Abstract
Glutathione peroxidases (GPX) catalyze the reduction of H2O2 or organic hydroperoxides to water or corresponding alcohols using reduced glutathione, which plays an essential role in ROS (reactive oxygen species) homeostasis and stress signaling. Thellungiella salsuginea (Eutrema salsugineum), a relative of Arabidopsis thaliana, displays an extremely high level of tolerance to salt, drought, cold and oxidative stresses. The enzymatic antioxidant systems may contribute to the stress tolerance of T. salsuginea. In the present study, we aimed at understanding the roles of the antioxidant enzymes in T. salsuginea by focusing on the GPX family. We identified the eight GPX genes in T. salsuginea, and the structure of the N-terminal domains indicated their putative chloroplastic, mitochondrial and cytoplasmic location. The exon-intron organization of these genes exhibited a conserved pattern among plant GPX genes. Multiple environmental stresses and hormone response related cis-acting elements were predicted in the promoters of TsGPX genes. The gene and protein expression profiles of TsGPXs in response to high level of salinity and osmotic stresses, in leaves and roots of T. salsuginea were investigated using real-time RT-PCR and western blotting analysis. Our result showed that different members of the GPX gene family were coordinately regulated under specific environmental stress conditions, and supported the important roles of TsGPXs in salt and drought stress response in T. salsuginea.
Keywords: Thellungiella salsuginea, glutathione peroxidase, salt stress, drought stress, gene family
1. Introduction
In cells, reactive oxygen species (ROS) such as superoxide (•O2 −), hydrogen peroxide (H2O2), and hydroxyl radical (•OH) are produced during basic metabolic processes like respiration and photosynthesis. In addition, environmental stresses can also promote ROS production, leading to oxidative stress [1]. Although ROS was demonstrated to play an important role in signal transduction when cells are exposed to unfavorable conditions [2], accumulated ROS may result in uncontrolled oxidation of various cellular components, leading to free radical-mediated destruction of the cell structure [3], such as membrane and protein modifications. Eukaryotic cells have evolved elaborate and complicated antioxidant systems, including enzymatic and non-enzymatic components, to maintain the homeostasis between ROS production and elimination. The enzymatic antioxidant systems include superoxide dismutase (SOD), catalase (CAT), glutathione peroxidases (GPX), and glutathione-S-transferase (GST).
As a large family of antioxidant enzymes, glutathione peroxidases (GPXs) reduce H2O2 and organic hydroperoxides to water and correspondingly alcohols using reduced glutathione (GSH), inhibit the ROS-induced damage to membrane and protein, and play crucial roles in protecting cells from oxidative damage [4]. GPXs have attracted extensive investigation in animals and human, and animal GPXs were divided into five classes: cytosolic GPX, gastro-intestinal GPX, plasma GPX, phospholipid hydroperoxide GPX (PHGPX), and seleno-independent epididymis GPX [5]. GPX1 was demonstrated to be an important indicator for cardiovascular diseases prognosis [6], however, our understanding of the structure and function of plant GPXs is limited.
At present, GPX genes from several plant species, such as Nicotiana sylvestris [7], Citrus sinensis [8], Avena fatua [9], Arabidopsis thaliana [10], Brassica campestris [11], Spinacia oleracea [12], Helianthus annuus [13], Pisum sativum [14], Lycopersicon esculentum [15], Oryza sativa [16], Triticum aestivum [17], and Panax ginseng [18], have been isolated and characterized, however, few genome-wide GPX family identification and characterization studies have been reported. Three GPX family studies were performed in A. thaliana [19], Lotus japonicas [20], and Populus trichocarpa [21]; these studies revealed that plant GPX family consists of multiple GPXs with distinct subcellular location and functions, and that members exhibit different tissue-specific expression patterns and environmental stress responses, functioning coordinately in ROS scavenging. These works also highlight the importance of GPX study at the genome-wide level.
Thellungiella salsuginea (Eutrema salsugineum), a Brassicaceae species closely related to A. thaliana [22], can survive in environments with high soil salinity, low temperature, or water deficiency [23,24]. Compared to A. thaliana, T. salsuginea was more tolerant not only to high salinity but also to oxidative stress [25], and the level of lipid hydroperoxides in T. salsuginea leaves under salt stress was remarkably lower than that of A. thaliana [26], indicating that the enzymatic antioxidant systems in T. salsuginea behaves more efficiently. However, the genetic basis underlying this physiological mechanism of T. salsuginea is still not clear. To date, there is no report on expression profiling of GPXs in T. salsuginea under abiotic stress conditions. In the present study, we identified eight GPX genes in T. salsuginea and analyzed their gene structure and promoter sequences. The potential subcellular locations of TsGPX genes were predicted. We examined the expression of TsGPX transcripts and proteins in T. salsuginea leaves and roots that were exposed to short-term salt and osmotic stresses.
2. Results and Discussion
2.1. Identification and Characterization of TsGPX Genes
We isolated and characterized the GPX genes of T. salsuginea. We searched the T. salsuginea ESTs in GenBank using the coding regions (CDs) of GPX1-8 from A. thaliana, a close relative of T. salsuginea. As a result, 7, 1, 1, 1, 2, 2, and 1 ESTs were found, which represent putative TsGPX1, TsGPX2, TsGPX3, TsGPX5, TsGPX6, TsGPX7, and TsGPX8, respectively (Table S1). We then assembled all ESTs representing the same TsGPX genes, and the full length CDs of TsGPX1, TsGPX2, TsGPX6, and TsGPX8 were obtained. Because the ESTs or the assembled sequences for TsGPX3, TsGPX5 and TsGPX7 did not cover the 3′ end of the corresponding TsGPX genes, we isolated the full length CDs of TsGPX3, TsGPX5, and TsGPX7 using 3′ RACE (Tables S2 and S3, Figure S1). Since the EST sequence for TsGPX4 was not available, the full length CDs of TsGPX4 was isolated by PCR using degenerate primers (Table S3) designed according to the cDNA sequence of AtGPX4.
We further examined all putative TsGPX genes in a recent released version of T. salsuginea genome in Phytozome (http://www.phytozome.net/). Thirteen TsGPX transcripts were found by blasting with the conserved amino acid sequence and the CDs of GPX1-8 as the query sequences (Table 1). The thirteen TsGPX transcripts were transcribed from nine genomic loci, indicating that several transcripts were products of alternative splicing. For example, Thhalv10000311m and Thhalv10000351m were transcribed from GPX1 locus; Thhalv10001645m and Thhalv10001644m were transcribed from the GPX3 locus; Thhalv10006271m, Thhalv10006247m, and Thhalv10006248m were transcribed from the GPX5 locus.
