Abstract
HIV-1 infection persists even after years of antiretroviral therapy (ART). Although ART can halt viral replication and thereby reduce viremia to clinically undetectable levels, proviral latency established within the host genome remains largely unaffected by ART and can replenish systemic infection following interruption of therapy. Pharmacologic strategies, which not only target viral replication but also deplete proviral infection, are required for successful clearance of HIV-1 infection. This review highlights the current understanding of molecular mechanisms that establish and maintain HIV-1 latency in its major reservoir, the resting memory CD4+ T cell. We also identify the molecular targets that might be exploited to induce HIV-1 expression, remove epigenetic restrictions, or enhance effective transcription. Finally, we discuss the potential pharmacologic approaches toward targeting viral persistence in different cellular and anatomical reservoirs to achieve a cure of HIV-1 infection.
Keywords: antiretroviral therapy, ART, viral reservoir, eradication
INTRODUCTION
The advent of antiretroviral therapy (ART) and the chronic suppression of human immunodeficiency virus type 1 (HIV-1) replication has been a major medical success, greatly increasing survival and improving quality of life. Successful ART results in clinically undetectable levels of plasma viremia (<50 copies ml−1), allowing immune reconstitution. However, once infection is established, it cannot be cleared by current ART. Persistent proviral infection in a small pool of latently infected cells is insensitive to ART, is not detected by immune surveillance, and provides a long-lived source of rebound viremia—the “reservoir” of HIV infection.
With a half-life of ~44 weeks, resting memory CD4+ T cells appear to constitute the major reservoir of viral persistence (1–6). Therefore, it has been estimated that life-long therapy (60 years) would be necessary to cure the disease. The latent HIV-1 infection in CD4+ T cells is established early in infection, even in patients who are treated with ART within the first week of infection (2). Although the frequency of infected resting memory CD4+ cells is low (estimated to total only 106–107 cells in an infected individual), prompt rebound of viremia will ensue if ART is interrupted and if these cells are allowed to express virus owing to antigenic stimulation or immune activation.
The mechanisms that lead to HIV-1 latency in CD4+ T cells are still not completely understood. There is ample evidence that multiple restrictions prevent the emergence of virus from latency, including both cellular and viral factors (7, 8). Moreover, infected resting memory CD4+ T cells may be able to undergo homeostatic proliferation, another factor that may contribute to the persistence of this reservoir, again a process inherent to the nature of memory cells (9). Nevertheless, intervention at the molecular level can lead to HIV-1 reactivation, which suggests that proviral latency is amenable to therapeutic intervention. Therefore, understanding the molecular mechanisms responsible for the establishment, maintenance, and reactivation of HIV-1 latency in this important reservoir is critical to the pursuit of successful eradication strategies.
HIV-1 infection occurs in a variety of anatomic compartments, including pharmacologically “privileged” sites such as the central nervous system (CNS) (10, 11) and sites in which drug metabolism is poorly understood such as the gut-associated lymphoid tissue (GALT) (12). The blood-brain barrier of the CNS limits the penetration of some antiretrovirals and therefore provides viral sanctuary (13, 14). Therapeutic strategies that target virus in these privileged reservoirs may be required to eradicate HIV-1 infection.
Macrophages have long been considered another drug-insensitive reservoir (15–17). However, owing to their shorter life span, their significance in long-term maintenance of persistent HIV-1 infection in patients receiving ART is not clear (18–20). Dendritic cells (DCs) and hematopoietic stem cells (HSCs) are also implicated as reservoirs or sources of low-level viremia, but the data conflict, and this review does not focus on this issue (21–24). In this review, we describe the molecular mechanisms that establish latency in resting CD4+ T cells and identify the viral and cellular components that serve as potential targets for pharmaceutical intervention. We also discuss approaches to target HIV-1 hidden in pharmacologically privileged sites, the main limitations in targeting persistent infection, and future perspectives.
ESTABLISHMENT OF HIV-1 LATENCY IN MEMORY CD4+ T CELLS
Quiescent CD4+ T cells are less permissive for HIV-1 infection, posing blocks at several stages of the HIV-1 life cycle. First, the low nucleotide pools in resting CD4+ T cells result in inefficient completion of viral reverse transcription (25). Furthermore, existence of active APOBEC3G (apolipoprotein B mRNA-editing enzyme catalytic polypeptide 3G) in resting CD4+ cells causes G-to-A hypermutation during reverse transcription, resulting in the production of defective viral genomes (26). Inefficient nuclear import of the viral preintegration complex (PIC) and the existence of a nuclear preintegration block in resting CD4+ T cells have also been reported (27, 28). The prevailing view is that latency is usually established in CD4+ cells when the virus infects an activated CD4+ T cell as it transitions to the resting memory T cell state. Quiescent infection of naïve CD4+ T cells is also less common (29) but could be established in the thymic environment, where cells are subjected to low-level activation and antiapoptotic factors are induced (30–32). Once cells return to quiescence in the G0 phase of the cell cycle, active transcription and virus production are repressed owing to the cumulative effect of several cellular restriction mechanisms. There is no distinguishing marker that identifies latently infected cells from uninfected T cells, posing a formidable challenge to viral eradication.
However, there may be some contribution of direct but inefficient infection of resting T cells (33). This may occur particularly in mucosal tissue; in the simian immunodeficiency virus (SIV) model, infected CD4+ T cells were discovered in the GALT, which lacked cellular activation markers (34). Another study in a rhesus macaque model of SIV infection reports direct infection of resting CD4+ T cells of the vaginal mucosa. In this setting, resting CD4+ T cells express high levels of the viral coreceptor CCR5 and sufficient intracellular nucleotide concentrations to support viral infection (35). It seems likely that abundant expression of CCR5 and suboptimal activation of human mucosal tissues, owing to its unique environment, may allow direct HIV infection as well, similar to what is seen in the monkey model (36). Signaling through the CCR5 coreceptor may also render resting memory CD4+ T cells permissive for direct infection and may explain why infection is almost always established by CCR5-tropic HIV-1 in humans (37, 38). Cytokines released as a result of cellular response may allow resting CD4+ cells to be permissive to HIV-1 infection (39, 40).
Given these observations, it seems possible that during early/acute HIV infection, a substantial number of CD4+ T cells that are not activated or that are suboptimally activated may become HIV infected. Although it seems likely that many of these cells would later become activated and/or enter the cell cycle because of the global immune activation induced during the ramp-up period of HIV viremia, a significant proportion of the latently infected CD4+ T cell reservoir may be established in the first months of HIV infection. In a small cohort, infection of such cells in patients treated early appeared to be unusually rare (41).
MAINTENANCE OF HIV-1 LATENCY IN MEMORY CD4+ T CELLS
HIV-1 latency can best be described as a lack of virion production and minimal proviral gene expression. Integration of HIV-1 proviral DNA into the host genome is a critical step in the viral life cycle. Therefore, inescapably, the chromatin environment at the site of integration plays an important role in regulation of viral gene expression. Regardless of the site of proviral integration, efficient and sustained viral transcription also requires the stimulatory action of the viral transactivator Tat, as well as certain host cellular factors. Diverse mechanisms restrain viral production. One of the principal approaches to therapies directed at ablating latent provirus seeks to identify the critical restrictions to viral production that, alone or together, can be targeted to purge quiescent proviral genomes.
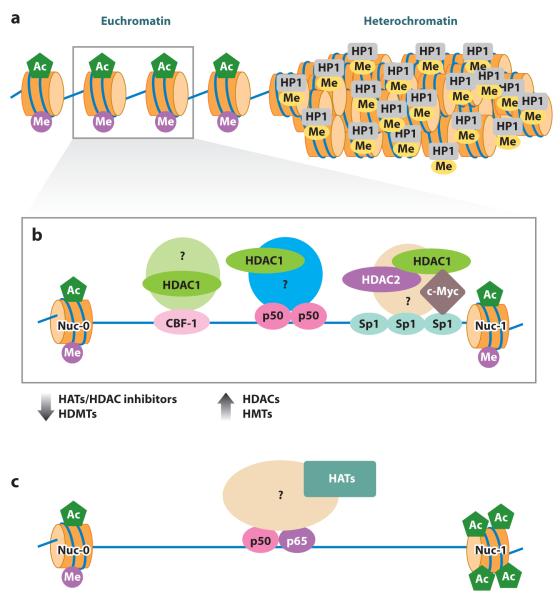
The chromatin environment influences the establishment and maintenance of proviral quiescence (42–47). Human genomic DNA is packaged into a higher-order chromatin structure by the interaction of DNA with histone octomers, and this process forms nucleosomes (Figure 1a). Once integrated, HIV-1 is subject to the same epigenetic mechanisms of regulation as host DNA and must overcome the physical barriers imposed by nucleosome structure for active gene transcription (48). Heterochromatin, which is more compact and structured than euchromatin, reduces the accessibility of transcription factors to the proviral promoter, thereby restricting the initiation of proviral transcription.
Figure 1.
