Abstract
Niemann–Pick C (NPC) disease is a fatal neurodegenerative disorder characterized by a lysosomal accumulation of cholesterol and other lipids within the cells of patients. Clinically identical forms of NPC disease are caused by defects in either of two different proteins: NPC1, a lysosomal–endosomal transmembrane protein and NPC2, a soluble lysosomal protein with cholesterol binding properties. Although it is clear that NPC1 and NPC2 are required for the egress of lipids from the lysosome, the precise roles of these proteins in this process is unknown. To gain insight into the normal function of NPC2 and to investigate its interactions, if any, with NPC1, we have generated a murine NPC2 hypomorph that expresses 0–4% residual protein in different tissues and have examined its phenotype in the presence and absence of NPC1. The phenotypes of NPC1 and NPC2 single mutants and an NPC1;NPC2 double mutant are similar or identical in terms of disease onset and progression, pathology, neuronal storage, and biochemistry of lipid accumulation. These findings provide genetic evidence that the NPC1 and NPC2 proteins function in concert to facilitate the intracellular transport of lipids from the lysosome to other cellular sites.
Deficiency of either NPC1 or NPC2 in Niemann–Pick C (NPC) disease results in the lysosomal storage of lipids including unesterified cholesterol, phospholipids, glycolipids, and sphingomyelin, highlighting the importance of these proteins in cellular lipid transport. However, the molecular role of these proteins in this process remains unknown. In general, NPC1 has been thought to act as a mediator for endosomal sorting of cholesterol and other lipids in a sterol-regulated manner (1) and it has been suggested that it may function as a transmembrane lipid pump (2). Despite containing a “sterol-sensing domain,” no direct interactions between NPC1 and lipids or other proteins have been identified to date. In contrast, NPC2 has been shown to directly bind sterols (3, §§), and the structural basis for this interaction has been investigated by using crystallographic (5) and mutagenic (6) approaches. Although NPC2 is also capable of facilitating the transfer of sterols between model donor and acceptor membranes,§§ little is known about its precise intracellular function.
To gain insight into the normal function of NPC2 and its interactions, if any, with NPC1, we have targeted the murine gene generating a severe hypomorph and have investigated the resulting phenotype in the presence and absence of NPC1. We find that the phenotype of NPC2-mutant mice is similar or identical to the NPC1 null mutant, albeit slightly delayed. In addition, we find that mice deficient in both proteins are essentially identical to the NPC1 mutant by all criteria examined here.
Materials and Methods
The NPC2 Targeting Construct. The sequence of the murine 129Sv NPC2 gene was determined from long-range PCR products. For gene targeting (Fig. 1A), a 7.5-kb PCR product that comprised part of exon 1, intron 1, and a part of exon 2 ending in an engineered in-frame stop codon (Cys42Stop) was inserted into the BamHI site of pTKLNL (provided by Richard Mortensen, University of Michigan, Ann Arbor). The 2.6-kb right arm, starting at codon Ser-54, is comprised of part of exon 2 and part of intron 2. Homologous recombination of this vector into the NPC2 gene was therefore designed to introduce a premature stop codon and replace 11 amino acids of NPC2 encoded by exon 2 with the lox-P flanked neo cassette. Targeting details and oligonucleotide sequences are available on request.
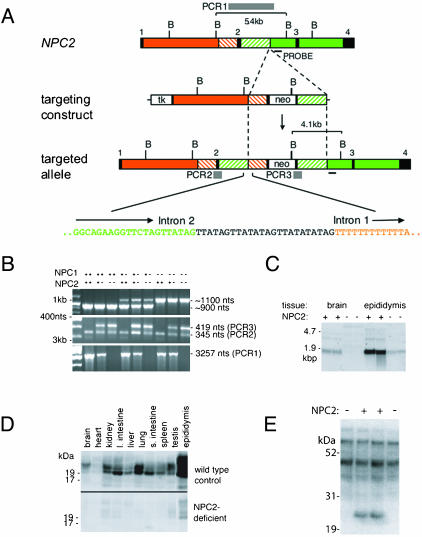
Fig. 1.