Table 1.
TsGPX genes and their genomic locations.
| Number | Sequence ID | Name | Location in genome |
|---|---|---|---|
| 1 | Thhalv10000311m | GPX1 | scaffold_15: 4940063–4941983 |
| 2 | Thhalv10000351m | GPX1 isoform | scaffold_15: 4940063–4941470 |
| 3 | Thhalv10017319m | GPX2 | scaffold_10: 6460958–6462801 |
| 4 | Thhalv10001645m | GPX3 | scaffold_22: 63411–66356 |
| 5 | Thhalv10001644m | GPX3 isoform | scaffold_22: 63411–66356 |
| 6 | Thhalv10001660m | GPX4 | scaffold_22: 1860223–1861484 |
| 7 | Thhalv10006271m | GPX5 | scaffold_19: 155838–157677 |
| 8 | Thhalv10006247m | GPX5 isoform | scaffold_19: 155838–157955 |
| 9 | Thhalv10006248m | GPX5 isoform | scaffold_19: 155838–157677 |
| 10 | Thhalv10028932m | GPX6 | scaffold_3: 3990214–3992118 |
| 11 | Thhalv10029228m | Not determined | scaffold_3: 3955295–3956113 |
| 12 | Thhalv10026852m | GPX7 | scaffold_1: 3456273–3457732 |
| 13 | Thhalv10023725m | GPX8 | scaffold_8: 965149–966667 |
Thhalv10029228m; a GPX6-like transcript; encoded a polypeptide identical to the 3′ part of TsGPX6 (Thhalv10028932m). Thhalv10029228m may be a new GPX gene in T. salsuginea, since there is no corresponding gene in A. thaliana and other plant species. It may only represent a falsely assembled genomic sequence, since no such EST was found in GenBank or the high-throughput transcriptome database established in our lab (unpublished data). Therefore, we excluded Thhalv10029228m from further analysis in the present study, and will address the existence and the function of this gene in future.
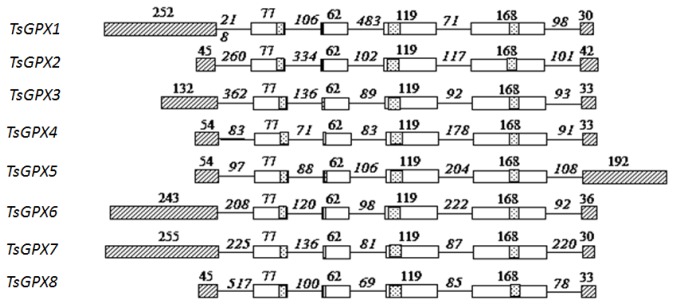
All the open reading frames (ORFs) of the eight TsGPX genes consisted of six exons and five introns (Figure 1). The same exon-intron organization was also observed for the eight GPX genes of A. thaliana (AtGPX) [19], the six GPX genes of poplar [21], and the five GPX genes of L. japonicus (LjGPX) [20], as well as some GPX genes of citrus [27] and wheat [17], suggesting a high degree of conservation among plant species in the exon-intron organization.
Figure 1.
Intron-exon organization of the eight TsGPX genes. The box represents exon, and the line represents intron. Dotted boxes show conserved domains, and slashed boxes represent exon 1 and exon 6. The numbers represent the length of the corresponding exon or intron.
We also observed a high degree of conservation among plant species in the exon lengths. Exons 2 to 5 have identical size for all the AtGPX and LjGPX genes, with the exception of exon 5 in LjGPX4. The lengths of exon 1 in TsGPX, AtGPX, and LjGPX are variable, since several GPX genes lack N-terminal signal peptides. The lengths of exon 6 in TsGPXs range from 30 to 42 bp, with the exception of TsGPX5, which is 192 bp in length (Figure 1). Although most TsGPX genes appear to be similar in exon length, the lengths of introns are highly variable; such features are also reported in GPX genes from other plant species, such as Arabidopsis and L. japonicas [19,20].
We noticed that, similar with the other plant GPXs, the three conserved domains, “NVASKCG”, “ILAFPCNQF”, and “KWNF (E/T/A) KFLV” [28], were located in the second, third, fourth, and fifth exon, respectively, in all the eight TsGPX genes.
Considering that plant GPX genes were reported to be involved in responses to various environmental stresses [10,17–19], we analyzed the promoter regions of the eight TsGPX genes to locate putative cis-acting regulatory elements related to stress and hormone signaling (Table 2) using PlantCARE software [29]. There were multiple cis-acting regulatory elements in each TsGPX promoters, with different ones in each promoter. The quantity of the cis-acting regulatory elements varied with the individual promoter. Almost all TsGPX promoters contained cis-acting elements that were responsive to methyl jasmonate (TsGPX2-8), five TsGPX promoters contained elements that are responsive to gibberellin (TsGPX1, 2, 4, 5, and 7), auxin (TsGPX1, 3, 4, 5, and 8) and drought (TsGPX1, 2, 3, 4, and 7), whereas fewer TsGPX promoters contained elements involved in response to salicylic acid (TsGPX1, 4, and 7), low-temperature (TsGPX1, 2, and 5), and ethylene (TsGPX6). We also analyzed the putative cis-acting regulatory elements in each AtGPX promoter (Table 2). Compared with the TsGPX promoters, relatively less stress and hormone response related cis-acting regulatory elements were located in AtGPX promoters.
Table 2.
Putative cis-acting regulatory elements related to stress and hormone response in TsGPX and AtGPX promoters.