Epigenetic regulation of HIV-1 latency and reactivation. (a) Chromatin structure showing a euchromatic region with acetylated histones ( green) and histone methylated on lysine 4 (H3K4, purple), and a heterochromatic region with repressive methyl marks on lysine 9 (H3K9, yellow) and HP1 (heterochromatin protein 1) recruitment. HIV-1 preferentially integrates into introns of transcriptionally active genes in the euchromatic regions. (b) Proviral latency in the euchromatic region is maintained by recruitment of histone deacetylases (HDACs) 1, 2, and 3 to the HIV-1 long terminal repeat (LTR). Various cellular transcription factors including C-promoter binding factor-1 (CBF-1), nuclear factor κB (NF-κB) p50 homodimer, and Sp1 can participate in recruitment of complexes that include HDAC1. The recruiting complexes such as LSF/YY1 are not shown. HDAC2 is probably recruited in association with HDAC1. Factors that recruit HDAC3 to the LTR are not clearly defined. HDACs maintain a hypoacetylated state of local histones and therefore maintain nucleosome 1 (Nuc-1) in a state that restricts the binding of preinitiation complexes at the viral promoter. Recruitment of histone methyltransferases (HMTs) EZH2 and SUV39H1 can methylate histone H3K9. Methylated H3K9 can be recognized by HP1γ, leading to the generation of heterochromatin, a restrictive chromatin environment for transcription initiation. (c) Histone acetylation is favored by HDAC inhibitors and induced by histone acetyltransferases (HATs) recruited to the LTR by various cellular factors including NF-κB (p50–p65) and nuclear factor of activated T cells (NFAT). This allows sufficient transcription of early viral genes such as tat that, in turn, subverts cellular restrictions, recruits elongation factors, and ensures robust production of viral RNA message. Histone demethylases (HDMTs) remove restrictive H3K9 methylation, relieving repression. Other abbreviations: Ac, acetylated; Me, methylated.
It is unclear, however, which integration events yield replication-competent latent proviral genomes, as the majority of integrants appear to encode defective genomes unable to produce replication-competent virus (29). Although most of these defective genomes likely result from hypermutation induced by cellular APOBEC3G, some integrants may take up residence in regions of the human genome that are highly restrictive to proviral expression.
HIV-1 preferentially integrates into the introns of transcriptionally active genes in resting memory CD4+ T cells (49). This likely results from the interactions of the viral PIC with host factors that are present at regions of active host gene expression. Consistent with this study, Bushman and colleagues (50) found that HIV-1 integrates in transcriptionally active genes during productive infection of cultured T cells. These observations, however, contradict cell line models of quiescent HIV-1 infection in which provirus was found preferentially integrated in heterochromatin (44, 51). This illustrates that cell line models may reflect the outcome of in vivo selection conditions and also that the heterochromatin environment may exert a predominant influence on proviral expression in the uncommon instances where HIV integration occurs in such sites.
Several cellular factors have been reported to interact with the PIC, thus driving the selection of integration site. Lens epithelium-derived growth factor (LEDGF)/p75 has been found to be an important cofactor in directing HIV-1 integration into active sites in the host genome (52, 53). The presence of LEDGF/p75 in Jurkat T cells favors integration in GC-rich regions, whereas the absence of LEDGF/p75 favors integration in AT-rich regions (54). The levels of LEDGF/p75 in different T cell populations may dictate integration into either heterochromatic or euchromatic regions. Indeed, a recent study showed that the integration sites of quiescent/inducible HIV-1 vectors in T cell lines could be associated with both heterochromatin and actively transcribed genes (55).
In this regard, it is important to note that the HIV-1 PIC interacts with the patient SE translocation (SET)/template-activating factor-Iβ (TAF-Iβ) complex, which prevents autointegration of virus into its own genome (56). SET also functions as a histone chaperone and is able to bind unacetylated, hypoacetylated, or repressively marked histones (57–59). SET/TAF-Iβ, therefore, may direct integration of HIV-1 proviral genome into a heterochromatic region.
Regardless of the site of proviral integration, two nucleosomes (Nuc-0 and Nuc-1) are positioned strictly with respect to cis-acting regulatory elements on the viral promoter. Nuc-0 is positioned upstream of the modulatory region, whereas Nuc-1 is immediately downstream of the HIV core promoter and cis-regulatory elements (+10 to +155). A growing list of evidence suggests that Nuc-1 restricts HIV-1 transcription in models of chronic HIV-1 infection and must be displaced during transcriptional activation (46, 60, 61). The ATP-dependent SWItch/Sucrose NonFermentable (SWI/SNF) complex, which can cause the remodeling of nucleosomes, allows efficient transcription of the HIV-1 genome (62–64). It therefore appears that epigenetic silencing at Nuc-1 contributes to HIV-1 latency, and its disruption is a prerequisite for activation of viral expression.
Histones can undergo many post-transcriptional modifications including phosphorylation, acetylation, carbonylation, ubiquitination, sumolylation, ADP-ribosylation, glycosylation, or methylation, any of which can cause changes in both histone structure and function. Broadly, chromatin acetylation by histone acetyltransferases (HATs) allows recruitment of transcriptional activators and transcription complexes, whereas histones deacetylated by histone deacetylases (HDACs) allow the recruitment of complexes that form repressive heterochromatin, resulting in transcriptional repression (Figure 1b,c). Deacetylation of histones at Nuc-1 of the HIV-1 long terminal repeat (LTR) has been demonstrated to pose a barrier to HIV expression following the cooperative recruitment of HDAC1 to the HIV-1 LTR by the host transcription factors yin yang 1 (YY1) and late SV40 factor (LSF) (45). This mechanism was demonstrated to be biologically relevant. Exposure of the resting CD4+ T cells of HIV-infected patients to small synthetic inhibitors (polyamides), which disrupted the binding of LSF to the LTR and therefore the HDAC recruitment at that site, led to the outgrowth of HIV (65). Many drugs that inhibit HDAC activity, such as trichostatin A (TSA) or valproic acid (VPA), induce HIV-1 transcription in latently infected cell lines and resting CD4+ T cells isolated from patients who do not have detectable plasma viremia (43, 61, 66).
Several identified transcription factors can recruit HDAC1 to the LTR including nuclear factor κB (NF-κB) p50 homodimers (67), activator protein 4 (AP-4) (68), C-promoter binding factor-1 (CBF-1) (69), and c-Myc in complex with Sp1 (70) (Figure 1b). Although there are four classes of HDACs, only class I HDACs—including HDAC1, HDAC2, and HDAC3—occupy the HIV-1 LTR as demonstrated by chromatin immunoprecipitation assay (ChIP) (71). This is in accordance with recent findings that class I HDACs are associated with active genes rather than silent genes and that their transient recruitment removes the acetyl group added by HATs, thus keeping the genes inactive (72). The concerted action of HATs and HDACs is critical to the regulation of poised genes.
HDAC1 and HDAC2 are usually co-localized in complexes at promoters. Cellular factors that recruit HDAC3 to the HIV-1 LTR have not been specifically identified. Ras-responsive binding factor 2 (RBF-2) in complex with the transcription factors TFII-I and RBEIII may recruit HDAC3 to the HIV-1 LTR (73, 74). The co-recruitment of HDAC1, HDAC2, and HDAC3 to the LTR suggests that blocking their function individually may not be therapeutically sufficient for efficient viral induction.
Nevertheless, targeting individual transcription factors specific to a particular HDAC recruitment can result in viral expression. We would reconcile this apparent contradiction by hypothesizing that HIV latency is a dynamic state, and that within a population of quiescent proviruses, inhibition of a single target (e.g., HDAC1) could allow expression of a fraction of the population of promoters. Therefore, although specific HDAC inhibitors (HDACis) might allow more selective induction of HIV expression, a considerable amount of study is still required to understand how such reagents should be best employed—alone, together, in series, and/or in combination with other reagents that target additional restrictions to proviral expression.
In this vein, it is important to note that the histone methyltransferases (HMTs) EZH2 and SUV39H1 have also been recently reported to regulate HIV-1 transcription by inducing histone H3 at lysine 9 (H3K9) methylation (75, 76). Binding of the heterochromatin-associated factor heterochromatin protein 1γ (HP1γ) to methylated H3K9 imposes further restrictions on the local chromatin environment (Figure 1). Pearson and colleagues (47) further corroborated those findings and showed that progressive iterative histone modifications drive a proviral promoter into latency in primary CD4+ T cells. Promoter regions of HIV DNA can also be methylated on cytosine residues (referred to as CpG islands) and inhibit viral transcription (77, 78). However, analysis of integrated HIV-1 genomes in resting CD4+ T cells obtained from patients shows infrequent methylation at the promoter region of the HIV LTR (77). Although evidence is accumulating that histone methylation regulates HIV expression, the role of DNA methylation in maintaining HIV-1 latency remains to be established. HIV DNA may be methylated when it is durably quiescent, but removal of the DNA methyl marks does not appear to be required for reactivation of expression (77).
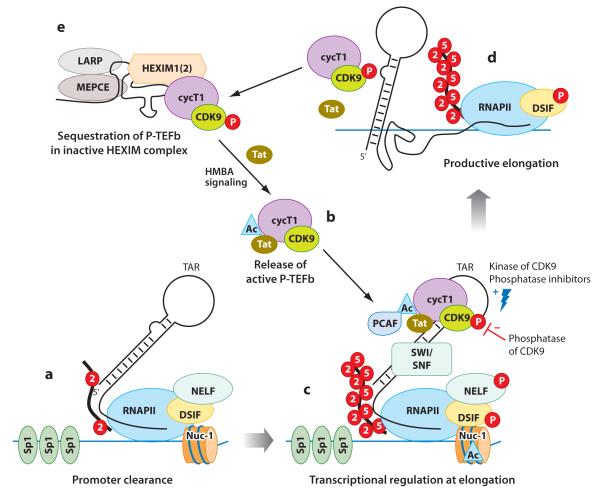
Another important restriction to HIV transcription occurs at the initiation and elongation steps of transcription (79, 80). The promoter of actively transcribed genes is characterized by occupancy of preassembled RNA polymerase II (RNAPII) complexes as well as histones marked with H3K4me3 and H3K9ac. The recruitment of positive transcription elongation factor b (P-TEFb) allows such stalled polymerases to overcome this block and initiate efficient transcription (79). As HIV-1 tends to integrate into the introns of actively transcribed genes, its transcription is more likely to be regulated in a manner similar to that of inducible genes (49). Indeed, the promoter and cis-regulatory regions of the HIV-1 LTR is accessible to binding of transcription initiation complexes (81). Initiation of HIV-1 transcription in resting CD4+ T cells is detectable but often results in an abortive transcript resulting from a block at transcriptional elongation (82). The presence of Nuc-1 likely contributes to this block. The removal of Nuc-1 as a result of chromatin remodeling allows sufficient transcription of early viral genes such as the viral transactivator Tat. Once Tat is expressed, it recruits P-TEFb to the HIV-1 promoter region and ensures sustained and enhanced production of viral transcripts.