Targeted disruption of the murine NPC2 gene (NPC2). (A) Structure of NPC2, the targeting construct, the targeted locus and the sequence at the NPC2 intron 2:left arm intron 1 junction. BglII sites; boxes: black shading, exons; unshaded, thymidine kinase (tk) and neo; orange, intron 1; green, introns 2 and 3; green and orange dashed, sequences duplicated in the targeted allele. The strategy for inactivating NPC2 was to insert a loxP-flanked neo cassette into exon 2 with the concomitant replacement of amino acid cysteine 42 with a stop codon and a deletion of amino acids 43–53. An aberrant recombination event, characterized by Southern blotting and sequencing of genomic PCR products, resulted in the insertion of the neo selection marker and part of the left arm containing intron 1 into intron 2 of NPC2. Twenty-four nucleotides of unknown derivation are inserted at the site of the intron 2 – intron 1 junction. Gray bars depict informative PCR products PCR1, PCR2, and PCR3 for NPC2 genotyping. (B) PCR genotyping of genomic mouse tail DNA samples representing all combinations of NPC1 and NPC2 genotype. (C) Northern blot analysis of total RNA purified from the brain and epididymis of 30-day-old wild-type and homozygous NPC2-targeted mice. (D) Immunodetection of NPC2 in different tissues from 30-day wild-type and homozygous NPC2-targeted mice. (E) Mannose-6-phosphate containing glycoproteins in epididymal extracts detected with 125I-labeled cation-independent mannose 6-phosphate receptor in a blot assay.
Generation of an NPC2-Targeted Mouse. All experiments and procedures involving live animals were conducted in compliance with animal protocols approved by the Robert Wood Johnson Medical School Institutional Animal Care and Use Committee. Euthanasia was performed by using 98 mg/ml sodium pentobarbital containing 12.5 mg/ml phenytoin (a 1:4 dilution of Euthasol; Delmarva Laboratories, Midlothian, VA) unless stated otherwise. W9.5 129Sv embryonic stem (ES) cells, originally derived by Colin Stewart (National Cancer Institute, Frederick, MD), were electroporated by using a Bio-Rad Gene Pulser with vector DNA linearized with NotI, and targeted clones were selected by using neomycin and gancyclovir. Screening was achieved by Southern blotting by using a probe that detected a 4.1-kb BglII product that was specific to the targeted allele (Fig. 1 A). One apparently targeted ES cell clone was identified, expanded, and used for microinjection. ES cells were injected into C57BL6 blastocysts that were transferred into pseudopregnant Swiss–Webster females by using standard techniques. Highly chimeric males were mated with C57BL6 females, generating a number of offspring that were confirmed to be heterozygotes by Southern blotting. BALB/c npcnih mice have been described (7–10).
Genotyping Mice by PCR. A consequence of aberrant recombination resulting in the insertion of the neo selection marker and part of the left arm containing intron 1 into intron 2 (Fig. 1A) is that PCR genotyping of the NPC2-targeted mice required two separate reactions. The product PCR3 (Fig. 1B) determines the presence or absence of a targeted allele but does not differentiate between heterozygotes and homozygous targeted mice. PCR3 and PCR2 (an internal control amplified from both targeted and wild-type alleles) were generated by using AmpliTaq Gold (Perkin–Elmer) and the following primers: TTTCCTCCCTAGTCAAACTCAACT, CATTCTCAGTATTGTTTTGCCAAG, and AGTGAGAATTATGGACCCAGACTC. Cycling conditions for this primer set are 94°C for 11 min, followed by 35 cycles of 94°C for 30 s, 53°C for 30 s, and 68°C for 1 min. PCR1 (Fig. 1A), which determines the presence or absence of a wild-type allele but does not differentiate between homozygous wild-type and heterozygous mice, is generated by using the primers GCACACGTAGAGCTCAGAGAATAA and TTCTCACCACCACTACTGTGTTTT with Ex Taq polymerase (Takara Shuzo, Kyoto) with 35 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 2 min. NPC1 was genotyped by using an established PCR protocol (9). Genotyping of mutant mice is illustrated in Fig. 1B.
Detection of NPC2 and Man6-P Glycoproteins by Western Blotting. Mice were killed, dissected, and organs frozen in liquid nitrogen and stored at –80°C for preparation of RNA (see below) or soluble protein extracts for Western blotting. Extracts were prepared by using a Polytron homogenizer with microgenerator (Brinkmann) in PBS containing 0.2% Tween 30, 5 mM β-glycerophosphate, 1 mM EDTA, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 1 mM Pefabloc (Roche Applied Science). Protein content was determined (11) and extracts were fractionated on Novex 10% NuPage Bis-Tris precast gels by using Mes electrophoresis buffer (Invitrogen). Proteins were transferred to nitrocellulose and NPC2 was detected by using an affinity purified rabbit polyclonal antibody and an I125-labeled goat–anti-rabbit secondary antibody. Residual levels of NPC2 expression in epididymis from NPC2-targeted mice were determined by comparing several dilutions of duplicate protein extracts from tissue derived from duplicate targeted mice with a standard curve created by serial dilution of extracts from wild-type mice. Blots were quantified by storage phosphor autoradiography by using a Typhoon 9400 (Amersham Pharmacia). Man6-P glycoproteins were detected by using an iodinated soluble form of the cation-independent mannose 6-phosphate receptor as described (12) by using extracts that were prepared, electrophoresed, and transferred to nitrocellulose as described for immunodetection.