| Environmental stress or hormone | Cis-acting regulatory elements | Sequence | TsGPX | AtGPX |
|---|---|---|---|---|
| Abscisic acid (ABA) | ABRE | TACGTG | TsGPX2, 4, 5, and 7 | AtGPX1, 4,5, and 8 |
|
| ||||
| Auxin | TGA-element | AACGAC | TsGPX1 and 8 | AtGPX2 and 8 |
| AuxRR-core | GGTCCAT | TsGPX3, 4, and 5 | - | |
|
| ||||
| Salicylic acid (SA) | TCA-element | CCATCTTTTT | TsGPX1 and 4 | AtGPX4 |
| GAGAAGAATA | TsGPX7 | AtGPX3 and 7 | ||
|
| ||||
| Gibberellin (GA) | TATC-box | TATCCCA | TsGPX1 | - |
| GARE-motif | TCTGTTG | TsGPX2 | - | |
| P-box | GCCTTTTGAGT | TsGPX4 and 5 | AtGPX5, and 7 | |
| GARE-motif | TCTGTTG | TsGPX5 | - | |
| AAACAGA | TsGPX7 | AtGPX3, 6, and 7 | ||
|
| ||||
| Methyl Jasmonate (MeJA) | CGTCA-motif | CGTCA | TsGPX2, 3, 4, 5, 6, 7, and 8 | AtGPX3, 4, 5, 7, and 8 |
| TGACG-motif | TGACG | TsGPX2, 3, 4, 5, 6, 7, and 8 | AtGPX3, 4, 5, 7, and 8 | |
|
| ||||
| Ethylene | ERE | ATTTCAAA | TsGPX6 | AtGPX1, 3 |
|
| ||||
| Drought inducibility | MBS | TAACTG | TsGPX1, 2, 3, 4, and 7 | AtGPX2, 5, 7 |
|
| ||||
| Low-temperature | LTR | CCGAAA | TsGPX1, 2, and 5 | AtGPX5 and 8 |
|
| ||||
| Anaerobic induction | ARE | TGGTTT | TsGPX2, 3, 6,and 8 | AtGPX2, 3, 7, and 8 |
2.2. Predicted Properties of TsGPX Proteins
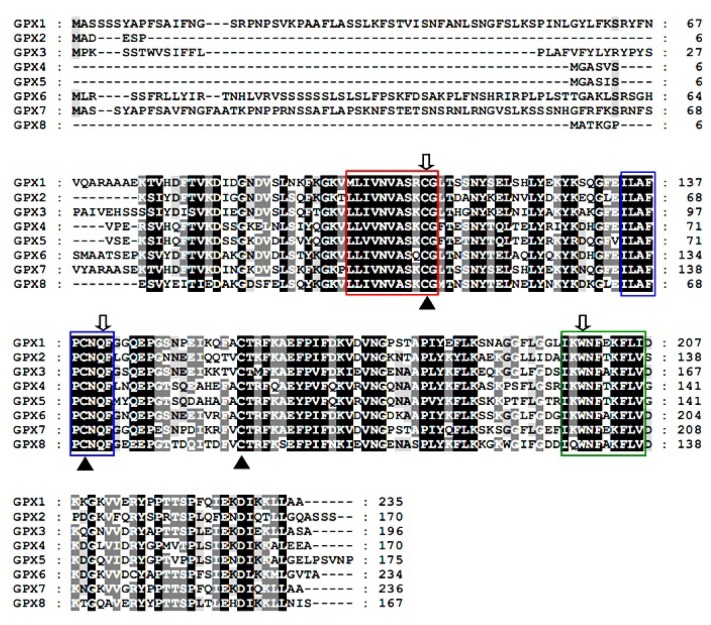
The derived amino acid sequences of the eight TsGPX proteins contain not only the three motifs present in most plant GPXs [28], but also other conserved sequences such as NY (T/K/S) (E/Q) L, CT (R/K/M) F (K/Q) (A/S) EFPIF, and PLY (K/E/Q) FLK (Figure 2). These motifs contain the three residues, C (Cys), Q (Gln) and W (Trp), which are proposed to be part of the catalytic site, and the three conserved Cys residues.
Figure 2.
Amino acid sequences of Thellungiella salsuginea glutathione peroxidase (TsGPX) proteins. The conserved three Cys residues of plant GPX proteins are indicated by triangles. Boxed sequences represent highly conserved domains in which residues forming a catalytic triad in GPX are marked with an arrow.
According to the alignment of TsGPX sequences shown in Figure 3 and the data presented in Table 3, the eight TsGPX proteins can be divided into three categories based on the amino acid sequence length: TsGPX1, TsGPX6 and TsGPX7 (c. 235 amino acids; 26 kDa); TsGPX2, TsGPX4, TsGPX5, and TsGPX8 (c. 170 amino acids; 19 kDa); and TsGPX3 (196 amino acids; 23 kDa). The poor homology among the N-terminal amino acid residues of TsGPX1, TsGPX6, and TsGPX7 indicated that these proteins bear signal peptides for organelle targeting (Figure 3). Protein subcellular localization prediction programs suggested that TsGPX1 and TsGPX7 has a chloroplastic N-terminal transit peptide and that TsGPX6 has an N-terminal transit peptide for targeting to mitochondria. TsGPX3 was predicted to be located in the secretory pathway (Table 3).
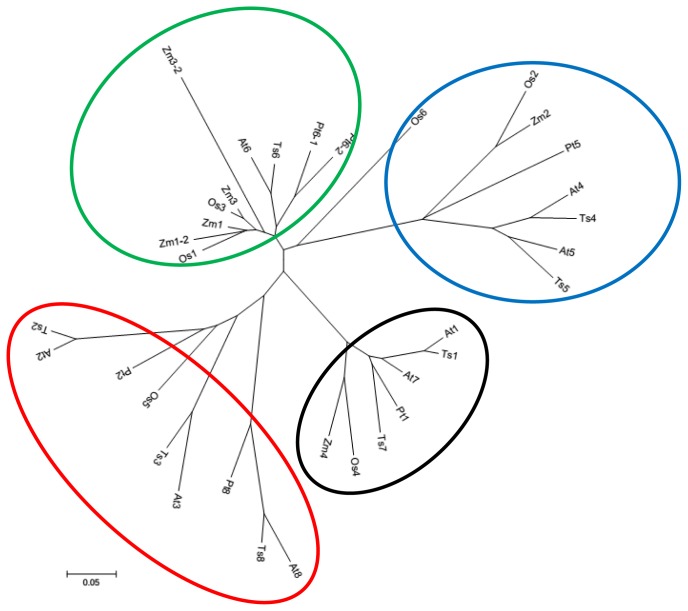
Figure 3.
Phylogenetic analyses of thirty four plant glutathione peroxidase (GPX) proteins. The tree was constructed using the neighbor-joining method of CLUSTALW, with 1000 bootstraps, and the bar indicates 0.05 substitutions per site. Each ellipse shows a clade. Abbreviations of plant species: Ts, Thellungiella salsuginea; At, Arabidopsis thaliana; Os, Oryza sativa; Zm, Zea mays, Pt, Populus trichocarpa. The plant GPXs include At1 (At2g25080), At2 (At2g31570), At3 (At2g43350), At4 (At2g48150), At5 (At3g63080), At6 (At4g11600), At7 (At4g31870), At8 (At1g63460), Pt1 (POPTR_0006s28120), Pt2 (POPTR_0007s02160), Pt5 (POPTR_0014s13490), Pt6-1 (POPTR_0001s09270), Pt6-2 (POPTR_0003s12620), Pt8 (POPTR_0001s09280), Os1 (Os04g0556300), Os2 (Os03g0358100), Os3 (Os02g0664000), Os4 (Os06g0185900), Os5 (Os11g18170), Os6 (A3AYS5_ORYSJ), Zm1 (Q6JAH6_MAIZE), Zm1-2 (B6SU31_MAIZE), Zm2 (B6T5N2_MAIZE), Zm3 (B4FRF0_MAIZE), Zm3-2 (AC204541), and Zm4 (B6U7S4_MAIZE).