The HIV-1 core promoter contains three binding sites for the general transcription factor Sp1, which is critical to formation of the PIC (Figure 2a). In the absence of Tat, transcription is initiated but stalled around position +59, well into the region occupied by Nuc-1. The block to elongation at this point is due to the presence of the Spt5 subunit of the DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) sensitivity-inducing factor (DSIF) and the negative elongation factor (NELF) (83, 84). However, this initial HIV-1 transcript encodes the stem-loop structure of TAR (trans-activation-responsive region), a docking site for Tat (85) (Figure 2a). Tat can recruit P-TEFb, a complex composed of cyclin T1 (cycT1) and cyclin-dependent kinase 9 (CDK9), to TAR. The CDK9-mediated phosphorylation of NELF relieves the transcriptional block, and the phosphorylation of DSIF converts it to a positive transcriptional elongation factor (83, 86–88) (Figure 2b,c). Tat also allows CDK9 to phosphorylate serine 2 (Ser2) and serine 5 (Ser5) of the carboxy-terminal domain (CTD) of RNAPII, prompting both efficient initiation and elongation (89) (Figure 2c,d). Recently, it was discovered that Tat recruits elongation factor ELL2 to P-TEFb for more coordinated activation of HIV-1 transcription (90). Tat also recruits acetyltransferase p300/CBP-associated factor (PCAF), which in turn acetylates Tat and enhances its ability to recruit P-TEFb to the promoter region (91, 92). In several cell line model systems, Tat promotes binding of several cellular factors, including the ATP-dependent remodeling complex SWI/SNF (62, 93) and the histone-modifying enzymes p300 and CREB binding protein (CBP) (94) as well as the histone chaperone protein nucleosome assembly protein 1 (hNAP1) (95). These transcription factors further assist in chromatin unfolding and efficient transcription. The lack of Tat or Tat-associated cellular factors appears to result in inefficient transcription and maintenance of HIV-1 latency.
Figure 2.
Regulation of transcriptional elongation by viral protein Tat. (a) HIV-1 transcription is initiated, but processivity of RNA polymerase II (RNAPII) is stalled at the +59 nucleotide region due to the concerted action of negative elongation factor (NELF), DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) sensitivity-inducing factor (DSIF), and nucleosome 1 (Nuc-1). These short transcripts encode TAR (trans-activation-response region). In the absence of Tat, transcription is largely nonprocessive. (b, c) Tat recruits positive transcription elongation factor b (P-TEFb) from the inactive hexamethylene bisacetamide–inducible protein 1 (HEXIM1) complex onto TAR. Acetylation of Tat at lysine 28 by acetyltransferases enhances its TAR-binding activity as well as kinase activity (91, 94). Acetylation of Tat also facilitates the release of P-TEFb from the inactive HEXIM complex. Hexamethylene bisacetamide (HMBA), a promising antilatency drug, similarly increases the pool of active P-TEFb available to the viral promoter. Following recruitment to TAR, cyclin-dependent kinase 9 (CDK9) is phosphorylated at threonine 186, which increases its kinase activity. Phosphatase inhibitors specific for this step are potential drug candidates. Active P-TEFb phosphorylates serine 2 (Ser2) and serine 5 (Ser5) of the carboxy-terminal domain (CTD) of RNAPII, therefore increasing both the initiation and the elongation of transcription. Phosphorylation of NELF by P-TEFb prompts its release, whereas phosphorylation of DSIF converts it to a positive elongation factor. Furthermore, Tat recruits the ATP-dependent remodeling complex SWI/SNF (SWItch/Sucrose NonFermentable complex) to the promoter to enhance nucleosome modeling and increase transcriptional elongation. (d, e) Phosphorylation of CDK9 also signals its sequestration into a large, inactive HEXIM1 complex. Acetylation of Tat at K50 leads to its dissociation from TAR, making it available for subsequent rounds of transcriptional activation. Other abbreviations: Ac, acetylated; cycT1, cyclin T1; LARP, La-related protein; MEPCE, methyl phosphate capping enzyme; P, phosphorylated; PCAF, p300/CBP-associated factor.
As P-TEFb is critical to regulation of transcription of mammalian genes, it is no surprise that its availability and activity are tightly regulated. P-TEFb is sequestered in an enzymatically inactive complex composed of hexamethylene bisacetamide–inducible protein 1 (HEXIM1) or HEXIM2, the 7SK small nuclear ribonucleoprotein, the La-related protein 7 (LARP7), and the methyl phosphate capping enzyme (MEPCE) (96–103) (Figure 2e). HEXIM1 binds to P-TEFb directly by interacting with the cycT1 subunit of P-TEFb, whereas 7SK serves as a scaffold to facilitate HEXIM1/P-TEFb binding. MEPCE and LARP7 provide stability to 7SK; therefore, their silencing results in enhanced transcription from cellular polymerase II promoters as well as from Tat-dependent HIV-1 promoter (101–103). Tat outcompetes HEXIM1 for P-TEFb binding and recruits it to TAR, inducing productive transcription (104). To add to this complexity, a recent report suggests that HEXIM1 can bind double-stranded RNA structures to TAR and thereby inhibit P-TEFb activity in the absence of Tat (105, 106).
The kinase activity of P-TEFb is regulated by complex post-translational modification as well. Phosphorylation of CDK9 at threonine 186 (Thr186) causes increased P-TEFb kinase activity, but it also results in increased sequestration into the inactive HEXIM1 complex (107, 108). Serine/threonine protein phosphatase PP2A and phosphatase PPM1 (also known as PP2Cα) dephosphorylate P-TEFb and negatively regulate its kinase function (109, 110). Acetylation of CDK9 at conserved lysines in the catalytic core modulates its kinase function and thus decreases its transcriptional activity (111). In contrast, acetylation of cycT1 triggers dissociation of P-TEFb from the HEXIM1 inhibitory complex, thereby increasing transcriptional activation (112).
A deficiency of host cellular factors in resting CD4+ T cells can also contribute to the maintenance of latency. For instance, resting CD4+ T cells express lower levels of cycT1 and CDK9, the components of P-TEFb, therefore increasing the innate barrier to HIV-1 transcription (113–115). Similarly, lower levels of polypyrimidine tract binding protein (PTB) in resting cells result in nuclear retention of multiply spliced transcripts of tat and rev, which are critical to viral expression (116). Conversely, signaling pathways that stimulate T cells and increase cycT1 and CDK expression increase HIV-1 transcription (113–115). High expression of Tat or PTB removes the requirement for cell activation to yield efficient virus production (116). In addition, host microRNA may impede HIV-1 production in resting cells, as the viral TAR RNA serves as a target for Dicer cleavage (117, 118). Therefore, resting cells provide an ideal environment for the maintenance of HIV-1 latency. It is a challenge to devise therapeutics that can induce the expression of virus in such a reservoir and allow the clearance of these persistently infected cells.
MODELS TO STUDY PHARMACEUTICAL APPROACHES TO TARGET HIV-1 LATENCY
Although we have made good progress in understanding the molecular mechanisms of HIV-1 latency and identifying the targets for pharmacologic intervention to induce viral transcription, the lack of relevant preclinical systems has hampered the progress of translational research. Currently, approaches to disrupt latency are evaluated in chronically infected cell line systems or ex vivo in primary cells. The establishment of latency in cell line systems is often linked to mutations in viral genes or to an effect specific to the site of integration—perhaps not uniformly representative of the quiescent nature of resting CD4+ T cells in patients.
Viral outgrowth using resting CD4+ T cells isolated from ART-treated aviremic HIV-infected patients is the gold-standard tool for screening and evaluating antilatency drug candidates, although it is a difficult and costly one. Latently infected cells are treated with a drug candidate to induce virus production, and viral progeny have to be amplified through co-culture with allogeneic, activated, CD8-depleted, peripheral blood mononuclear cells over a 2-week period (119, 120).
Although assays of resting CD4+ T cells obtained from patients are an excellent system for validating the antilatency activity of drug candidates, a whole-animal system is needed for a more complete evaluation of antilatency strategies. A small animal model that recapitulates HIV-1 latency on ART will greatly enhance studies including dose-finding studies and combinatorial approaches to purge latent reservoirs. Moreover, human studies are slow and difficult and are likely to confer some risk to patients who are otherwise clinically stable. Development of a small animal model of latency is therefore crucial to rigorously test the efficacy of novel approaches and their effects on tissue reservoirs.
The SCID-hu (Thy/Liv) mouse model provides the cellular thymic microenvironment necessary for the generation of latently infected naïve cells (30, 121). However, resting memory CD4+ T cell infection—the major source of persistent infection in humans—is absent in SCID-hu (Thy/Liv) mice. The SIV-infected macaque is another important model, in which the existence of persistently infected resting memory CD4+ T cells has been demonstrated (122).