Northern Blotting and Real-Time PCR. Total RNA was purified from frozen tissue by using RNeasy spin columns (Qiagen, Chatsworth, CA), fractionated by denaturing agarose gel electrophoresis, and transferred to Genescreen Plus nylon membrane (Perkin–Elmer). NPC2 mRNA was detected by using a 32P-labeled probe generated with random hexamers from a 565-nt BamHI fragment of an NPC2 cDNA clone (I.M.A.G.E.: 1451353). Quantitation of NPC2 expression by duplicate independent real-time PCR experiments was achieved with the Brilliant SYBR Green QRT-PCR kit (Stratagene) and an MX4000 Multiplex Quantitative PCR System (Stratagene) by using oligos TATCCACGATGCGTTTTCTG and TCAGGCTCAGGAATAGGGAA that generate a 280-nt NPC2 RT-PCR fragment from total RNA. An NPC2 cDNA clone (I.M.A.G.E.: 390611) was used to generate standard curves.
Cholesterol Measurement by Gas Chromatography. Cholesterol measurements were performed as described (13). After the addition of the internal standard coprostanol, cholesterol, and the plant sterols campesterol, sitostanol, and sitosterol were extracted with n-hexane after the saponification of 0.1–0.2 g of minced liver by refluxing for 1 h at70°C in 1 M ethanolic NaOH. Trimethylsilyl ether derivatives were prepared by the addition of Sil Prep (Alltech Associates, Deerfield, IL). Aliquots were then injected into a Hewlett–Packard 6890 gas chromatograph (Agilent Technologies, Palo Alto, CA) equipped with a flame ionization detector and a 25-m CP-Sil 5CB capillary column (Varian, Palo Alto, CA). The instrument was operated with He as the carrier gas at 1 ml/min, and the system was calibrated by injecting known amounts of a mixture of coprostanol, cholesterol, and sitosterol.
Lipid Biochemistry. Fifty-day-old mice were killed and dissected, and frozen tissues were stored at –80°C. Brain analyses were carried out on whole hemispheres after dissecting out the cerebellum and brainstem. Brain and liver lipids were extracted in chloroform–methanol 1:2 (vol/vol) as described (14). The total lipid extract was desalted and separated into two fractions (neutral lipids and acidic lipids) by using reverse-phase Bond Elut C18 columns (15) purchased from Varian. All other methods, including quantitative methods, were as described (14, 16).
Pathology. Mice at 7 and 11 weeks of age were deeply anesthetized and perfused with 4% paraformaldehyde in 0.1 M phosphate buffer. Brains were removed and fixed for an additional 24hat4°C, followed by storage in 0.1 M phosphate buffer. Brains were sectioned on a Leica vibratome at 30 μm and stained for ganglioside and cholesterol storage and for Purkinje cell loss/degeneration (17). Antibodies to GM2 and GM3 gangliosides were kindly provided by Progenics (Tarrytown, NY) and S. Hakomori (University of Washington, Seattle), respectively. Antibodies to calbindin and parvalbumin were purchased from Sigma, as was filipin for demonstration of free cholesterol. Tissue sections were examined on a Olympus (Melville, NY) research microscope and photographed with a MagnaFire (Optronics, Goleta, CA) digital camera. Brain samples for ultrastructural study were postfixed in osmium, dehydrated, and embedded in Epon-aryldite. Ultrathin sections were cut from the plastic embedded blocks, stained with uranyl acetate and lead citrate, and examined on a Philips (Eindhoven, The Netherlands) CM10 electron microscope.
Results
Targeted Disruption of NPC2. The targeting construct for disruption of the murine NPC2 gene and examples of PCR genotyping are shown in Fig. 1 A and B. In mice homozygous for the targeted NPC2 allele, correctly spliced mRNA was present at greatly reduced levels compared to normal controls (Fig. 1C; 2.1% and 4.5% in brain and epididymis, respectively, by real-time PCR). Consistent with this finding, quantitative immunodetection revealed 3.5 ± 0.9% of normal NPC2 protein levels in epididymis with little or no expression in most other tissues examined (Fig. 1D). A 20- to 25-kDa glycoprotein containing mannose-6 phosphate, presumably representing NPC2, was absent in epididymis (Fig. 1E) and other tissues (data not shown). Together, these data indicate that the NPC2-targeted mouse is a severe hypomorph rather than a null, and this is consistent with the gene disruption: originally planned as a disruption of exon 2, aberrant recombination resulted in the insertion of the neo selection marker and part of the left arm containing intron 1 into intron 2 (Fig. 1 A). In an NPC type 2 patient, a mutation within intron 2 also causes misplicing, so that correctly spliced NPC2 mRNA was at ≈6% of normal levels (18). Compared to patients inheriting two null alleles, homozygosity for this splice junction mutation resulted in a later onset and slower progression of disease.