Table 3.
The physical and chemical properties of TsGPXs.
| Name | Length (aa) | Molecular mass (kDa) | pI | TP * | Subcellular localization * | Transmembrane domain * |
|---|---|---|---|---|---|---|
| TsGPX1 | 235 | 25.902 | 9.94 | 73 | chloro | no |
| TsGPX2 | 170 | 19.020 | 5.48 | – | cyt | no |
| TsGPX3 | 196 | 23.258 | 7.33 | – | SP, CM, ER | yes |
| TsGPX4 | 170 | 19.147 | 9.08 | – | cyt | no |
| TsGPX5 | 175 | 19.612 | 9.79 | – | cyt | no |
| TsGPX6 | 234 | 25.937 | 9.35 | 58 | mito | no |
| TsGPX7 | 236 | 26.227 | 10.30 | 75 | chloro | no |
| TsGPX8 | 167 | 18.990 | 4.75 | – | cyt | no |
Transit peptides (number of amino acid residues) and subcellular localizations (mito, mitochondria; chloro, chloroplasts; cyt, cytosol; SP, secretary pathway; ER, endoplasmic reticulum; CM, cytoplasmic membrane) of TsGPX proteins, as predicted by the TARGETP and PSORT programs.
The amino acid sequences of the eight TsGPXs, along with eight AtGPXs, six ZmGPXs, six OsGPXs, and six PtGPXs, were used to build an unrooted phylogenetic tree (Figure 3). The tree is composed of four clades. The black clade included TsGPX1, TsGPX7, AtGPX1, AtGPX7, and PtGPX1. The red clade included TsGPX2, TsGPX3, and TsGPX8, as well as the corresponding Arabidopsis GPXs, and PtGPX2 and PtGPX8. The blue clade included TsGPX4, TsGPX5, AtGPX4, AtGPX5, and PtGPX5. The green clade included TsGPX6, AtGPX6, PtGPX6-1, and PtGPX6-2. From this amino acid sequence comparison, it is clear that Thellungiella GPXs are more closely related to Arabidopsis GPXs than to rice and Populus GPXs.
Although TsGPX3 has a relatively larger molecular weight, it was clustered in the same clade as TsGPX2 and TsGPX8, two GPXs with smaller molecular weights, which indicate that TsGPX2 and TsGPX8 are more closely related to TsGPX3 than to TsGPX4 and TsGPX5. The three largest TsGPXs clustered into two clades; TsGPX1 and TsGPX7, two GPXs targeted to chloroplast, were clustered in the black clade, while the mitochondria targeted TsGPX6 was clustered in the green clade. These results are consistent with a previous study [30], and support our subcellular localization prediction for TsGPXs.
2.3. Expression Analyses of GPX Genes in T. salsuginea Plants Exposed to High Salinity and Osmotic Treatment
Considering that multiple stress-response cis-acting elements were predicted in the promoters of Ts GPX genes, we investigated the expression profiles of TsGPX in T. salsuginea exposed to high salinity and osmotic stress treatments at the transcriptional level using quantitative RT-PCR technology (Figure 4).
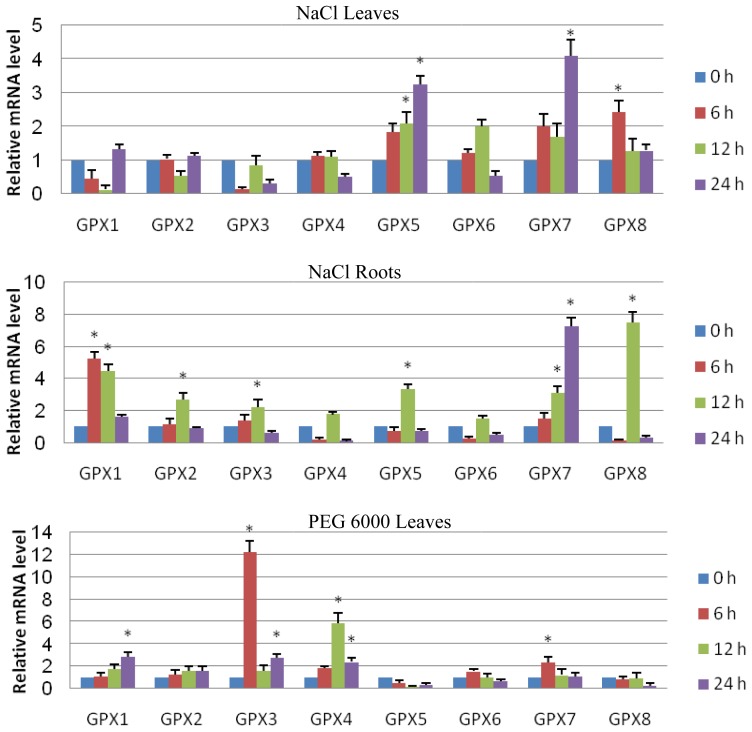
Figure 4.
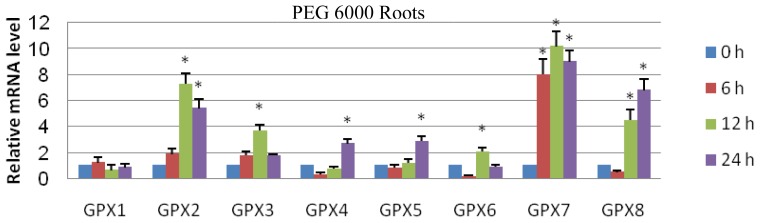
Time-course expression analysis of Thellungiella salsuginea glutathione peroxidase (TsGPX) genes in leaves or roots of T. salsuginea exposed to NaCl (300 mM) or PEG 6000 (20%). mRNA levels were normalized with respect to TsTubulin and are expressed relative to the values at 0 h (control), which were given an arbitrary value of 1. Data represent the means ± SE of at least three replicates. Asterisks denote significant up-regulation (ratio > 2.0) or down-regulation (ratio < 0.5).