We have recently demonstrated that combination ART in HIV-1-infected hu-Rag2−/− γc−/− mice recapitulates some aspects of ART in humans. Complete suppression of viremia on ART and viral rebound following discontinuation of ART were observed, suggesting the presence of persistent infection in this model (123). Resting memory CD4+ T cells constitute the predominant human T cell population in hu-Rag2−/− γc−/− mice. Studies to definitively quantitate the frequency of latent infection in resting memory CD4+ T cells in hu-Rag2−/− γc−/− mice are now under way.
PHARMACOLOGIC STRATEGIES TO TARGET HIV-1 LATENCY IN CD4+ T CELLS
Eradication of HIV-1 is an extremely challenging goal. HIV-1 persists, and reservoirs are maintained despite intensification of ART (124, 125). The unrelenting efforts of the scientific community to understand the molecular mechanisms of establishment, maintenance, and disruption of latency have provided clues that may allow the development of strategies for viral eradication. A central assumption of most current strategies is that the reactivation of HIV expression within the reservoir cells will make the infected cells vulnerable to elimination by host immune response and viral cytopathic effects. At the same time, viral spread must be prevented by antiretrovirals of sufficient efficacy and potency. ART that is sufficient to suppress plasma viremia may not be sufficient to completely block spread of viral infection within tissues.
As maintenance of HIV-1 latency is multifactorial, combination approaches targeting multiple blocks are likely to be required for complete elimination of HIV-1. Here we suggest several therapeutic targets that could be subject to pharmacologic intervention.
Epigenetic Regulation of HIV-1 Latency
Class I HDACs are critical to deacetylation of histone at Nuc-1 of the HIV-1 LTR and to maintenance of HIV-1 latency in chronically infected cell lines (43, 61, 66). Inhibition of HDAC enzymatic activity also leads to induction of virus expression in resting CD4+ T cells of HIV-1-infected patients (126). Selective inhibitors of class I HDACs 1, 2, and 3 were effective at inducing viral expression from patients’ cells (119). In contrast, class II–specific HDACis—including those of HDACs 4, 5, 7, and 9—were poor in inducing virus. More importantly, HDACi treatment of resting CD4+ T cells did not result in T cell activation and therefore is unlikely to augment spread of virus after induction of the expression of latent HIV-1 (119).
HDACis offer several pharmacologic advantages. Weak HDACis have been used for treating various human ailments for years with good tolerance, and a pipeline of more potent inhibitors is in development for uses in oncology and other therapeutic areas. Moreover, the lipophilic nature of most small-molecule HDACis suggests that they could access anatomic reservoirs, such as brain microglia.
The success of VPA, an approved drug to treat epilepsy, in inducing latent infection in ex vivo primary cells prompted its evaluation for eliminating the HIV-1 reservoir in HIV-1-infected patients. A significant decline in resting CD4+ T cell infection was observed in three out of four patients treated with VPA who were also receiving intensified ART (127). In the absence of ART intensification, however, only four of eleven patients receiving VPA showed a significant depletion of resting cell infection (128). Other studies involving VPA did not measure a decline in resting cell infection (129, 130).
A clinical trial using the more potent class I–specific inhibitor suberoylanilide hydroxamic acid (SAHA), a drug approved to treat cutaneous leukemia, is in development. The hope is to demonstrate proof of concept that a clinically achievable exposure to an HDACi might disrupt latent HIV-1 infection.
H3K9 and H3K27 methylation are associated with a restrictive chromatin environment at the HIV-1 LTR. Factors that stimulate HIV-1 transcription result in demethylation at these sites (131). HMTs, in concert with HDACs, appear to contribute to epigenetic silencing of HIV-1 transcription. Histone methyltransferase inhibitors (HMTis) are therefore new pharmacologic candidates to induce the expression of latent HIV-1. However, these reagents have only begun to be studied for this purpose, despite their application in cancer chemotherapy. Three specific inhibitors of lysine methyltransferase are available: chaetocin, 3-deazaneplanocin A, and BIX-01294. A recent report suggests that BIX-01294 can induce HIV-1 expression in a cell line model of latency. Further study of such compounds should be done.
The role of DNA CpG methylation alone in establishing latency remains controversial at best. DNA CpG methylation has recently been shown to repress HIV-1 transcription in a chronically infected cell line model (77, 78). However, the HDACi SAHA was the most effective reagent for induction of HIV-1 expression in heavily methylated LTRs. In some experiments, whereas HIV-1 quiescence was associated with methylated LTR DNA, SAHA induced expression of the LTR without demethylating these sites. Nevertheless, the removal of DNA methylation by the DNA cytosine methylation inhibitor 5-aza-21-deoxycytidine led to induction of HIV-1 expression in some settings and warrants further in-depth studies in primary cells and other model systems (77).
Activation of Positive Transcription Elongation Factor b
Restriction of the nuclear level of active P-TEFb also contributes to proviral latency. Because of the critical role of P-TEFb in controlling mammalian gene expression, it is tightly regulated in active and inactive complexes by tethering to HEXIM1/2. HIV-1 Tat efficiently recruits active P-TEFb to the HIV-1 promoter region to remove the block at transcriptional elongation (104). P-TEFb, however, can activate HIV-1 transcription in the absence of Tat (132). Hexamethylene bisacetamide (HMBA) can induce the release of P-TEFb from HEXIM1 and enhance HIV-1 expression without NF-κB and Tat (133, 134). P-TEFb might be recruited to the HIV-1 promoter by several transcription factors including Sp1 and NF-κB or by Brd4 in the absence of Tat (133, 135, 136). Agents such as HMBA might offer another approach to induce the expression of quiescent persistent HIV infection.
Post-translational modification of HEXIM1 as well as P-TEFb is important for P-TEFb release from inhibitory complexes and full activation (108, 112, 134). The cellular protein phosphatase PP1 may be involved in the release of P-TEFb in both Tat-dependent and -independent manners (137, 138). The C terminus of CDK9 has several phosphorylation sites at serine and threonine residues (Ser347, Ser354, and Ser357; Thr350, Thr354, and Thr186). The phosphorylation of these residues is important for nuclear transport and modulation of P-TEFb kinase activity (107, 109, 139). Phosphorylation of CDK9 at Thr29 is associated with inhibition of kinase activity at the HIV-1 promoter (140). It is obvious that CDK9 undergoes a complex cycle of phosphorylation and dephosphorylation for promoter recruitment and full activation. PP1 and PP2A have been implicated in these processes. Global inhibition of PP2A by pharmacologic inhibitors such as okadaic acid has produced mixed results, inducing HIV-1 expression in some chronically infected cell lines while blunting its expression in others (141, 142). More careful research is needed to fully understand how, when, and which phosphatase causes the activation of P-TEFb.
Induction of Protein Kinase C Signaling Pathway
Protein kinase C (PKC) signaling plays an important role in the activation of nuclear factor of activated T cells (NFAT), NF-κB, and activator protein 1 (AP-1), steps that are essential for T cell activation. The HIV-1 LTR contains binding sites for these factors, which have a proven role in activating viral expression. NF-κB in particular plays multiple roles in HIV-1 transcription: (a) chromatin repressive p50/p50 homodimers are replaced with p50/p65 heterodimers, displacing HDAC (67); (b) p50/p65 recruits HATs such as p300 and CBP to the promoter, in a process that could play an important role in acetylating histones and Tat (46, 143); and (c) p50/p65 recruits P- TEFb to the promoter to regulate transcriptional elongation (136). Williams and colleagues (144) noted that sustained activation of NF-κB was efficient in induction of HIV-1 latency. Several pharmacologic PKC agonists including a jatrophane diterpene (named SJ23B) (145), 12-deoxyphorbol 13-phenylacetate (dPP) (146), and prostratin (147) induce proviral expression efficiently in latently infected CD4+ T cells.
The PKC agonist prostratin is effective at inducing expression of virus from both latently infected cell lines and primary cells (147–149). The combination of prostratin with an HDACi resulted in the synergistic induction of virus in chronically infected cell lines and latently infected patient cells (150, 151). Prostratin downregulates expression of the HIV receptors CD4 and CXCR4, therefore inhibiting de novo HIV infection (149). Prostratin is currently extracted from a medicinal plant, Homolanthus nutans, and toxicities in preclinical studies have delayed human testing of the drug. A recent breakthrough involves the synthetic production of prostratin, which may allow rapid evaluation of drug parameters such as pharmacokinetic properties and toxicity studies (152).
A more specific inducer of NF-κB that does not appear to affect NFAT and PKC has been recently reported (153). The compound, 5-hydroxynaphthalene-1,4-dione (HN), induced latent infection in primary cells without inducing T cell activation. Because HN is a quinone, it produced reactive oxygen species (ROS) and may not be fit for clinical studies. Nevertheless, the results suggest that screening compounds that specifically activate NF-κB without globally activating T cells will yield interesting pharmacologic candidates to eliminate persistent infection.
Inhibiting Survival of Memory Cells Following Homeostatic Proliferation
Interleukin 7 (IL-7) induces viral expression from quiescent naïve cells and latently infected resting cells from patients via the JAK/STAT signaling pathway; NFAT is a key mediator of this effect (154, 155). Recently, recombinant IL-7 was also tested in clinical trials for human use and led to an increase in T cell population (156). Chomont and colleagues (9), however, suggested that central memory (TCM) and transitional memory (TTM) are the major cellular reservoirs in HIV-1-infected patients, and that homeostatic proliferation of central memory cells may expand the infected cell reservoir via mitosis. Depletion of CD4+ T cells in chronically infected patients results in increased plasma IL-7 levels, which promote proliferation and survival of the TTM subset. IL-7 induces Bcl-2, a key antiapoptotic protein, ensuring survival of proliferated cells. Recently, Abbott Laboratories (Abbott Park, Illinois) has developed Bcl-2 inhibitors ABT-737 and ABT-263, under clinical study for cancer therapy (157). Such a Bcl-2 inhibitor might overcome this block of apoptosis to ensure death of proliferating HIV-1-infected T cells.
