Abstract
Ataxia–telangiectasia (A–T) is a syndrome of cancer susceptibility, immune dysfunction, and neurodegeneration that is caused by mutations in the A–T-mutated (ATM) gene. ATM has been implicated as a critical regulator of cellular responses to DNA damage, including the activation of cell cycle checkpoints and induction of apoptosis. Although defective cell cycle-checkpoint regulation and associated genomic instability presumably contribute to cancer susceptibility in A–T, the mechanism of neurodegeneration in A–T is not well understood. In addition, although ATM is required for the induction of the p53 transcriptional program in response to DNA damage, the identities of the relevant transcription factors that mediate ATM-dependent changes in gene expression remain largely undetermined. In this article, we describe a signal transduction pathway linking ATM directly to the Ca2+/cAMP response element-binding protein, CREB, a transcription factor that regulates cell growth, homeostasis, and survival. ATM phosphorylated CREB in vitro and in vivo in response to ionizing radiation (IR) and H2O2 on a stress-inducible domain. IR-induced phosphorylation of CREB correlated with a decrease in CREB transactivation potential and reduced interaction between CREB and its transcriptional coactivator, CREB-binding protein (CBP). A CREB mutant containing Ala substitutions at ATM phosphorylation sites displayed enhanced transactivation potential, resistance to inhibition by IR, and increased binding to CBP. We propose that ATM-mediated phosphorylation of CREB in response to DNA damage modulates CREB-dependent gene expression and that dysregulation of the ATM–CREB pathway may contribute to neurodegeneration in A–T.
Ataxia–telangiectasia (A–T) is a recessive genetic syndrome characterized by immune deficiency, cancer susceptibility, and cerebellar degeneration (1). A–T is caused by mutations in ATM, which encodes a protein kinase belonging to the phosphoinositide 3-kinase-related kinase gene superfamily (2). At the cellular level, ATM-deficient cells grow poorly in culture, are genetically unstable and display exquisite sensitivity to ionizing radiation (IR) and radiomimetic drugs (1). A–T cells are also characteristically defective in the G1–S, intraS, and G2–M cell cycle checkpoints after γ-irradiation (3), which is believed to contribute to genomic instability and cancer susceptibility.
The checkpoint-signaling functions of ATM are achieved by means of the coordinated phosphorylation of polypeptide substrates, including p53, BRCA1, NBS1, and CHK2, which transmit signals to the DNA repair, apoptosis, and cell cycle machinery (3). Far less is known regarding the mechanism of cerebellar degeneration in A–T. Purkinje and granule neurons, which are most severely affected in human A–T, are not grossly abnormal in ATM–/–mice (4, 5). However, brains from ATM–/–mice display subtle developmental defects and are abnormally resistant to IR-induced apoptosis (6, 7). It has been proposed that neurodegeneration in human A–T is due to the developmental escape of genomically damaged neurons, which are destined to degenerate (6). A second hypothesis posits that neurodegeneration occurs as the result of increased oxidative stress (8, 9). In support of this hypothesis, brains from ATM–/–mice display markers of oxidative stress including increased levels of heme oxygenase, thioredoxin, and reactive oxygen species (10–14). The basis for enhanced oxidative stress in the absence of ATM has not been determined, but it may be a consequence of persistent DNA damage (11).
Identification of ATM-dependent signaling pathways that regulate neuron homeostasis or survival is imperative for understanding the neurodegenerative process in A–T. One relatively well characterized stress-response and neuron-survival factor is the Ca2+/cAMP response element-binding protein, CREB. CREB is a basic leucine zipper family transcription factor that plays key roles in neuronal development, function, and survival (15). CREB transcriptional activity is up-regulated in response to many cellular stimuli, including cAMP, Ca2+, hypoxia, UV light, and growth factors (16, 17). The canonical CREB activation pathway involves stimulus-induced phosphorylation of CREB on Ser-133 by PKA or other Ser-133 kinases (18). The phosphorylation of Ser-133 promotes an interaction between the kinase-inducible domain (KID) of CREB and the transcriptional coactivator CREB-binding protein (CBP), which potentiates the expression of CREB target genes (19–22). CREB activity is also subject to negative regulation. Phosphorylation of Ser-142 by calmodulin kinase II (CaMKII) or casein kinase 2 inhibits CREB transcriptional activity and destabilizes the interaction between the KID and CBP (23–25).