Short-term exposure of T. salsuginea plants to salt stress (300 mM NaCl) induced or repressed the expression of some members of TsGPX family genes, and the effect varied with tissues, i.e., leaves and roots. In leaves, three TsGPX genes (TsGPX5, TsGPX7, and TsGPX8) were highly induced by salt stress at least at one time-point during salt treatment, with the highest transcription level of over 4-fold induced (Figure 4). While in roots, six TsGPX genes (TsGPX1, TsGPX2, TsGPX3, TsGPX5, TsGPX7, and TsGPX8) were highly induced by salt stress, with the highest transcription level of over 7-fold induced (Figure 4). Three TsGPX genes, TsGPX5, TsGPX7, and TsGPX8, were highly induced by NaCl stress in both leaves and roots, suggesting that they may play important roles in high salinity tolerance in T. salsuginea plants.
The expression levels of TsGPX family genes were also regulated under short-term osmotic treatment. In leaves, four TsGPX genes (TsGPX1, TsGPX3, TsGPX4, and TsGPX7) were highly induced by osmotic stress at least at one time-point during treatment, with the highest transcription level of over 12-fold (Figure 4). While in roots, almost all TsGPX genes (except TsGPX1) were highly induced by osmotic stress, with the highest transcription level of over 10-fold (Figure 4). Three TsGPX genes, TsGPX3, TsGPX4, and TsGPX7, were highly induced by osmotic stress in both leaves and roots, suggesting that they may be important in osmotic stress tolerance in T. salsuginea plants.
According to the above results, TsGPX7 may be the most important GPX member for salt and osmotic response in T. salsuginea, since it was up-regulated by salt and osmotic stress in both leaves and roots. Similarly, the TsGPX3, TsGPX5, and TsGPX8 may also play important roles in salt and osmotic response in T. salsuginea plants. The functions of these TsGPXs in stress tolerance will be investigated in future studies using a transgenic approach.
T. salsuginea is an Arabidopsis-related extremophile, and analysis of the difference in gene expression between T. salsuginea and A. thaliana may promote our understanding of how plants cope with abiotic stresses [22,23]. To achieve this goal, we compared the gene expression data of GPX genes of T. salsuginea to the corresponding data of A. thaliana in the AtGenExpress (Table S4) [31]. Under salt stress, GPX5, GPX6, GPX7, and GPX8 were significantly up-regulated (ratio > 2) in T. salsuginea leaves, while GPX2, GPX6, and GPX8 were significantly up-regulated (ratio > 1.5) in A. thaliana leaves; GPX1, GPX2, GPX3, GPX5, GPX7, and GPX8 were significantly up-regulated in T. salsuginea roots, whereas GPX1, GPX2, GPX4, GPX6, and GPX7 were significantly up-regulated in A. thaliana roots. GPX5 were found to response to salt stress in T. salsuginea, but not in A. thaliana, supporting our hypothesis that TsGPX5 is essential in salt stress response in T. salsuginea.
Under osmotic stress, GPX1, GPX3, GPX4, and GPX7 were significantly up-regulated in T. salsuginea leaves, while only GPX3 and GPX6 were significantly up-regulated in A. thaliana leaves; All GPX genes, except GPX1, were significantly up-regulated in T. salsuginea roots, whereas in A. thaliana roots, only GPX1 was significantly up-regulated, although GPX6 and GPX7 were also up-regulated (ratio > 1.4).
The above results reveal that the TsGPX family genes exhibit different stress regulated expression patterns from that of A. thaliana and more GPX genes were induced under salt and osmotic stress conditions in T. salsuginea. This is consistent with the cis-acting elements shown in Table 2, in which less environmental stress and hormone response related cis-acting regulatory elements were found in AtGPX promoters. Our results support the hypothesis about the high stress tolerance of T. salsuginea, and that differences in salt tolerance mechanisms between salt-sensitive glycophytes, such as A. thaliana, and salt-tolerant halophytes, such as T. salsuginea, are suggested to result from changes in the regulation of the same basic set of genes involved in salt tolerance [32,33].
However, the GPX gene expression difference between T. salsuginea and A. thaliana should be explained cautiously, since the experiments are carried out in different conditions, and the stress treatment methods were also not identical.
2.4. Protein Expression Analyses of GPX Genes in T. salsuginea Plants Exposed to High Salinity and Osmotic Treatments
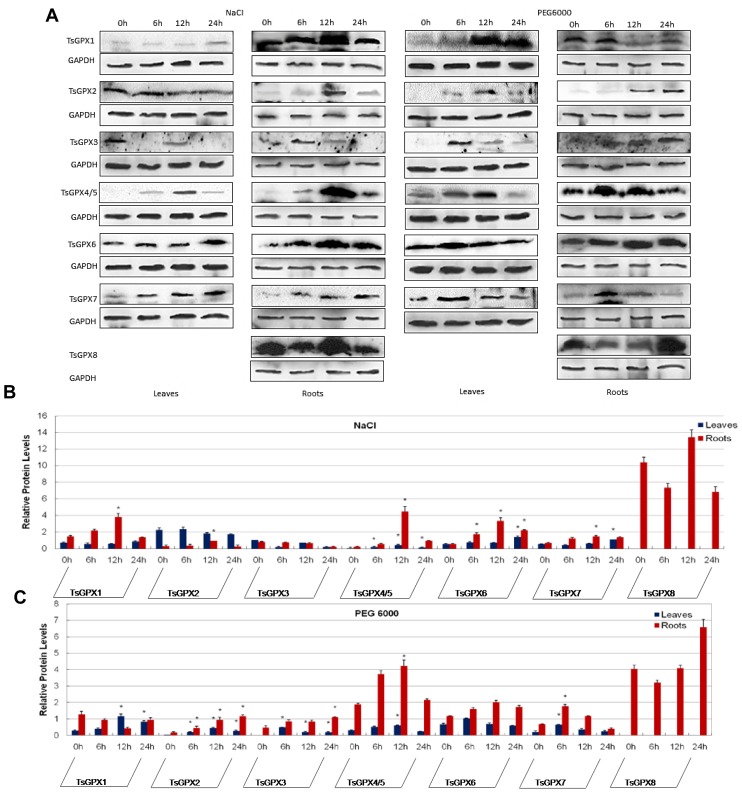
We further conducted Western blotting analysis to investigate the protein expression level of TsGPX genes under stress conditions, since the gene expression level is not always correlated with protein abundance [34]. The antisera against unique polypeptide sequence of each TsGPX proteins (Table S5) were prepared through rabbit immunization. It is noteworthy that, because of the high sequence similarity between TsGPX4 and TsGPX5, one antiserum was prepared and used for detecting both proteins. The TsGPX8 cannot be detected in leaves, which may be due to its low abundance in leaves.