Transmission of NPC Mutations. NPC1;NPC2 double mutant mice and controls of all genotypes were generated by mating double heterozygotes. Transmission of the double mutation was less than expected (2.85% vs. 6.25%; Table 1), which is suggestive of partial embryonic or perinatal lethality. The cause of such lethality is not known, but it is worth noting that decreased transmission of the NPC1 allele was observed previously (8, 19). It is possible that the underlying cause of decreased transmission of the double mutation is the same as NPC1 but is exacerbated by the accompanying NPC2 deficiency.
Table 1. Transmission frequencies of NPC1;NPC2 genotypes from double heterozygote [Npc1+/–/Npc2+/–] matings.
| Number of animals per genotype (% found:% expected)
|
|||
|---|---|---|---|
| NPC2-/- | NPC2+/- | NPC2+/+ | |
| NPC1-/- | 30 (2.8:6.25) | 117 (11.1:12.5) | 49 (4.7:6.25) |
| NPC1+/- | 130 (12.3:12.5) | 260 (24.7:25.0) | 117 (11.1:12.5) |
| NPC1+/+ | 96 (9.1:6.25) | 155 (14.7:12.5) | 99 (9.4:6.25) |
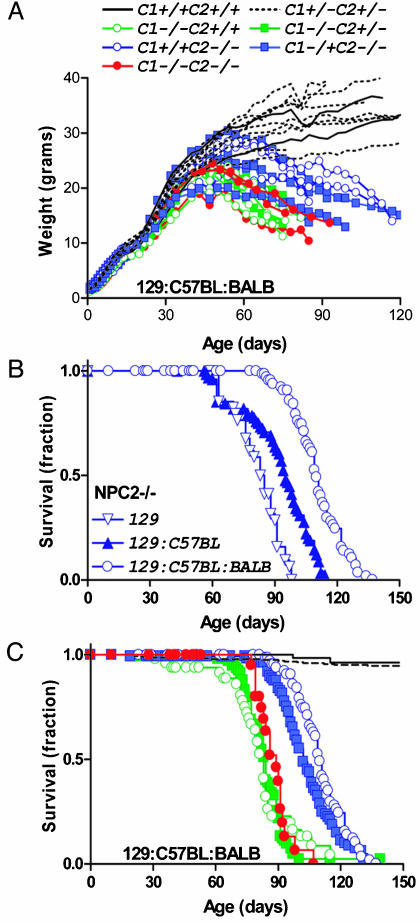
Growth and Survival of NPC Mutants. Growth of NPC2 hypomorph animals increased normally until ≈55 days, after which weights dropped rapidly until the time of natural death or euthanasia due to morbidity [males, Fig. 2A; relative weight loss for affected females was similar (data not shown)]. A continuous tremor was first detected at ≈55 days and was eventually accompanied by ataxia and overall locomotor dysfunction. Death of NPC2 hypomorph mice occurred between ≈90 and 130 days and was influenced by genetic background (Fig. 2B), which may indicate the presence of a strain-specific modifier(s) for the NPC2 phenotype, as has been proposed for NPC1 (20, 21). Disease onset was later and progression less rapid in NPC2 hypomorph mice compared with NPC1 mutants of similar genetic background (Fig. 2 A and C), most likely reflecting the fact that the NPC2-targeted mice are severe hypomorphs rather than null mutants. The double mutant mice are indistinguishable from the single NPC1 mutant in terms of gross neurological symptoms and time-course of disease onset and progression (Fig. 2 A and C). Survival of wild-type and double heterozygote mice was indistinguishable (Fig. 2C), and double heterozygotes observed as long as 40 months exhibited no neurological symptoms. Heterozygosity for NPC2 did not appear to have an effect on NPC1–/– animals (median survival, 84 and 83 days for NPC2+/+ and NPC2–/+, respectively; Fig. 2C). In the NPC2–/– animals, the NPC1 genotype may have had a small effect (median survival, 110 and 102 days for NPC1+/+ and NPC1–/+, respectively; Fig. 2C).
Fig. 2.
Growth and survival of NPC2 hypomorph mice in the presence or absence of NPC1. (A) Growth of male NPC mutant mice. (B) Survival of NPC2 hypomorph mice in different genetic backgrounds. (C) Survival of NPC2 hypomorph mice in the presence or absence of NPC1. Symbols are as in A; control, n = 11 (two NPC1+/+;NPC2+/+, nine NPC1+/–;NPC2+/–); NPC1 mutant n = 7 (five NPC1–/–;NPC2+/– and two NPC1+/+); NPC2 mutant n = 6 (three NPC1+/+;NPC2–/– and three NPC1+/–;NPC2–/–) and NPC1-NPC2 double mutant, n = 3.