CREB is a bona fide neuron survival factor. Overexpression of dominant-negative CREB mutants causes death of cultured neurons and genetic disruption of CREB in mice results in axonal growth defects and perinatal lethality (26–28). Furthermore, conditional disruption of CREB and the CREB-related factor, CREM, causes apoptosis of postmitotic neurons (29). The antiapoptosis functions of CREB are attributable to CREB-dependent transcription of prosurvival genes such as Bcl-2 and neuron growth factors (26, 30, 31).
Despite extensive literature investigating CREB activation, little is known regarding the regulation of CREB in response to overt DNA-damaging stimuli. Given that genetic inactivation of CREB and ATM yield neuronal phenotypes, we were intrigued by the possibility that ATM functions upstream of CREB in a DNA damage-induced signaling pathway. We now present evidence that ATM regulates CREB phosphorylation and transactivation potential directly in response to IR and oxidative stress by means of a Ser-133 phosphorylation-independent mechanism. These findings have implications for understanding how ATM modulates gene expression in response to stress stimuli, and they may provide a link between CREB dysregulation and neurodegeneration in A–T.
Materials and Methods
Cell Culture and Antisera. K562 and human embryonic kidney (HEK) 293T cells were maintained in RPMI medium 1640 and MEM containing 10% FCS, respectively. G-361 osteosarcoma cells, L40, and AT59 lymphoblasts were provided by Yosef Shiloh (Tel Aviv University, Tel Aviv). C3ABR, AT1, and AT3 lymphoblasts were provided by Martin Lavin (Queensland Institute of Medical Research, Queensland, Australia). All lymphoblast cultures were maintained in RPMI medium 1640/10% FCS. For the generation of α-pS-121 antisera, rabbits were immunized with a keyhole limpet hemocyanin-conjugated CREB phosphopeptide (CSVTDpSQKRR) derived from amino acids 117–125 of human CREB. The resulting antiserum was affinity-purified as described (32). Other Abs used in this study include α-CREB (Cell Signaling Technology, Beverly, MA), α-pS-133 (Upstate Biotechnology, Lake Placid, NY), α-ATM (Ab-3, Calbiochem), and α-GST (Sigma).
Plasmid Constructs. The CREB:pSPORT6 plasmid was obtained by means of the image est sequencing consortium (Invitrogen). GST–KIX was a gift from Jennifer Nyborg (Colorado State University, Fort Collins) and was expressed and purified as described (33). FLAG-CBP was kindly provided by Pang Yao (Duke University, Durham, NC). Histidine-tagged CREB (HIS-CREB) was generated by subcloning a full-length human CREB cDNA into the SalI-HindIII sites of pET 28a (Novagen). The HIS-CREB proteins were purified by nickel chromatography according to the manufacturer's instructions. Transient transfection of HEK 293T cells was performed by using calcium phosphate DNA precipitation.
Protein Analysis and Kinase Assays. ATM kinase assays were performed as described (34). Extracts were immunoprecipitated with 1 μg of control IgG or α-ATM at 4°C for 1 h. The immunoprecipitates were washed and incubated with 1 μg of HIS-CREB substrate per reaction. GST–KIX pull-down assays were performed as described (33). Briefly, HEK 293T cell extracts were prepared by using lysis buffer (35), and 100 μg of extract was incubated with 20 μg of GST–KIX immobilized on glutathione agarose beads in 1 ml of PBS containing 0.1% Nonidet P-40 and protease inhibitors (5 μg/ml leupeptin/10 μg/ml pepstatin/5 μg/ml aprotinin) at 4°C for 4 h. The beads were washed three times with PBS containing 0.5% Nonidet P-40 and eluted by boiling in 2 × SDS sample loading buffer. For CREB–CBP coimmunoprecipitations, HEK 293T cells were cotransfected with FLAG-CBP (4 μg) and CREB (0.4 μg) expression plasmids. At 24 h later, cells were exposed to 20 Gy of IR or mock irradiated and harvested 1 h later in coimmunoprecipitation buffer (50 mM Tris, pH 7.5/100 mM NaCl/0.5% Tween-20/0.2% Nonidet P-40) with protease inhibitors. Extracts were immunoprecipitated by using FLAG M2-agarose affinity gel (Sigma), washed in coimmunoprecipitation buffer containing 200 mM NaCl, and analyzed by SDS/PAGE and immunoblotting with α-CREB and α-FLAG Abs.