Short-term salt stress affected the protein abundance of some members of the TsGPX family. In leaves, TsGPX4/TsGPX5, TsGPX6, and TsGPX7 showed an up-regulated pattern at least at one time-point during salt treatment, reaching their peaks at 24 h (TsGPX6 and TsGPX7), or at 12 h (TsGPX4/TsGPX5) (Figure 5). While in roots, almost all TsGPXs (except TsGPX3) were induced at least at one time-point during salt treatment, reaching their peaks at 12 h.
Figure 5.
Western blotting and quantification. (A) Western blotting analysis of Thellungiella salsuginea glutathione peroxidase (TsGPX) in leaves or roots of T. salsuginea before (0) and at 6-, 12-, and 24-h after NaCl or PEG 6000 treatment. Anti-OsGAPDH antibody was used as a control for loading error; (B) Bar graph for the quantitative comparison among TsGPX protein expression levels during NaCl treatment; (C) Bar graph for the quantitative comparison among TsGPX protein expression levels during PEG 6000 treatment. TsGPX- and OsGAPDH-specific bands were quantified by ImageJ software and TsGPX protein expression levels were normalized to GAPDH values. Mean values ± SD of 3 independent experiments are shown. Asterisks denote significant up-regulation (ratio > 2.0) or down-regulation (ratio < 0.5).
Western blotting analysis also revealed that the protein expressions of TsGPX2, TsGPX3, TsGPX4/TsGPX5, and TsGPX7 were induced under short-term osmotic treatment in both leaves and roots, although their abundance peaks emerged at different time-points. For example, the abundance of TsGPX7 reached peak at 6 h in both leaves and roots, while TsGPX2 reached peak at 12 h in leaves and 24 h in roots.
Quantitative real-time RT-PCR is a widely used method for monitoring gene expression at the transcriptional level, while Western blotting is a widely accepted analytical technique to detect specific proteins in a sample of tissue extract. The regulation patterns of TsGPXs in T. salsuginea plants under stress conditions as revealed by both approaches is consistent in general, with some minor differences. For example, the TsGPX3, TsGPX5, TsGPX7 and TsGPX8, four GPXs shown to be important for salt and osmotic response in real time RT-PCR analyses, were found to be up-regulated in protein level in most of the stress conditions. It is also noteworthy that the antiserum used in the present study cannot detect TsGPX8 protein in leaves, while the gene expression of TsGPX8 was determined readily by the quantitative real-time RT-PCR, suggesting that the protein abundance of TsGPX8 may be very low in leaves of T. salsuginea.
3. Experimental Section
3.1. Plant Materials and Stress Treatments
Thellungiella salsuginea (Shandong ecotype) seedlings were grown in pots containing a mixture of vermiculite and perlite (1:1, v/v) under fluorescent light (120 μmol m−2 s−1, 16 h light/8 h dark) at 25 °C and 35%–50% relative humidity in a growth chamber. The plantlets were watered in a three-day interval with half strength of Hoagland’s solution.
Five-week-old plants were used as the starting materials for stress treatments. The salt treatment was applied by watering the seedling with 300 mM NaCl in half strength Hoagland solution, and the osmotic treatment was applied by watering the seedling with 20% PEG 6000 solution. The leave and root samples were collected 6, 12, and 24 h after treatment. The seedlings before treatment were used as the control (0 h). The tissue samples were immediately frozen in liquid nitrogen, and then stored at −80 °C for extracting RNA and protein. At least three biological repeats from each treatment were prepared for real-time PCR and western blotting analysis.
To monitor the water status of the stressed plants, the relative water contents (RWC) or the water potentials of the leaves were measured according to a method described previously [35], or with a WP4 dewpoint potentiometer (Decagon Devices, Pullman, WA, USA), respectively (Figures S2 and S3).
3.2. Isolation of the Eight TsGPX Genes
We searched the T. salsuginea ESTs in the GenBank using the CDs of GPX1-8 from A. thaliana. The ESTs representing TsGPX genes were assembled using software DNASTAR (DNASTAR Inc., Madison, WI, USA). The primers used for isolating full length CDs of TsGPX gene were designed using Primer3 (http://primer3.ut.ee/). Total RNA from T. salsuginea seedlings was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The potential contaminating genomic DNA was treated with RQ1 RNase-free DNase (Promega, Madison, WI, USA). The cDNA was synthesized using BcaBEST RNA PCR Kit (Takara, Shiga, Japan) according to the manufacturer’s protocol. 3′ RACE were performed to isolate full length CDs of some members of TsGPX gene family using 3′ Full RACE Core Set Ver.2.0 kit (TaKaRa, Shiga, Japan). All primers are listed in Tables S2 and S3.
3.3. Sequence Alignments, Phylogenetic Analysis, and Other Bioinformatics Analysis
Molecular weight (MW) and isoelectric point (pI) predictions for each deduced TsGPXs were performed using the Compute pI/Mw tool (http://www.expasy.org/tools/protparam.html). The deduced sequences of eight TsGPXs were aligned with the DNAMAN program (version 4.0; Lynnon Corporation, Quebec, Canada) to identify conserved domains. These TsGPXs were also used, together with the sequences of the homologue proteins from other green plants, to construct an unrooted phylogenetic tree by the neighbour joining method with the ClustalW1.8 [36] and MEGA 6.0.5 [37] programs. Intron-exon organization of TsGPX genes were analyzed using an online tool, Spidey (http://www.ncbi.nlm.nih.gov/spidey/). Predictions of transit peptides and subcellular localization were conducted with the programs TargetP1.1 [38] and PSORT [39].
For promoter analysis, we first used Neural Network Promoter Prediction [40] (http://www.fruitfly.org/seq_tools/promoter.html) to locate the transcription start site in the 2 kb upstream region from translation start code ATG, and then predicted cis-acting elements using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [29].
3.4. Quantitative Real-Time RT-PCR
Approximately 1 μg of DNase I-digested total RNA was converted into single-stranded cDNA using M-MLV Reverse Transcriptase (Promega, Madison, WI, USA). The cDNA were diluted 50-fold with deionized water before use as a template in real-time PCR. The quantitative reaction was performed on a MyiQ2 two-color real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA) using the SsoFast EvaGreen Supermix (Bio-Rad Laboratories, Hercules, CA, USA). The reaction mixture (20 μL) contained 2× SsoFast EvaGreen Supermix, 0.9 μM each of the forward and reverse primers, and 1 μL of cDNA template. PCR amplification was carried out under the following conditions: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 10 s. Three independent biological replicates for each sample and three technical replicates of each biological replicate were analyzed in quantitative real-time PCR analysis. The gene expressions of TsGPXs were normalized against an internal reference gene, TsTubulin (Accession number: BY821407). The relative gene expression was calculated according to the 2−ΔΔCt method [41]. All primers used in this study are listed in Table S6. The specificity of primers was verified using amplicon dissociation curves and gel-electrophoretic analysis. We also sequenced the PCR products of each pair of primer to validate their specificity.