TARGETING HIV-1 IN THE CENTRAL NERVOUS SYSTEM
HIV-1 infection in the CNS is established shortly after primary infection (10). HIV-1-infected CD4+ T cells and monocytes/macrophages primarily serve to traffic HIV-1 across the blood-brain barrier (158, 159). The rapid decay of HIV-1 in cerebrospinal fluid (CSF) parallels the kinetics of CD4+ T cells in the periphery in asymptomatic patients on ART, suggesting that plasma lymphocytes could be a source of viral load in CSF. Alternatively, monocytes originating from the bone marrow could repopulate perivascular, meningeal, and choroid plexus in the CNS, a “Trojan horse” mechanism responsible for disseminating HIV-1 to the brain (160, 161). Perivascular macrophages and microglia in particular are considered a primary source of HIV-1 production in the brain (162). Microglia, which are long-lived cells, are a potential source of persistent infection (163).
Epigenetic regulation of gene expression has been implicated in the maintenance of viral latency in microglia. The transcription factor chicken ovalbumin upstream promoter transcription factor interacting protein 2 (CTIP2) recruits HDAC1 or HDAC2 to the viral promoters, causing histone deacetylation and generating a repressive environment not conducive for gene expression (164, 165). Furthermore, the interaction of CTIP2 with SUV39H1 causes histone H3 methylation, which may further restrict HIV-1 expression in microglia (165). Targeting these cells with a combination of HDACis and HMT inhibitors, as discussed earlier, will be a logical way to target this latent infection.
In chronically infected patients, virus detected in the CSF originates from both plasma lymphocytes and local CNS tissue, suggesting a mixing of virus between the CNS and peripheral circulation (166). This is particularly important because it implies that virus from the CNS compartment could seed new infections in peripheral blood. Delayed virus decay is sometimes observed in the CSF in patients on ART with sustained viral replication even during long-term ART (13, 14). This could be related to the exclusion of some drugs from certain anatomic compartments. Therefore, efforts to eradicate persistent HIV-1 infection in the CNS must seek to enhance delivery of anti-HIV or antilatency drugs in CNS tissue.
Nanoparticles, which are solid colloidal particles typically in the size range of 100–300 nm, offer an attractive vehicle for delivering antiretrovirals and antilatency drugs to the CNS to target persistent HIV-1 infection. The therapeutic agent(s) could be entrapped or chemically linked to the surface of nanoparticles for delivery. To overcome the challenge of penetrating the tight endothelial junctions of the blood-brain barrier, nanoparticles could be conjugated to specific ligands such as thiamine, transferrin, or glucose for efficient delivery. The expression of receptors for these ligands on neuronal tissues is likely to increase their binding and therefore increase receptor-mediated uptake across the blood-brain barrier. Moreover, receptor-mediated transcytosis offers a targeted delivery, which reduces undue exposure to other organs.
In a proof-of-concept study, our group has recently shown that small (2-nm) gold nanoparticles can be derivatized to deliver multiple functional units. These particles, coated with a TAK-779 homolog, were effective inhibitors of HIV-1 entry (167). Tat-conjugated nanoparticles encapsulated with ritonavir, a protease inhibitor, enhanced sustained brain delivery of drugs without influencing the integrity of the blood-brain barrier (168). Targeted delivery of antiretrovirals and/or antilatency drugs may be necessary to effectively target latency in sanctuary sites.
PERSPECTIVES AND FUTURE DIRECTIONS
The eradication of persistent infection is a difficult challenge, as the mechanisms of HIV-1 latency are still not completely understood. The study of mechanisms of HIV-1 latency in primary cells has been hampered owing to the rare nature of infection; fewer than one out of one million cells are latently infected. The difficulty and importance of this challenge have led to more concerted efforts from the scientific community, resulting in a consistent gain in the understanding of HIV-1 latency. Together with early and effective ART to reduce the latent reservoir, therapies that target HIV-1 latency, cryptic replication, and sanctuary sites are needed to eradicate HIV-1 infection.
It is now increasingly clear that epigenetic restrictions pose an initial hurdle to viral transcription and cause maintenance of viral latency. A better understanding of the epigenetic regulation of HIV-1 latency and identification of pharmacologic targets will open avenues to rational therapeutic approaches for clearing infection. It is also possible that low-level expression of virus from cells may not prompt immune-mediated killing or apoptotic/necrotic killing. Other reagents or strategies (e.g., immunotherapy) to clear cells induced to express HIV antigens may be required.
HIV-1 latency is a result of several restrictions imposed by both cellular and viral factors and therefore requires a multiprong approach to disrupt it. HDACis appear promising, but adding molecules such as HMBA that induce HIV-1 expression by removing the block at transcriptional elongation or adding the NF-κB inducer prostratin might potentiate their effects. Other obstacles, such as inadequate cytoplasmic transport of transcribed viral mRNAs, might prove critical and require specific programs to therapeutically breach this barrier.
The complete eradication of HIV-1 will not be achieved until virus hidden in sanctuary sites can also be targeted. The design of better drugs and delivery systems is required; nanotechnology offers an attractive tool to achieve this goal. The development of an animal model of latency is also critical to allow rigorous testing of novel pharmacologic approaches before their application in HIV-1-infected patients. Development of such models will greatly accelerate our progress toward clinical trials aimed at eradicating HIV infection in patients. Eradication of HIV-1 latency requires a prolonged scientific commitment to understanding the molecular mechanism of persistence, to safe and effective drug discovery, to rational design of therapeutic approaches, and to testing in adequate model systems before clinical application.
ACKNOWLEDGMENTS
We thank Dr. Nancy Archin and Kara Keedy for critical reading of this manuscript. This work was supported by National Institutes of Health grants R21-AI081613 to S.K.C. and grants AI082608, MH085597, and DA030156 to D.M.M.
Footnotes
DISCLOSURE STATEMENT
D.M.M. receives research funding and honoraria from Merck Research Laboratories and has submitted a patent application for the use of HDACis in HIV therapy.
LITERATURE CITED
- 1.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA. 1997;94:13193–97. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. Early establishment of a pool of latently infected, resting CD4+ T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 1998;95:8869–73. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 2003;9:727–28. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 4.Blankson JN, Persaud D, Siliciano RF. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 2002;53:557–93. doi: 10.1146/annurev.med.53.082901.104024. [DOI] [PubMed] [Google Scholar]
- 5.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 6.Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. USA. 2008;105:3879–84. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Santo JP. Lung Kruüpple-like factor: a quintessential player in T cell quiescence. Nat. Immunol. 2001;2:667–68. doi: 10.1038/90598. [DOI] [PubMed] [Google Scholar]
- 8.Haaland RE, Yu W, Rice AP. Identification of LKLF-regulated genes in quiescent CD4+ T lymphocytes. Mol. Immunol. 2005;42:627–41. doi: 10.1016/j.molimm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis LE, Hjelle BL, Miller VE, Palmer DL, Llewellyn AL, et al. Early viral brain invasion in iatrogenic human immunodeficiency virus infection. Neurology. 1992;42:1736–39. doi: 10.1212/wnl.42.9.1736. [DOI] [PubMed] [Google Scholar]
- 11.Lambotte O, Chaix ML, Gasnault J, Goujard C, Lebras P, et al. Persistence of replication-competent HIV in the central nervous system despite long-term effective highly active antiretroviral therapy. AIDS. 2005;19:217–18. doi: 10.1097/00002030-200501280-00018. [DOI] [PubMed] [Google Scholar]
- 12.Chun TW, Nickle DC, Justement JS, Meyers JH, Roby G, et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J. Infect. Dis. 2008;197:714–20. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- 13.Gisolf EH, Enting RH, Jurriaans S, de Wolf F, dan der Ende ME, et al. Cerebrospinal fluid HIV-1 RNA during treatment with ritonavir/saquinavir or ritonavir/saquinavir/stavudine. AIDS. 2000;14:1583–89. doi: 10.1097/00002030-200007280-00014. [DOI] [PubMed] [Google Scholar]
- 14.Solas C, Lafeuillade A, Halfon P, Chadapaud S, Hittinger G, Lacarelle B. Discrepancies between protease inhibitor concentrations and viral load in reservoirs and sanctuary sites in human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 2003;47:238–43. doi: 10.1128/AAC.47.1.238-243.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collman R, Hassan NF, Walker R, Godfrey B, Cutilli J, et al. Infection of monocyte-derived macrophages with human immunodeficiency virus type 1 (HIV-1): Monocyte-tropic and lymphocyte-tropic strains of HIV-1 show distinctive patterns of replication in a panel of cell types. J. Exp. Med. 1989;170:1149–63. doi: 10.1084/jem.170.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orenstein JM, Fox C, Wahl SM. Macrophages as a source of HIV during opportunistic infections. Science. 1997;276:1857–61. doi: 10.1126/science.276.5320.1857. [DOI] [PubMed] [Google Scholar]
