhibits the influenza virus replication through cyclophilin A-dependent and -independent pathways. PLoS One. 2012;7:e37277. doi: 10.1371/journal.pone.0037277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luban J., Bossolt K.L., Franke E.K., Kalpana G.V., Goff S.P. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 76.Pfefferle S., Schopf J., Kogl M., Friedel C.C., Muller M.A., Carbajo-Lozoya J., Stellberger T., von Dall’Armi E., Herzog P., Kallies S., et al. The SARS-coronavirus-host interactome: Identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7:e1002331. doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qing M., Yang F., Zhang B., Zou G., Robida J.M., Yuan Z., Tang H., Shi P.Y. Cyclosporine inhibits flavivirus replication through blocking the interaction between host cyclophilins and viral NS5 protein. Antimicrob. Agents Chemother. 2009;53:3226–3235. doi: 10.1128/AAC.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Towers G.J., Hatziioannou T., Cowan S., Goff S.P., Luban J., Bieniasz P.D. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 2003;9:1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- 79.Bouchard M.J., Puro R.J., Wang L., Schneider R.J. Activation and inhibition of cellular calcium and tyrosine kinase signaling pathways identify targets of the HBx protein involved in hepatitis B virus replication. J. Virol. 2003;77:7713–7719. doi: 10.1128/JVI.77.14.7713-7719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watashi K. Alisporivir, a cyclosporin derivative that selectively inhibits cyclophilin, for the treatment of HCV infection. Curr. Opin. Investig. Drugs. 2010;11:213–224. [PubMed] [Google Scholar]
- 81.Membreno F.E., Espinales J.C., Lawitz E.J. Cyclophilin inhibitors for hepatitis C therapy. Clin. Liver Dis. 2013;17:129–139. doi: 10.1016/j.cld.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 82.Lucifora J., Esser K., Protzer U. Ezetimibe blocks hepatitis B virus infection after virus uptake into hepatocytes. Antiviral Res. 2013;97:195–197. doi: 10.1016/j.antiviral.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 83.Dong Z., Ekins S., Polli J.E. Structure-activity relationship for FDA approved drugs as inhibitors of the human sodium taurocholate cotransporting polypeptide (NTCP) Mol. Pharm. 2013;10:1008–1019. doi: 10.1021/mp300453k. [DOI] [PMC free article] [PubMed] [Google Scholar]