Luciferase Assays. Gal4-CREB luciferase assays were performed by using the PathDetect trans-reporting system (Stratagene). The Gal4-CREB plasmid (pFA2-CREB) expresses the N-terminal 280 aa of CREB fused to the Gal4 DNA binding domain. HEK 293T cells were cotransfected with 50 ng of WT or mutant Gal4-CREB expression vectors and 1 μg of GAL4-LUC. At 24 h later, the cells were exposed to IR (20 Gy) or mock irradiated. Luciferase activity was measured 1 h later by using a Moonlight luminometer (Pharmingen). Presented results were normalized for β-galactosidase expression and represent the averaged values of at least three independent experiments.
Results
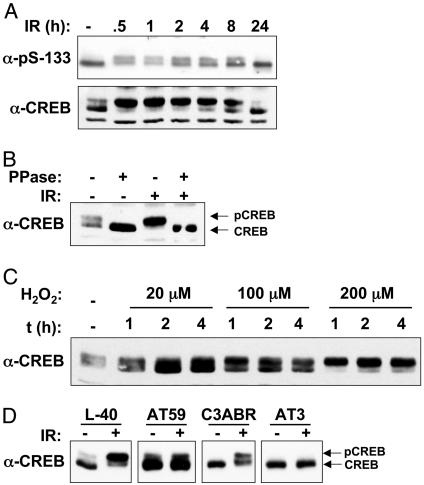
CREB Is Phosphorylated in Response to IR and H2O2. To explore the potential relationship between ATM and CREB, we initially tested whether CREB was phosphorylated on Ser-133 after exposure to IR, an archetypal ATM-activating stimulus (3). K562 cell extracts were prepared at various times after exposure to 20 Gy of IR and subjected to immunoblot analysis by using an Ab specific for Ser-133-phosphorylated CREB (α-pS-133). CREB from untreated cell extracts migrated as a major species of ≈40 kDa and a minor species of 42 kDa on SDS/PAGE gels (Fig. 1A). After exposure to IR, a greater fraction of CREB migrated as the slow-mobility form, although the overall band intensities did not change. The induction of the electrophoretic mobility shift was maximal by 30 min after IR and declined to unstimulated levels by 24 h. The IR-induced electrophoretic mobility shift was revealed also by using a α-CREB Ab (Fig. 1 A Lower), was detectable with IR doses as low as 2 Gy (see Fig. 6, which is published as supporting information on the PNAS web site), and was fully reversed upon phosphatase treatment of the cell extracts, indicating that the observed mobility shift was a consequence of phosphorylation (Fig. 1B). H2O2 also induced the time- and dose-dependent phosphorylation of CREB (Fig. 1C). Based on these results, we conclude that CREB is phosphorylated on a site distinct from Ser-133 in K562 cells after IR or H2O2 treatment.
Fig. 1.
ATM-dependent phosphorylation of CREB in response to IR and oxidative stress. (A) K562 cells were exposed to 20 Gy of IR and then harvested at the indicated times. Cell extracts were analyzed by SDS/PAGE and immunoblotting by using an Ab specific for the Ser-133 phosphorylation site of CREB (α-pS-133, Upper) or α-CREB (Lower). (B) IR-induced CREB electrophoretic mobility shift is abolished by phosphatase treatment. Cell extracts prepared from unirradiated or γ-irradiated K562 cells were treated with λ phosphatase (PPase) or sham treated and then analyzed by immunoblotting with α-CREB. (C) Phosphorylation of CREB in response to H2O2. K562 cells were treated with increasing concentrations of H2O2 for the indicated times. Cell extracts were prepared and analyzed by immunoblotting with α-CREB. (D) Phosphorylation of CREB in ATM deficient lymphoblasts. Cells expressing WT ATM (L40, C3ABR) or mutant ATM (AT59, AT3) were left untreated or exposed to 10 Gy of IR. Cell extracts were prepared and analyzed 1 h later by SDS/PAGE and immunoblotting with α-CREB.