- 17.Ho DD, Rota TR, Hirsch MS. Infection of monocyte/macrophages by human T lymphotropic virus type III. J. Clin. Investig. 1986;77:1712–15. doi: 10.1172/JCI112491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swingler S, Mann AM, Zhou J, Swingler C, Stevenson M. Apoptotic killing of HIV-1-infected macrophages is subverted by the viral envelope glycoprotein. PLoS Pathog. 2007;3:1281–90. doi: 10.1371/journal.ppat.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharova N, Swingler C, Sharkey M, Stevenson M. Macrophages archive HIV-1 virions for dis-semination in trans. EMBO J. 2005;24:2481–89. doi: 10.1038/sj.emboj.7600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swingler S, Mann A, Jacque J, Brichacek B, Sasseville VG, et al. HIV-1 Nef mediates lymphocyte chemotaxis and activation by infected macrophages. Nat. Med. 1999;5:997–103. doi: 10.1038/12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lore K, Smed-Sorensen A, Vasudevan J, Mascola JR, Koup RA. Myeloid and plasmacytoid dendritic cells transfer HIV-1 preferentially to antigen-specific CD4+ T cells. J. Exp. Med. 2005;201:2023–33. doi: 10.1084/jem.20042413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otero M, Nunnari G, Leto D, Sullivan J, Wang FX, et al. Peripheral blood dendritic cells are not a major reservoir for HIV type 1 in infected individuals on virally suppressive HAART. AIDS Res. Hum. Retroviruses . 2003;19:1097–103. doi: 10.1089/088922203771881194. [DOI] [PubMed] [Google Scholar]
- 23.Stanley SK, Kessler SW, Justement JS, Schnittman SM, Greenhouse JJ, et al. CD34+ bone marrow cells are infected with HIV in a subset of seropositive individuals. J. Immunol. 1992;149:689–97. [PubMed] [Google Scholar]
- 24.Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 2006;80:6441–57. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyerhans A, Vartanian JP, Hultgren C, Plikat U, Karlsson A, et al. Restriction and enhancement of human immunodeficiency virus type 1 replication by modulation of intracellular deoxynucleoside triphosphate pools. J. Virol. 1994;68:535–40. doi: 10.1128/jvi.68.1.535-540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–14. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 27.Stevenson M, Stanwick TL, Dempsey MP, Lamonica CA. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–60. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: Molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–22. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 29.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–88. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 30.Brooks DG, Kitchen SG, Kitchen CM, Scripture-Adams DD, Zack JA. Generation of HIV latency during thymopoiesis. Nat. Med. 2001;7:459–64. doi: 10.1038/86531. [DOI] [PubMed] [Google Scholar]
- 31.Yuan J, Crittenden RB, Bender TP. c-Myb promotes the survival of CD4+CD8+ double-positive thymocytes through upregulation of Bcl-xL. J. Immunol. 2010;184:2793–804. doi: 10.4049/jimmunol.0902846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dzhagalov I, Dunkle A, He YW. The anti-apoptotic Bcl-2 family member Mcl-1 promotes T lymphocyte survival at multiple stages. J. Immunol. 2008;181:521–28. doi: 10.4049/jimmunol.181.1.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swiggard WJ, Baytop C, Yu JJ, Dai J, Li C, et al. Human immunodeficiency virus type 1 can establish latent infection in resting CD4+ T cells in the absence of activating stimuli. J. Virol. 2005;79:14179–88. doi: 10.1128/JVI.79.22.14179-14188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, Duan L, Estes JD, Ma ZM, Rourke T, et al. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:1148–52. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z, Schuler T, Zupancic M, Wietgrefe S, Staskus KA, et al. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science. 1999;286:1353–57. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]
- 36.Zhang ZQ, Wietgrefe SW, Li Q, Shore MD, Duan L, et al. Roles of substrate availability and infection of resting and activated CD4+ T cells in transmission and acute simian immunodeficiency virus infection. Proc. Natl. Acad. Sci. USA. 2004;101:5640–45. doi: 10.1073/pnas.0308425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin YL, Mettling C, Portales P, Reant B, Clot J, Corbeau P. G-protein signaling triggered by R5 human immunodeficiency virus type 1 increases virus replication efficiency in primary T lymphocytes. J. Virol. 2005;79:7938–41. doi: 10.1128/JVI.79.12.7938-7941.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y, Yoder A. Chemokine coreceptor signaling in HIV-1 infection and pathogenesis. PLoS Pathog. 2009;5:e1000520. doi: 10.1371/journal.ppat.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Unutmaz D, KewalRamani VN, Marmon S, Littman DR. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 1999;189:1735–46. doi: 10.1084/jem.189.11.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chun TW, Engel D, Mizell SB, Ehler LA, Fauci AS. Induction of HIV-1 replication in latently infected CD4+ T cells using a combination of cytokines. J. Exp. Med. 1998;188:83–91. doi: 10.1084/jem.188.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chun TW, Justement JS, Moir S, Hallahan CW, Maenza J, et al. Decay of the HIV reservoir in patients receiving antiretroviral therapy for extended periods: implications for eradication of virus. J. Infect. Dis. 2007;195:1762–64. doi: 10.1086/518250. [DOI] [PubMed] [Google Scholar]
- 42.Van Lint C. Role of chromatin in HIV-1 transcriptional regulation. Adv. Pharmacol. 2000;48:121–60. doi: 10.1016/s1054-3589(00)48005-1. [DOI] [PubMed] [Google Scholar]
- 43.Quivy V, Van Lint C. Diversity of acetylation targets and roles in transcriptional regulation: the human immunodeficiency virus type 1 promoter as a model system. Biochem. Pharmacol. 2002;64:925–34. doi: 10.1016/s0006-2952(02)01152-8. [DOI] [PubMed] [Google Scholar]
- 44.Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 2001;20:1726–38. doi: 10.1093/emboj/20.7.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coull JJ, Romerio F, Sun JM, Volker JL, Galvin KM, et al. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 2000;74:6790–99. doi: 10.1128/jvi.74.15.6790-6799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lusic M, Marcello A, Cereseto A, Giacca M. Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J. 2003;22:6550–61. doi: 10.1093/emboj/cdg631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, et al. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 2008;82:12291–303. doi: 10.1128/JVI.01383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 49.Han Y, Lassen K, Monie D, Sedaghat AR, Shimoji S, et al. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 2004;78:6122–33. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–29. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 51.Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22:1868–77. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, et al. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–64. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 53.Hombrouck A, De Rijck J, Hendrix J, Vandekerckhove L, Voet A, et al. Virus evolution reveals an exclusive role for LEDGF/p75 in chromosomal tethering of HIV. PLoS Pathog. 2007;3:e47. doi: 10.1371/journal.ppat.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, et al. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–89. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 55.Lewinski MK, Bisgrove D, Shinn P, Chen H, Hoffmann C, et al. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 2005;79:6610–19. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan N, Cherepanov P, Daigle JE, Engelman A, Lieberman J. The SET complex acts as a barrier to autointegration of HIV-1. PLoS Pathog. 2009;5:e1000327. doi: 10.1371/journal.ppat.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kutney SN, Hong R, Macfarlan T, Chakravarti D. A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Iβ in integrating chromatin hypoacetylation and transcriptional repression. J. Biol. Chem. 2004;279:30850–55. doi: 10.1074/jbc.M404969200. [DOI] [PubMed] [Google Scholar]
- 58.Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–30. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 59.Kato K, Miyaji-Yamaguchi M, Okuwaki M, Nagata K. Histone acetylation-independent transcription stimulation by a histone chaperone. Nucleic Acids Res. 2007;35:705–15. doi: 10.1093/nar/gkl1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verdin E, Paras P, Jr, Van Lint C. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 1993;12:3249–59. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Lint C, Emiliani S, Ott M, Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 1996;15:1112–20. [PMC free article] [PubMed] [Google Scholar]
- 62.Henderson A, Holloway A, Reeves R, Tremethick DJ. Recruitment of SWI/SNF to the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 2004;24:389–97. doi: 10.1128/MCB.24.1.389-397.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Treand C, du Chene I, Bres V, Kiernan R, Benarous R, et al. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J. 2006;25:1690–99. doi: 10.1038/sj.emboj.7601074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizutani T, Ishizaka A, Tomizawa M, Okazaki T, Yamamichi N, et al. Loss of the Brm-type SWI/SNF chromatin remodeling complex is a strong barrier to the Tat-independent transcriptional elongation of human immunodeficiency virus type 1 transcripts. J. Virol. 2009;83:11569–80. doi: 10.1128/JVI.00742-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ylisastigui L, Coull JJ, Rucker VC, Melander C, Bosch RJ, et al. Polyamides reveal a role for repression in latency within resting T cells of HIV-infected donors. J. Infect. Dis. 2004;190:1429–37. doi: 10.1086/423822. [DOI] [PubMed] [Google Scholar]
- 66.Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM. Coaxing HIV-1 from resting CD4 T cells: Histone deacetylase inhibition allows latent viral expression. AIDS. 2004;18:1101–8. doi: 10.1097/00002030-200405210-00003. [DOI] [PubMed] [Google Scholar]
- 67.Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. NF-κB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25:139–49. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Imai K, Okamoto T. Transcriptional repression of human immunodeficiency virus type 1 by AP-4. J. Biol. Chem. 2006;281:12495–505. doi: 10.1074/jbc.M511773200. [DOI] [PubMed] [Google Scholar]