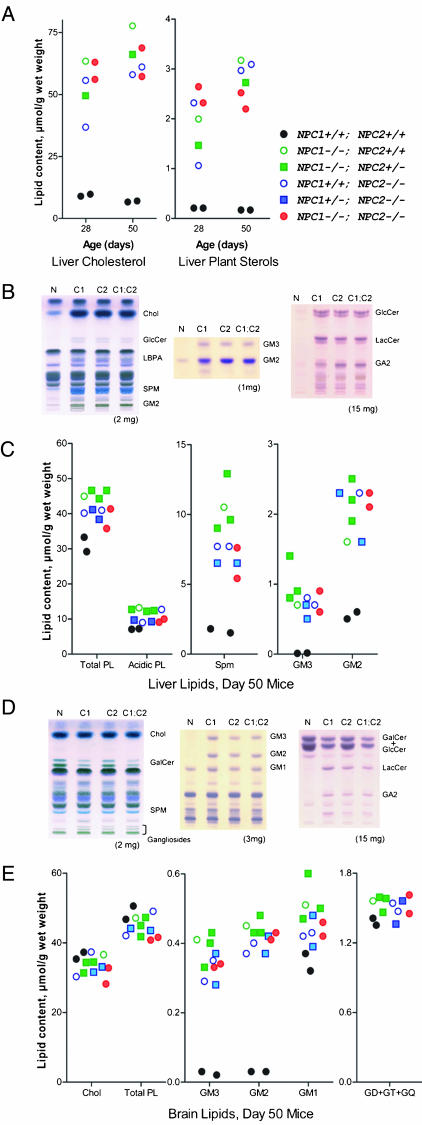
Lipid Biochemistry. In NPC disease, other lipids in addition to cholesterol also accumulate, including phospholipids, gangliosides, and neutral glycolipids, with different storage profiles in extraneural organs and in brain (reviewed in refs. 22 and 23). The profiles of stored lipids in NPC1, NPC2, and NPC1;NPC2 mutant mice were qualitatively and quantitatively very similar in both liver and brain (Fig. 3). In liver, total cholesterol and dietary plant sterols were increased ≈6-fold and ≈10-fold, respectively, at 28 days and ≈10-fold and ≈17-fold, respectively, at 50 days (Fig. 3A). At 50 days, there are also marked elevations in sphingomyelin, lyso(bis)phosphatidic acid, gangliosides GM2 and GM3, glucosylceramide (GlcCer), lactosylceramide (LacCer), and asialo-GM2 (GA2) (Fig. 3 B and C). In brain, abnormalities in total lipid levels were essentially restricted to glycolipids (Fig. 3 D and E). [Cholesterol is stored in the brain, but total levels do not increase, probably reflecting compensatory losses due to demyelination and neuronal death (24) and possible imbalance between neuronal cell bodies and distal axons (25)]. Both GM2 and GM3 were elevated 11- to 15-fold in all three mutant mice (Fig. 3E), and there were also increases in glucosylceramide (data not shown), lactosylceramide, and GA2 (Fig. 3D). Reflecting the general loss of myelin lipids, galactosylceramide was reduced (Fig. 3D).
Fig. 3.
Lipid accumulation in NPC mutant mice. (A) Liver cholesterol and dietary plant sterols (campesterol, sitosterol, and sitostanol) determined by gas chromatography at 28 and 50 days. (B and C) Thin-layer chromatography (TLC) and quantitation of lipids in liver. (D and E) TLC and quantitation of lipids in brain. (B and D) Left to right: main lipids, gangliosides, neutral glycolipids. Note: in the TLC of neutral glycolipids, GalCer resolves into two bands depending on fatty acid composition and in D; the upper GalCer band and GlcCer are not resolved. The tissue weight equivalent is indicated below. Wt, wild type control; C1, C2, and C1;C2, NPC1, NPC2, and NPC1;NPC2 mutant mice, respectively. Chol, cholesterol; GlcCer, glucosylceramide; LBPA, lyso(bis)phosphatidic acid; SPM, sphingomyelin; LacCer, lactosylceramide; GalCer, galactosylceramide; GD+GT+GQ, di-, tri-, and tetrasialogangliosides. Mice were in a mixed C57BL6/129SvEv/BALBc background.
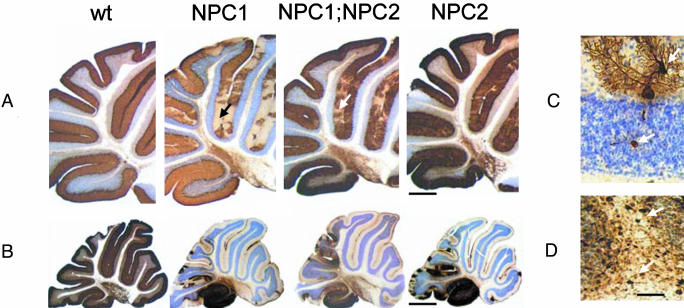
Pathology. Lipid storage occurs in nearly every organ of affected individuals, but the clinical hallmarks of NPC disease are mostly attributable to intraneuronal storage, dendritic and axonal alterations, and progressive neurodegeneration (17). In the NPC1 mouse, a striking neuropathologic change is that of a progressive and eventually almost total loss of cerebellar Purkinje cells. The NPC2 mutant exhibited a very similar, albeit slightly delayed, pattern of Purkinje cell degeneration compared to the NPC1 and NPC1;NPC2 mutants, which were essentially indistinguishable (Fig. 4 A and B).