We next examined the ATM dependence of IR-induced CREB phosphorylation. In contrast to cell lines expressing WT ATM (L40, C3ABR), the IR-induced electrophoretic mobility shift of CREB was absent in the ATM-deficient cell lines AT59 and AT3 (Fig. 1D). In AT59 cells, a fraction of CREB migrated as the slow-mobility form in the absence of IR. However, the abundance of this species did not increase after IR exposure, indicating that ATM is required for the inducible component of CREB phosphorylation. The A–T cell lines also displayed a complete defect in H2O2-induced CREB phosphorylation (Fig. 6B). Finally, both IR and H2O2-induced CREB phosphorylation were inhibited by the ATM inhibitor caffeine (Fig. 6C). These findings strongly suggest that functional ATM is required for the phosphorylation of CREB in response to IR and oxidative stress.
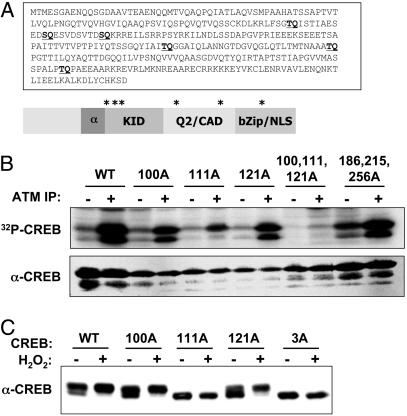
ATM Phosphorylates CREB in Vitro. CREB contains six Ser/Thr-Gln (SQ/TQ) motifs that may serve as phospho-acceptor sites for ATM and ATR kinase family members (Fig. 2A). Three of these residues (Thr-100, Ser-111, and Ser-121) are located near the N-terminal boundary of the KID, which spans amino acids 100–160 of CREB (21). To determine whether CREB is phosphorylated by ATM in vitro, we used a HIS-CREB fusion protein substrate in ATM kinase assays. HIS-CREB was phosphorylated upon incubation with α-ATM immunoprecipitates but not immunoprecipitates prepared by using control IgG (Fig. 2B). Experiments using ectopically expressed, hemagglutinin-tagged WT and kinase-inactive ATM revealed that a functional ATM kinase domain was essential for in vitro phosphorylation of CREB (see Fig. 7, which is published as supporting information on the PNAS web site). Individual Ala substitutions at Thr-100, Ser-111, or Ser-121 inhibited ATM-catalyzed phosphate incorporation, and the mutation of Ser-111 had a particularly strong inhibitory effect, reducing HIS-CREB phosphorylation by ≈70% (Fig. 2B). Mutation of Thr-100, Ser-111, and Ser-121 in combination reduced HIS-CREB phosphorylation to near-background levels, whereas mutation of Thr-186, Thr-215, and Thr-256 individually (data not shown) or in combination (Fig. 2B) had little effect. We conclude that ATM phosphorylates CREB on Thr-100, Ser-111, and Ser-121 and that Ser-111 represents the major in vitro phosphorylation site.
Fig. 2.
ATM phosphorylates CREB in vitro.(A) Primary amino acid sequence and schematic depiction of CREB. Locations of potential ATM phosphorylation sites are shown in bold and indicated by asterisks. (B) ATM kinase assay. Detergent extracts from G-361 cells were immunoprecipitated by using control rabbit IgG (–) or α-ATM Abs (+). The immunoprecipitates were incubated with either a WT HIS–CREB fusion protein substrate (WT) or a HIS–CREB substrate containing the indicated single or combination Ala substitutions. Kinase reactions were performed in the presence of [γ-32]ATP, and the reaction products were analyzed by SDS/PAGE and autoradiography. (C) Effects of ATM phosphorylation site mutations on CREB electrophoretic mobility. HEK 293T cells were transfected with expression plasmids encoding WT CREB (WT); CREB bearing single Ala substitutions at Thr-100, Ser-111, or Ser-121; or a CREB3A mutant containing Ala substitutions at all three sites. At 24 h after transfection, the cells were treated with H2O2 or vehicle for 1 h. Cell extracts were then immunoblotted with α-CREB.