- 69.Tyagi M, Karn J. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 2007;26:4985–95. doi: 10.1038/sj.emboj.7601928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang G, Espeseth A, Hazuda DJ, Margolis DM. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J. Virol. 2007;81:10914–23. doi: 10.1128/JVI.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Keedy KS, Archin NM, Gates AT, Espeseth A, Hazuda DJ, Margolis DM. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J. Virol. 2009;83:4749–56. doi: 10.1128/JVI.02585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Z, Zang C, Cui K, Schones DE, Barski A, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–31. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Malcolm T, Kam J, Pour PS, Sadowski I. Specific interaction of TFII-I with an upstream element on the HIV-1 LTR regulates induction of latent provirus. FEBS Lett. 2008;582:3903–8. doi: 10.1016/j.febslet.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 74.Wen YD, Cress WD, Roy AL, Seto E. Histone deacetylase 3 binds to and regulates the multi-functional transcription factor TFII-I. J. Biol. Chem. 2003;278:1841–47. doi: 10.1074/jbc.M206528200. [DOI] [PubMed] [Google Scholar]
- 75.du Chene I, Basyuk E, Lin YL, Triboulet R, Knezevich A, et al. Suv39H1 and HP1γare responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 2007;26:424–35. doi: 10.1038/sj.emboj.7601517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Friedman J, Pearson R, Karn J. Direct evidence that histone trimethylation is required for HIV proviral silencing during the establishment of latency. Proc. Conf. Retrovir. Oppor. Infect., 16th, Montreal, Can., Feb. 8–11; 2009. Abstr. 264. [Google Scholar]
- 77.Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, et al. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 2009;5:e1000554. doi: 10.1371/journal.ppat.1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 2009;5:e1000495. doi: 10.1371/journal.ppat.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–45. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–45. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Demarchi F, D’Agaro P, Falaschi A, Giacca M. In vivo footprinting analysis of constitutive and inducible protein-DNA interactions at the long terminal repeat of human immunodeficiency virus type 1. J. Virol. 1993;67:7450–60. doi: 10.1128/jvi.67.12.7450-7460.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lassen KG, Bailey JR, Siliciano RF. Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. J. Virol. 2004;78:9105–14. doi: 10.1128/JVI.78.17.9105-9114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ping YH, Rana TM. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 2001;276:12951–58. doi: 10.1074/jbc.M006130200. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Z, Klatt A, Gilmour DS, Henderson AJ. Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J. Biol. Chem. 2007;282:16981–88. doi: 10.1074/jbc.M610688200. [DOI] [PubMed] [Google Scholar]
- 85.Herrmann CH, Rice AP. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 1995;69:1612–20. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–62. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 87.Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 2004;24:787–95. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bourgeois CF, Kim YK, Churcher MJ, West MJ, Karn J. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 2002;22:1079–93. doi: 10.1128/MCB.22.4.1079-1093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, et al. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 2000;20:5077–86. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.He N, Liu M, Hsu J, Xue Y, Chou S, et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell. 2010;38:428–38. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, et al. HIV-1 Tat transcriptional activity is regulated by acetylation. EMBO J. 1999;18:6106–18. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bres V, Kiernan R, Emiliani S, Benkirane M. Tat acetyl-acceptor lysines are important for human immunodeficiency virus type-1 replication. J. Biol. Chem. 2002;277:22215–21. doi: 10.1074/jbc.M201895200. [DOI] [PubMed] [Google Scholar]
- 93.Mahmoudi T, Parra M, Vries RG, Kauder SE, Verrijzer CP, et al. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J. Biol. Chem. 2006;281:19960–68. doi: 10.1074/jbc.M603336200. [DOI] [PubMed] [Google Scholar]
- 94.Benkirane M, Chun RF, Xiao H, Ogryzko VV, Howard BH, et al. Activation of integrated provirus requires histone acetyltransferase: p300 and P/CAF are coactivators for HIV-1 Tat. J. Biol. Chem. 1998;273:24898–905. doi: 10.1074/jbc.273.38.24898. [DOI] [PubMed] [Google Scholar]
- 95.Vardabasso C, Manganaro L, Lusic M, Marcello A, Giacca M. The histone chaperone protein Nucleosome Assembly Protein-1 (hNAP-1) binds HIV-1 Tat and promotes viral transcription. Retrovirology. 2008;5:8. doi: 10.1186/1742-4690-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barboric M, Kohoutek J, Price JP, Blazek D, Price DH, Peterlin BM. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J. 2005;24:4291–303. doi: 10.1038/sj.emboj.7600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, et al. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 2003;23:4859–69. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–25. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 99.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–82. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 100.Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–29. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Markert A, Grimm M, Martinez J, Wiesner J, Meyerhans A, et al. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9:569–75. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.He N, Jahchan NS, Hong E, Li Q, Bayfield MA, et al. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell. 2008;29:588–99. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell. 2007;27:262–74. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Barboric M, Yik JH, Czudnochowski N, Yang Z, Chen R, et al. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 2007;35:2003–12. doi: 10.1093/nar/gkm063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Q, Cooper JJ, Altwerger GH, Feldkamp MD, Shea MA, Price DH. HEXIM1 is a promiscuous double-stranded RNA-binding protein and interacts with RNAs in addition to 7SK in cultured cells. Nucleic Acids Res. 2007;35:2503–12. doi: 10.1093/nar/gkm150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007;35:4347–58. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 2005;280:28819–26. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 108.Chen R, Yang Z, Zhou Q. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 2004;279:4153–60. doi: 10.1074/jbc.M310044200. [DOI] [PubMed] [Google Scholar]
- 109.Wang Y, Dow EC, Liang YY, Ramakrishnan R, Liu H, et al. Phosphatase PPM1A regulates phosphorylation of Thr-186 in the Cdk9 T-loop. J. Biol. Chem. 2008;283:33578–84. doi: 10.1074/jbc.M807495200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ammosova T, Washington K, Debebe Z, Brady J, Nekhai S. Dephosphorylation of CDK9 by protein phosphatase 2A and protein phosphatase-1 in Tat-activated HIV-1 transcription. Retrovirology. 2005;2:47. doi: 10.1186/1742-4690-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sabo A, Lusic M, Cereseto A, Giacca M. Acetylation of conserved lysines in the catalytic core of cyclin-dependent kinase 9 inhibits kinase activity and regulates transcription. Mol. Cell. Biol. 2008;28:2201–12. doi: 10.1128/MCB.01557-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cho S, Schroeder S, Kaehlcke K, Kwon HS, Pedal A, et al. Acetylation of cyclin T1 regulates the equilibrium between active and inactive P-TEFb in cells. EMBO J. 2009;28:1407–17. doi: 10.1038/emboj.2009.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sung TL, Rice AP. Effects of prostratin on Cyclin T1/P-TEFb function and the gene expression profile in primary resting CD4+ T cells. Retrovirology. 2006;3:66. doi: 10.1186/1742-4690-3-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Garriga J, Peng J, Parreno M, Price DH, Henderson EE, Grana X. Upregulation of cyclin T1/CDK9 complexes during T cell activation. Oncogene. 1998;17:3093–102. doi: 10.1038/sj.onc.1202548. [DOI] [PubMed] [Google Scholar]
- 115.Herrmann CH, Carroll RG, Wei P, Jones KA, Rice AP. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J. Virol. 1998;72:9881–88. doi: 10.1128/jvi.72.12.9881-9888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006;2:e68. doi: 10.1371/journal.ppat.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huang J, Wang F, Argyris E, Chen K, Liang Z, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 2007;13:1241–47. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 118.Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, et al. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol. Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Archin NM, Keedy KS, Espeseth A, Dang H, Hazuda DJ, Margolis DM. Expression of latent human immunodeficiency type 1 is induced by novel and selective histone deacetylase inhibitors. AIDS. 2009;23:1799–806. doi: 10.1097/QAD.0b013e32832ec1dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol. Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 121.Brooks DG, Hamer DH, Arlen PA, Gao L, Bristol G, et al. Molecular characterization, reactivation, and depletion of latent HIV. Immunity. 2003;19:413–23. doi: 10.1016/s1074-7613(03)00236-x. [DOI] [PubMed] [Google Scholar]
- 122.Dinoso JB, Rabi SA, Blankson JN, Gama L, Mankowski JL, et al. A simian immunodeficiency virus-infected macaque model to study viral reservoirs that persist during highly active antiretroviral therapy. J. Virol. 2009;83:9247–57. doi: 10.1128/JVI.00840-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Choudhary SK, Rezk NL, Ince WL, Cheema M, Zhang L, et al. Suppression of human immunodeficiency virus type 1 (HIV-1) viremia with reverse transcriptase and integrase inhibitors, CD4+ T-cell recovery, and viral rebound upon interruption of therapy in a new model for HIV treatment in the humanized Rag2−/− γc−/− mouse. J. Virol. 2009;83:8254–58. doi: 10.1128/JVI.00580-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gandhi RT, Bosch RJ, Aga E, Albrecht M, Demeter LM, et al. No evidence for decay of the latent reservoir in HIV-1-infected patients receiving intensive enfuvirtide-containing antiretroviral therapy. J. Infect. Dis. 2010;201:293–96. doi: 10.1086/649569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dinoso JB, Kim SY, Wiegand AM, Palmer SE, Gange SJ, et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA. 2009;106:9403–8. doi: 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res. Hum. Retroviruses. 2009;25:207–12. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, et al. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet. 2005;366:549–55. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Archin NM, Eron JJ, Palmer S, Hartmann-Duff A, Martinson JA, et al. Valproic acid without intensified antiviral therapy has limited impact on persistent HIV infection of resting CD4+ T cells. AIDS. 2008;22:1131–35. doi: 10.1097/QAD.0b013e3282fd6df4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Siliciano JD, Lai J, Callender M, Pitt E, Zhang H, et al. Stability of the latent reservoir for HIV-1 in patients receiving valproic acid. J. Infect. Dis. 2007;195:833–36. doi: 10.1086/511823. [DOI] [PubMed] [Google Scholar]