Fig. 4.
Neurodegenerative changes affecting cerebellar Purkinje cells. Calbindin-stained sections of cerebellum from wild-type (Wt), NPC1, NPC1;NPC2, and NPC2 mutant mice at (A) 7 and (B) 11 wk of age. (C) A calbindin-stained Purkinje cell in an NPC2 hypomorph mouse cerebellum showing neurodegenerative changes (dendritic and axonal swellings, upper and lower arrows, respectively) typical of all three disease models. (D) Parvalbumin–immunoreactive axonal enlargements (spheroids) in the deep white matter of a cerebellar folium (arrows) of an NPC2 hypomorph mouse that are typical for all three models. Nissl counterstain. Mice were in a mixed C57BL6/129SvEv/BALBc background. [Bars = 500 μm(A), 1,100 μm(B), and 80 μm(D).]
Alterations in neuronal morphology are commonly observed in lysosomal storage diseases (26), including NPC1 disease (17). Parvalbumin staining of a surviving Purkinje cell in the 11-week NPC2 mice reveals a distended region of storage within the dendritic tree and an axonal spheroid within the underlying granule cell layer (Fig. 4C). Examination of cerebellar white matter at a lower magnification reveals the presence of significant numbers of parvalbumin-positive axonal spheroids from Purkinje cells in the process of dying (Fig. 4D).
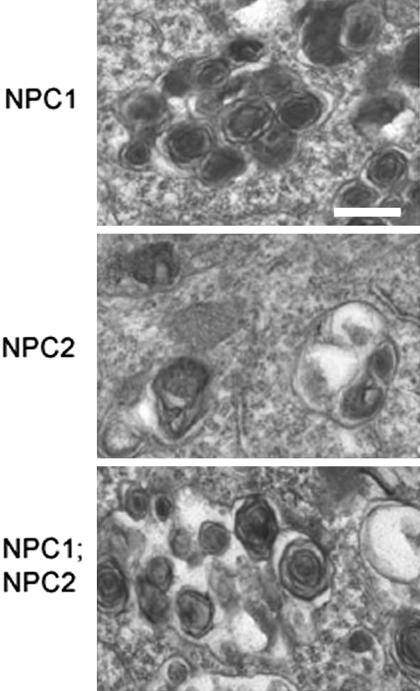
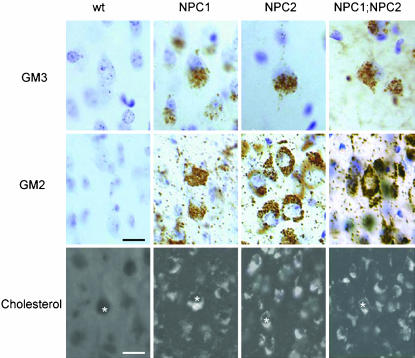
At the cellular level, lipid storage in the three mutant genotypes was identical. Electron microscopic analysis of neurons from NPC1, NPC2, and NPC1;NPC2 mutant mice revealed heterogeneous lamellar neuronal inclusions as seen in NPC patients (Fig. 5). Filipin staining revealed storage of unesterified cholesterol in all three mutants within the neocortex, dentate gyrus, hippocampus, and cerebellum (Fig. 6; data not shown) as observed earlier for NPC1 (17). In addition to free cholesterol, neurons in NPC1 mice undergo substantial accumulation of GM2 and GM3 gangliosides; again, the pattern of storage was the same among NPC1, NPC2, and NPC1;NPC2 mutant animals (Fig. 6). At the subcellular level, distribution of GM2, GM3, and cholesterol (27) (S.U.W., data not shown) appear identical in NPC1 and NPC2 mutant mice.
Fig. 5.
Ultrastructure of neuronal storage material by electron microscopy. (Bar = 0.25 μm.) Mice were 7 wk old and in a mixed C57BL6/129SvEv/BALBc background.
Fig. 6.
Lipid storage in NPC mutants. Examples of individual neurons shown by * over nucleus. Mice were 7 wk old and in a mixed C57BL6/129SvEv/BALBc background. [Bars = 25 μm (GM2 and GM3 ganglioside immunocytochemistry) and 15 μm (cholesterol detection by filipin staining)].