3.5. Antibody Preparation and Western Blotting Analysis
The antisera against TsGPXs were prepared by immunizing rabbits with the polypeptides representing individual TsGPXs (Table S5). One antiserum was prepared and used for detecting both TsGPX4 and TsGPX5, due to their high level of sequence similarity. The titers of antisera against TsGPX1, GPX2, GPX6, GPX4/GPX5, GPX7, and GPX8 were no less than 51,200, and the titer of TsGPX3 was 12,800. We further validated their efficacy using the E. coli expressed TsGPXs.
The rosette leaf proteins were extracted using a trichloroacetic acid/acetone method as described previously [42]. The protein concentration was measured using Bradford methods (Bioteke, Beijing, China). Bands on the membrane were finally visualized using a Pro-Light HRP Chemiluminescence Kit (TIANGEN, Beijing, China) and captured using a Tanon 5500 chemiluminescence Gel imaging system (Tanon, Shanghai, China). Anti-OsGAPDH antibody (Beijing Protein Innovation, Beijing, China) was used as the internal reference. TsGPX- and OsGAPDH-specific bands were quantified by ImageJ software (http://rsbweb.nih.gov/ij/).
4. Conclusions
In conclusion, we performed a genome-wide identification of the GPX family genes in T. salsuginea. The gene and protein expression profiles of TsGPXs in response to high salinity and osmotic stress, in leaves and roots of T. salsuginea were investigated using quantitative real-time RT-PCR and Western blotting analysis, and the difference in stress-induced expression patterns of GPX family genes were analyzed. Our results showed that in different tissues, different members of the GPX gene family were coordinately regulated under specific environmental stress conditions. The results presented in this study therefore provide evidence at both the transcriptional and protein levels that support the conclusion that GPXs, such as, GPX7, play important roles in salt and osmotic stress response in T. salsuginea.
Supplementary Information
Acknowledgments
This research was financially supported in part by the grants from the National Natural Science Foundation of China (grant No. 31070361 and 31370356), the Key Project of Chinese Ministry of Education (grant No. 210266); “985 Project” of Minzu University of China and 111 Project for Minzu University of China (grant No. B08044).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Author Contributions
Yinjun Zhou and Fei Gao conceived and designed the experiments, supervised the project, interpreted the data, and wrote the manuscript. Tingting Ma and Jing Chen conducted the quantitative real-time RT-PCR. Jing Chen, Huayun Li, Zhanglei Li, Ning Wang and Zichen Zhang performed the Western blotting analysis. Tingting Ma and Huayun Li conducted the bioinformatics analysis.
References
- 1.Grene R. Oxidative stress and acclimation mechanisms in plants. The Arabidopsis Book. 2002;1:e0036. doi: 10.1199/tab.0036.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foyer C.H., Noctor G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dat J., Vandenabeele S., Vranová E., van Montagu M., Inzé D., van Breusegem F. Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. 2000;57:779–795. doi: 10.1007/s000180050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ursini F., Maiorino M., Brigelius-Flohé R., Aumann K.D., Roveri A., Schomburg D., Flohé L. Diversity of glutathione peroxidases. Methods Enzymol. 1995;252:38–53. doi: 10.1016/0076-6879(95)52007-4. [DOI] [PubMed] [Google Scholar]
- 5.Herbette S., Roeckel-Drevet P., Drevet J.R. Seleno-independent glutathione peroxidases: More than simple antioxidant scavengers. FEBS J. 2007;274:2163–2180. doi: 10.1111/j.1742-4658.2007.05774.x. [DOI] [PubMed] [Google Scholar]
- 6.Espinola-Klein C., Rupprecht H.J., Bickel C., Schnabel R., Genth-Zotz S., Torzewski M., Lackner K., Munzel T., Blankenberg S. AtheroGene Investigators. Glutathione peroxidase-1 activity, atherosclerotic burden, and cardiovascular prognosis. Am. J. Cardiol. 2007;99:808–812. doi: 10.1016/j.amjcard.2006.10.041. [DOI] [PubMed] [Google Scholar]
- 7.Criqui M.C., Jamet E., Parmentier Y., Marbach J., Durr A., Fleck J. Isolation and characterization of a plant cDNA showing homology to animal glutathione peroxidases. Plant Mol. Biol. 1992;18:623–627. doi: 10.1007/BF00040684. [DOI] [PubMed] [Google Scholar]
- 8.Holland D., Ben-Hayyim G., Faltin Z., Camoin L., Strosberg A.D., Eshdat Y. Molecular characterization of salt-stress associated protein in citrus: protein and cDNA sequence homology to mammalian glutathione peroxidases. Plant Mol. Biol. 1993;21:923–927. doi: 10.1007/BF00027124. [DOI] [PubMed] [Google Scholar]
- 9.Johnson R.R., Cranston H.J., Chaverra M.E., Dyer W.E. Characterization of cDNA clones for differentially expressed genes in embryos of dormant and non-dormant Avenua fatua (L.) caryopses. Plant Mol. Biol. 1995;28:113–122. doi: 10.1007/BF00042043. [DOI] [PubMed] [Google Scholar]
- 10.Gaber A., Ogata T., Maruta T., Yoshimura K., Tamoi M., Shigeoka S. The involvement of Arabidopsis glutathione peroxidase 8 in the suppression of oxidative damage in the nucleus and cytosol. Plant Cell Physiol. 2012;53:1596–1606. doi: 10.1093/pcp/pcs100. [DOI] [PubMed] [Google Scholar]
- 11.Eshdat Y., Holland D., Faltin Z., Ben-Hayyim G. Plant glutathione peroxidases. Plant Physiol. 1997;100:234–240. [Google Scholar]
- 12.Sugimoto M., Furui S., Suzuki Y. Molecular cloning and characterization of a cDNA encoding putative phospholipid hydroperoxide glutathione from spinach. Biosci. Biotechnol. Biochem. 1997;61:1379–1381. doi: 10.1271/bbb.61.1379. [DOI] [PubMed] [Google Scholar]
- 13.Roeckel-Drevet P., Gagne G., de Labrouhe T.D., Dufaure J.P., Nicolas P., Drevet J.R. Molecular characterization, organ distribution and stress-mediated induction of two glutathione peroxidase-encoding mRNAs in sunflower (Helianthus annuus) Physiol. Plant. 1998;103:385–394. [Google Scholar]
- 14.Mullineaux P.M., Karpinski S., Jimenez A., Clearly S.P., Robinson C., Creissen G.P. Identification of cDNAs encoding plastid-targeted glutathione peroxidase. Plant J. 1998;13:375–379. doi: 10.1046/j.1365-313x.1998.00052.x. [DOI] [PubMed] [Google Scholar]