- 130.Sagot-Lerolle N, Lamine A, Chaix ML, Boufassa F, Aboulker JP, et al. Prolonged valproic acid treatment does not reduce the size of latent HIV reservoir. AIDS. 2008;22:1125–29. doi: 10.1097/QAD.0b013e3282fd6ddc. [DOI] [PubMed] [Google Scholar]
- 131.Tyagi M, Pearson RJ, Karn J. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 2010;84(13):6425–37. doi: 10.1128/JVI.01519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Klichko V, Archin N, Kaur R, Lehrman G, Margolis D. Hexamethylbisacetamide remodels the human immunodeficiency virus type 1 (HIV-1) promoter and induces Tat-independent HIV-1 expression but blunts cell activation. J. Virol. 2006;80:4570–79. doi: 10.1128/JVI.80.9.4570-4579.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Choudhary SK, Archin NM, Margolis DM. Hexamethylbisacetamide and disruption of human immunodeficiency virus type 1 latency in CD4+ T cells. J. Infect. Dis. 2008;197:1162–70. doi: 10.1086/529525. [DOI] [PubMed] [Google Scholar]
- 134.Contreras X, Barboric M, Lenasi T, Peterlin BM. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 2007;3:1459–69. doi: 10.1371/journal.ppat.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang Z, Yik JH, Chen R, He N, Jang MK, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–45. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 136.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell. 2001;8:327–37. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 137.Chen R, Liu M, Li H, Xue Y, Ramey WN, et al. PP2B and PP1α cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22:1356–68. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ammosova T, Jerebtsova M, Beullens M, Lesage B, Jackson A, et al. Nuclear targeting of protein phosphatase-1 by HIV-1 Tat protein. J. Biol. Chem. 2005;280:36364–71. doi: 10.1074/jbc.M503673200. [DOI] [PubMed] [Google Scholar]
- 139.Garber ME, Mayall TP, Suess EM, Meisenhelder J, Thompson NE, Jones KA. CDK9 autophos-phorylation regulates high-affinity binding of the human immunodeficiency virus type 1 Tat–P-TEFb complex to TAR RNA. Mol. Cell. Biol. 2000;20:6958–69. doi: 10.1128/mcb.20.18.6958-6969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhou M, Huang K, Jung KJ, Cho WK, Klase Z, et al. Bromodomain protein Brd4 regulates human immunodeficiency virus transcription through phosphorylation of CDK9 at threonine 29. J. Virol. 2009;83:1036–44. doi: 10.1128/JVI.01316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vlach J, Garcia A, Jacque JM, Rodriguez MS, Michelson S, Virelizier JL. Induction of Sp1 phosphorylation and NF-κB-independent HIV promoter domain activity in T lymphocytes stimulated by okadaic acid. Virology. 1995;208:753–61. doi: 10.1006/viro.1995.1207. [DOI] [PubMed] [Google Scholar]
- 142.Epie N, Ammosova T, Turner W, Nekhai S. Inhibition of PP2A by LIS1 increases HIV-1 gene expression. Retrovirology. 2006;3:65. doi: 10.1186/1742-4690-3-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell. 2002;9:625–36. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 144.Williams SA, Kwon H, Chen LF, Greene WC. Sustained induction of NF-κB is required for efficient expression of latent human immunodeficiency virus type 1. J. Virol. 2007;81:6043–56. doi: 10.1128/JVI.02074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bedoya LM, Marquez N, Martinez N, Gutierrez-Eisman S, Alvarez A, et al. SJ23B, a jatrophane diterpene, activates classical PKCs and displays strong activity against HIV in vitro. Biochem. Pharmacol. 2009;77:965–78. doi: 10.1016/j.bcp.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 146.Bocklandt S, Blumberg PM, Hamer DH. Activation of latent HIV-1 expression by the potent anti-tumor promoter 12-deoxyphorbol 13-phenylacetate. Antiviral Res. 2003;59:89–98. doi: 10.1016/s0166-3542(03)00034-2. [DOI] [PubMed] [Google Scholar]
- 147.Kulkosky J, Culnan DM, Roman J, Dornadula G, Schnell M, et al. Prostratin: Activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood. 2001;98:3006–15. doi: 10.1182/blood.v98.10.3006. [DOI] [PubMed] [Google Scholar]
- 148.Korin YD, Brooks DG, Brown S, Korotzer A, Zack JA. Effects of prostratin on T-cell activation and human immunodeficiency virus latency. J. Virol. 2002;76:8118–23. doi: 10.1128/JVI.76.16.8118-8123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Biancotto A, Grivel JC, Gondois-Rey F, Bettendroffer L, Vigne R, et al. Dual role of prostratin in inhibition of infection and reactivation of human immunodeficiency virus from latency in primary blood lymphocytes and lymphoid tissue. J. Virol. 2004;78:10507–15. doi: 10.1128/JVI.78.19.10507-10515.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Burnett JC, Lim KI, Calafi A, Rossi JJ, Schaffer DV, Arkin AP. Combinatorial latency reactivation for HIV-1 subtypes and variants. J. Virol. 2010;84:5958–74. doi: 10.1128/JVI.00161-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Reuse S, Calao M, Kabeya K, Guiguen A, Gatot JS, et al. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PLoS One. 2009;4:e6093. doi: 10.1371/journal.pone.0006093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wender PA, Kee JM, Warrington JM. Practical synthesis of prostratin, DPP, and their analogs, adjuvant leads against latent HIV. Science. 2008;320:649–52. doi: 10.1126/science.1154690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yang HC, Xing S, Shan L, O’Connell K, Dinoso J, et al. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J. Clin. Investig. 2009;119:3473–86. doi: 10.1172/JCI39199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang FX, Xu Y, Sullivan J, Souder E, Argyris EG, et al. IL-7 is a potent and proviral strain-specific inducer of latent HIV-1 cellular reservoirs of infected individuals on virally suppressive HAART. J. Clin. Investig. 2005;115:128–37. doi: 10.1172/JCI22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lehrman G, Ylisastigui L, Bosch RJ, Margolis DM. Interleukin-7 induces HIV type 1 outgrowth from peripheral resting CD4+ T cells. J. Acquir. Immune Defic. Syndr. 2004;36:1103–4. doi: 10.1097/00126334-200408150-00015. [DOI] [PubMed] [Google Scholar]
- 156.Sereti I, Dunham RM, Spritzler J, Aga E, Proschan MA, et al. IL-7 administration drives T cell cycle entry and expansion in HIV-1 infection. Blood. 2009;113:6304–14. doi: 10.1182/blood-2008-10-186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Vogler M, Dinsdale D, Dyer MJ, Cohen GM. Bcl-2 inhibitors: small molecules with a big impact on cancer therapy. Cell Death Differ. 2009;16:360–67. doi: 10.1038/cdd.2008.137. [DOI] [PubMed] [Google Scholar]
- 158.Nottet HS, Persidsky Y, Sasseville VG, Nukuna AN, Bock P, et al. Mechanisms for the transendothelial migration of HIV-1-infected monocytes into brain. J. Immunol. 1996;156:1284–95. [PubMed] [Google Scholar]
- 159.Koenig S, Gendelman HE, Orenstein JM, Dal Canto MC, Pezeshkpour GH, et al. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science. 1986;233:1089–93. doi: 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- 160.Williams K, Alvarez X, Lackner AA. Central nervous system perivascular cells are immunoregulatory cells that connect the CNS with the peripheral immune system. Glia. 2001;36:156–64. doi: 10.1002/glia.1105. [DOI] [PubMed] [Google Scholar]
- 161.Hickey WF, Vass K, Lassmann H. Bone marrow–derived elements in the central nervous system: an immunohistochemical and ultrastructural survey of rat chimeras. J. Neuropathol. Exp. Neurol. 1992;51:246–56. doi: 10.1097/00005072-199205000-00002. [DOI] [PubMed] [Google Scholar]
- 162.Williams KC, Corey S, Westmoreland SV, Pauley D, Knight H, et al. Perivascular macrophages are the primary cell type productively infected by simian immunodeficiency virus in the brains of macaques: implications for the neuropathogenesis of AIDS. J. Exp. Med. 2001;193:905–15. doi: 10.1084/jem.193.8.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Suh HS, Kim MO, Lee SC. Inhibition of granulocyte-macrophage colony-stimulating factor signaling and microglial proliferation by anti-CD45RO: role of Hck tyrosine kinase and phosphatidylinositol 3-kinase/Akt. J. Immunol. 2005;174:2712–19. doi: 10.4049/jimmunol.174.5.2712. [DOI] [PubMed] [Google Scholar]
- 164.Rohr O, Lecestre D, Chasserot-Golaz S, Marban C, Avram D, et al. Recruitment of Tat to heterochromatin protein HP1 via interaction with CTIP2 inhibits human immunodeficiency virus type 1 replication in microglial cells. J. Virol. 2003;77:5415–27. doi: 10.1128/JVI.77.9.5415-5427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Marban C, Redel L, Suzanne S, Van Lint C, Lecestre D, et al. COUP-TF interacting protein 2 represses the initial phase of HIV-1 gene transcription in human microglial cells. Nucleic Acids Res. 2005;33:2318–31. doi: 10.1093/nar/gki529. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 166.Ellis RJ, Gamst AC, Capparelli E, Spector SA, Hsia K, et al. Cerebrospinal fluid HIV RNA originates from both local CNS and systemic sources. Neurology. 2000;54:927–36. doi: 10.1212/wnl.54.4.927. [DOI] [PubMed] [Google Scholar]
- 167.Bowman MC, Ballard TE, Ackerson CJ, Feldheim DL, Margolis DM, Melander C. Inhibition of HIV fusion with multivalent gold nanoparticles. J. Am. Chem. Soc. 2008;130:6896–97. doi: 10.1021/ja710321g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rao KS, Reddy MK, Horning JL, Labhasetwar V. TAT-conjugated nanoparticles for the CNS delivery of anti-HIV drugs. Biomaterials. 2008;29:4429–38. doi: 10.1016/j.biomaterials.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]