Discussion
In investigating the relative functions of NPC1 and NPC2, our reasoning was that if these two proteins play independent and unrelated molecular roles in lipid transport, then the phenotype of a combined NPC1;NPC2 mutant mouse would be expected to be more severe than that of either individual mutant. Alternatively, if the function of NPC1 and NPC2 were closely linked (for example, if each facilitated nonredundant sequential steps in a central lipid transport pathway), then the phenotype of the double-mutant mouse should approximate that of the more severe of either individual mutant. We show here that the phenotypes of the NPC1 mutant and NPC1;NPC2 double mutant appear indistinguishable, and that the phenotype of the NPC2 mutant is very similar albeit with slightly later onset and slower progression. These results provide genetic evidence for a cellular pathway for the transport of lysosomal lipids in which both proteins participate and in which neither can compensate for loss of the other.
Interestingly, treatment of NPC1 mutants with the inhibitor of glucosylceramide synthesis, N-butyldeoxynojirimycin, delayed neurological dysfunction and increased lifespan, highlighting the importance of glycosphingolipids in neuropathogenesis (4). If parallel ongoing studies with the NPC2-hypomorph (S.U.W., unpublished data) provide similar results, this may be interpreted as providing further evidence for closely linked functions between NPC1 and NPC2.
The respective roles of each protein in a pathway and the nature of their functional or physical interactions remain to be elucidated. However, a model compatible with our data is that NPC2 might transport free cholesterol (generated, for example, by the action of acid lipase on lipoprotein-derived cholesteryl ester) through the aqueous lumen of the endosome-lysosome to the delimiting membrane. Here, or in derivative vesicular compartments, NPC1 might detect changes in cholesterol levels and regulate the movement of luminal lipids by either altering vesicular transport or by participating directly in lipid transport across membrane bilayers. The availability of the mouse models described here and derivative cell lines will allow many approaches to test such hypotheses. Approaches that have been informative with NPC1 in the past and that may provide useful information with respect to the relative roles of NPC1 and NPC2 include genetic studies of NPC mutant mice crossed with other mutants, physiological studies of lipid synthesis and metabolism in the whole animal, and, most directly, cell-based studies of lipid uptake and intracellular transport.
Our results do not rule out the possibility of additional, more specialized roles for NPC1 and NPC2 in the context of a complex organism. For example, the synergistic effect of combined NPC1 and NPC2 deficiencies on embryonic/perinatal survival suggests this might be the case and may also point toward genetic components within the mixed C57BL6/129SvEv/BALBc background that influence early viability. However, regardless of possible specialized roles, the primary conclusion of this study is that NPC1 and NPC2 have nonredundant functions in a common pathway for lipid transport. Additional proteomic and genetic approaches will be required to elucidate this pathway and other potential functions of NPC1 and NPC2. Ongoing efforts to generate the NPC2 hypomorph and a true null in an isogenic BALBc background represent an important next step toward achieving these aims.
Acknowledgments
We thank Folashade Adeshuko, Vivienne Halili, Jody Litrenta, and Gloria Stephney for technical help and Thomas Ludwig (Columbia University, New York), Arnold Rabson (Center for Advanced Biotechnology and Medicine), and Hsin-Ching Lin (Center for Advanced Biotechnology and Medicine) for helpful advice or reagents. This work was supported by the Ara Parseghian Medical Research Foundation (P.L. and S.U.W.), the National Institutes of Health (M.M.S.), the Institut National de la Santé et de la Recherche Médicale (INSERM) (M.T.V.), and Vaincre les Maladies Lysosomales (M.T.V. and L.V.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: NPC, Niemann–Pick type C.
Footnotes
Baker, C. S., Magargee, S. F. & Hammerstedt, R. H. (1993) Biol. Reprod. 48, 86 (abstr.).