- 15.Depege D., Drevet J., Boyer N. Molecular cloning and characterization of tomato cDNA encoding glutathione peroxidase-like proteins. Eur. J. Biochem. 1998;253:445–451. doi: 10.1046/j.1432-1327.1998.2530445.x. [DOI] [PubMed] [Google Scholar]
- 16.Passaia G., Fonini L.S., Caverzan A., Jardim-Messeder D., Christoff A.P., Gaeta M.L., de Araujo Mariath J.E., Margis R., Margis-Pinheiro M. The mitochondrial glutathione peroxidase GPX3 is essential for H2O2 homeostasis and root and shoot development in rice. Plant Sci. 2013;208:93–101. doi: 10.1016/j.plantsci.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 17.Zhai C.Z., Zhao L., Yin L.J., Chen M., Wang Q.Y., Li L.C., Xu Z.S., Ma Y.Z. Two wheat glutathione peroxidase genes whose products are located in chloroplasts improve salt and H2O2 tolerances in Arabidopsis. PLoS One. 2013;8:e73989. doi: 10.1371/journal.pone.0073989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y.J., Jang M.G., Noh H.Y., Lee H.J., Sukweenadhi J., Kim J.H., Kim S.Y., Kwon W.S., Yang D.C. Molecular characterization of two glutathione peroxidase genes of Panax ginseng and their expression analysis against environmental stresses. Gene. 2014;535:33–41. doi: 10.1016/j.gene.2013.10.071. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez Milla M.A., Maure A., Rodriguez Huete A., Gustafson J.P. Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 2003;36:602–615. doi: 10.1046/j.1365-313x.2003.01901.x. [DOI] [PubMed] [Google Scholar]
- 20.Romas J., Matamoros M.A., Naya L., James E.K., Rouhier N., Sato S., Tabata S., Becana M. The glutathione peroxidase gene family of Lotus japonicus: Characterization of genomic clones, expression analyses and immunolocalization in legumes. New Phytol. 2009;181:103–114. doi: 10.1111/j.1469-8137.2008.02629.x. [DOI] [PubMed] [Google Scholar]
- 21.Navrot N., Collin V., Gualberto J., Gelhaye E., Hirasawa M., Rey P., Knaff D.B., Issakidis E., Jacquot J.P., Rouhier N. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 2006;142:1364–1379. doi: 10.1104/pp.106.089458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch M.A., German D.A. Taxonomy and systematics are key to biological information: Arabidopsis, Eutrema (Thellungiella), Noccaea and Schrenkiella (Brassicaceae) as examples. Front. Plant Sci. 2013;4:267. doi: 10.3389/fpls.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batelli G., Oh D.H., D’Urzo M.P., Orsini F., Dassanayake M., Zhu J.K., Bohnert H.J., Bressan R.A., Maggio A. Using Arabidopsis-related model species (ARMS): Growth, genetic transformation, and comparative genomics. Methods Mol. Biol. 2014;1062:27–51. doi: 10.1007/978-1-62703-580-4_2. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y.P., Giorgi F.M., Lohse M., Kvederaviciute K., Klages S., Usadel B., Meskiene I., Reinhardt R., Hincha D.K. Transcriptome sequencing and microarray design for functional genomics in the extremophile Arabidopsis relative Thellungiella salsuginea (Eutrema salsugineum) BMC Genomics. 2013;14:793. doi: 10.1186/1471-2164-14-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taji T., Seki M., Satou M., Sakurai T., Kobayashi M., Ishiyama K., Narusaka Y., Narusaka M., Zhu J.K., Shinozaki K. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol. 2004;135:1697–1709. doi: 10.1104/pp.104.039909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.M’rah S., Ouerghi Z., Eymery F., Rey P., Hajji M., Grignon C., Lachaâl M. Efficiency of biochemical protection against toxic effects of accumulated salt differentiates Thellungiella halophila from Arabidopsis thaliana. J. Plant Physiol. 2007;164:375–384. doi: 10.1016/j.jplph.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 27.Avsian-Kretchmer O., Gueta-Dahan Y., Lev-Yadun S., Ben-Hayyim G. The salt-stress signal transduction pathway that activates the gpx1 promoter is mediated by intracellular H2O2, different from the pathway induced by extracellular H2O2. Plant Physiol. 2004;135:1685–1696. doi: 10.1104/pp.104.041921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Churin Y., Schilling S., Börner T. A gene family encoding glutathione peroxidase homologues in Hordeum vulgare (barley) FEBS Lett. 1999;459:33–38. doi: 10.1016/s0014-5793(99)01208-9. [DOI] [PubMed] [Google Scholar]
- 29.Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., van de Peer Y., Rouzé P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margis R., Dunand C., Teixeira F.K., Margis-Pinheiro M. Glutathione peroxidase family—An evolutionary overview. FEBS J. 2008;275:3959–3970. doi: 10.1111/j.1742-4658.2008.06542.x. [DOI] [PubMed] [Google Scholar]
- 31.Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 32.Kant S., Kant P., Raveh E., Barak S. Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila. Plant Cell Environ. 2006;29:1220–1234. doi: 10.1111/j.1365-3040.2006.01502.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhu J.K. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 34.Lan P., Li W., Schmidt W. Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell. Proteomics. 2012;11:1156–1166. doi: 10.1074/mcp.M112.020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyer J.S., James R.A., Munns R., Condon A.G., Passioura J.B. Osmotic adjustment may lead to anomalously low estimates of relative water content in wheat and barley. Funct. Plant Biol. 2008;35:1172–1182. doi: 10.1071/FP08157. [DOI] [PubMed] [Google Scholar]
- 36.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 39.Nakai K., Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reese M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001;26:51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 41.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Gao F., Zhou Y., Zhu W., Li X., Fan L., Zhang G. Proteomic analysis of cold stress-responsive proteins in Thellungiella rosette leaves. Planta. 2009;230:1033–1046. doi: 10.1007/s00425-009-1003-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.