References
- 1.Ioannou, Y. A. (2001) Nat. Rev. Mol. Cell Biol. 2, 657–668. [DOI] [PubMed] [Google Scholar]
- 2.Davies, J. P., Chen, F. W. & Ioannou, Y. A. (2000) Science 290, 2295–2298. [DOI] [PubMed] [Google Scholar]
- 3.Okamura, N., Kiuchi, S., Tamba, M., Kashima, T., Hiramoto, S., Baba, T., Dacheux, F., Dacheux, J. L., Sugita, Y. & Jin, Y. Z. (1999) Biochim. Biophys. Acta 1438, 377–387. [DOI] [PubMed] [Google Scholar]
- 4.Zervas, M., Somers, K. L., Thrall, M. A. & Walkley, S. U. (2001) Curr. Biol. 11, 1283–1287. 86. [DOI] [PubMed] [Google Scholar]
- 5.Friedland, N., Liou, H.-L., Lobel, P. & Stock, A. M. (2003) Proc. Natl. Acad. Sci. USA 100, 2512–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ko, D. C., Binkley, J., Sidow, A. & Scott, M. P. (2003) Proc. Natl. Acad. Sci. USA 100, 2518–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pentchev, P. G., Gal, A. E., Booth, A. D., Omodeo-Sale, F., Fouks, J., Neumeyer, B. A., Quirk, J. M., Dawson, G. & Brady, R. O. (1980) Biochim. Biophys. Acta 619, 669–679. [DOI] [PubMed] [Google Scholar]
- 8.Morris, M. D., Bhuvaneswaran, C., Shio, H. & Fowler, S. (1982) Am. J. Pathol. 108, 140–149. [PMC free article] [PubMed] [Google Scholar]
- 9.Loftus, S. K., Morris, J. A., Carstea, E. D., Gu, J. Z., Cummings, C., Brown, A., Ellison, J., Ohno, K., Rosenfeld, M. A., Tagle, D. A., et al. (1997) Science 277, 232–235. [DOI] [PubMed] [Google Scholar]
- 10.Shio, H., Fowler, S., Bhuvaneswaran, C. & Morris, M. D. (1982) Am. J. Pathol. 108, 150–159. [PMC free article] [PubMed] [Google Scholar]
- 11.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 12.Valenzano, K. J., Kallay, L. M. & Lobel, P. (1993) Anal. Biochem. 209, 156–162. [DOI] [PubMed] [Google Scholar]
- 13.Tint, G. S., Pentchev, P., Xu, G., Batta, A. K., Shefer, S., Salen, G. & Honda, A. (1998) J. Inherit. Metab. Dis. 21, 853–863. [DOI] [PubMed] [Google Scholar]
- 14.Fujita, N., Suzuki, K., Vanier, M. T., Popko, B., Maeda, N., Klein, A., Henseler, M., Sandhoff, K. & Nakayasu, H. (1996) Hum. Mol. Genet. 5, 711–725. [DOI] [PubMed] [Google Scholar]
- 15.Kyrklund, T. (1987) Lipids 22, 274–277. [DOI] [PubMed] [Google Scholar]
- 16.Liu, Y., Wu, Y. P., Wada, R., Neufeld, E. B., Mullin, K. A., Howard, A. C., Pentchev, P. G., Vanier, M. T., Suzuki, K. & Proia, R. L. (2000) Hum. Mol. Genet. 9, 1087–1092. [DOI] [PubMed] [Google Scholar]
- 17.Zervas, M., Dobrenis, K. & Walkley, S. U. (2001) J. Neuropathol. Exp. Neurol. 60, 49–64. [DOI] [PubMed] [Google Scholar]
- 18.Millat, G., Chikh, K., Naureckiene, S., Sleat, D. E., Fensom, A. H., Higaki, K., Elleder, M., Lobel, P. & Vanier, M. T. (2001) Am. J. Hum. Genet. 69, 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erickson, R. P., Aviles, R. A., Zhang, J., Kozloski, M. A., Garver, W. S. & Heidenreich, R. A. (1997) Mamm. Genome 8, 355–356. [DOI] [PubMed] [Google Scholar]
- 20.Zhang, J. & Erickson, R. P. (2000) Mamm. Genome 11, 69–71. [DOI] [PubMed] [Google Scholar]
- 21.Miyawaki, S., Yoshida, H., Mitsuoka, S., Enomoto, H. & Ikehara, S. (1986) J. Hered. 77, 379–384. [DOI] [PubMed] [Google Scholar]
- 22.Patterson, M. C., Vanier, M. T., Suzuki, S., Morris, J. A., Carstea, E., Neufeld, E. B., Blanchette-Mackie, J. E. & Pentchev, P. G. (2001) in The Metabolic and Molecular Bases of Inherited Disease (McGraw–Hill, New York), 8th Ed., pp. 3611–3634.
- 23.Vanier, M. T. (1999) Neurochem. Res. 24, 481–489. [DOI] [PubMed] [Google Scholar]
- 24.Xie, C., Turley, S. D., Pentchev, P. G. & Dietschy, J. M. (1999) Am. J. Physiol. 276, E336–E344. [DOI] [PubMed] [Google Scholar]
- 25.Karten, B., Vance, D. E., Campenot, R. B. & Vance, J. E. (2002) J. Neurochem. 83, 1154–1163. [DOI] [PubMed] [Google Scholar]
- 26.Walkley, S. U. (1998) Brain Pathol. 8, 175–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Litrenta, J., Adeshuko, F., Zhou, S., Stephney, G., Wiseman, J., Dobrenis, K., Sleat, D., Lobel, P. & Walkley, S. (2003) 2003 Abstract Viewer/Itinerary Planner (Society for Neuroscience Washington, DC), Online Program No. 537.15 (http://sfn.scholarone.com/itin2003/index.html).