CREB Is Phosphorylated on Ser-111 and Ser-121 in Intact Cells. To examine initially whether CREB is phosphorylated on one or more ATM phosphorylation sites in vivo, we transfected HEK 293T cells with plasmids encoding WT CREB (CREBWT) or CREB containing individual or combination Ala substitutions at Thr-100, Ser-111, or Ser-121, and we then examined the H2O2-induced electrophoretic mobility shift of the overexpressed proteins. Most overexpressed CREBWT migrated as the hyperphosphorylated form on SDS/PAGE gels, and exposure to H2O2 caused a further increase in the phosphorylated fraction (Fig. 2C). Ala substitutions at Thr-100 (CREB100A) and Ser-121 (CREB121A) partially suppressed the basal level of hyperphosphorylated CREB, but they had no effect on the H2O2-induced mobility shift (Fig. 2C). In contrast, a Ser-111→Ala mutation abolished the H2O2-induced mobility shift and this effect was recapitulated by using a CREB3A mutant containing Ala substitutions at all three phosphorylation sites. These findings suggest that CREB is phosphorylated on Thr-100, Ser-111, and Ser-121 in vivo, and that phosphorylation of Ser-111 is required for the H2O2-induced mobility shift.
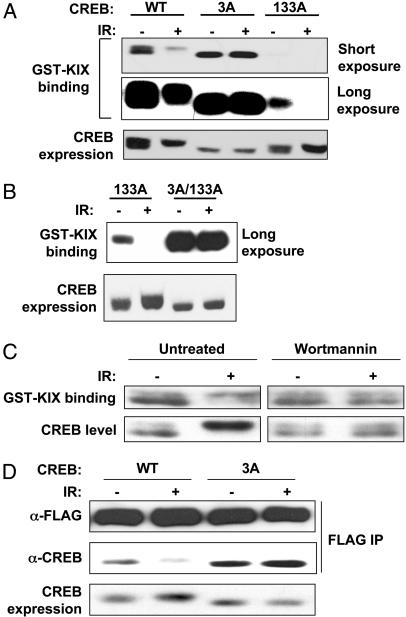
ATM Phosphorylation Sites Antagonize CREB–CBP Complex Formation. The three ATM phosphorylation sites in CREB lie within the KID, which binds to CBP through a 94-aa region termed the KIX domain, in a Ser-133 phosphorylation-dependent manner (25). To determine whether the ATM sites modulate CREB–CBP interactions, we used a GST–KIX pull-down assay that measures the binding of soluble CREB proteins to GST–KIX immobilized on glutathione agarose beads (25). HEK 293T cells were transfected with CREBWT, CREB3A, or CREB133A expression plasmids and exposed to IR or mock irradiated. Cell extracts were then incubated with GST–KIX beads, and the bound proteins were analyzed by SDS/PAGE and immunoblotting with α-CREB. The GST–KIX pull-down assay revealed that CREBWT bound to GST–KIX, whereas CREB133A containing an Ala substitution at Ser-133 showed reduced affinity (Fig. 3A). In contrast, the KIX-binding capacity of CREB3A was elevated 4- to 5-fold relative to CREBWT (Fig. 3A and data not shown). The CREBWT and CREB3A proteins exhibited comparable reactivity with the α-pS-133 Ab, suggesting that differential binding to GST–KIX was not a consequence of differences in Ser-133 phosphorylation (data not shown).
Fig. 3.
ATM phosphorylation sites antagonize CREB–CBP complex formation. (A) Analysis of CREB–KIX interactions by GST–KIX pull-down assay. HEK 293T cells were transfected with vector, CREBWT, CREB3A, or CREB133A expression plasmids. Equal amounts of cell extracts from irradiated (20 Gy, 1 h) or unirradiated cells were incubated with GST–KIX agarose beads. Bound proteins were eluted and analyzed by immunoblotting with α-CREB. Short and long exposures of the film are shown. Expression levels of the transfected CREB proteins are shown in Bottom.(B) IR-dependent inhibition of CREB133A–KIX interactions requires intact ATM phosphorylation sites. HEK 293T cells were transfected with CREB133A or CREB3A/133A containing Ala substitutions at Ser-133 and all three ATM phosphorylation sites. GST–KIX pull-down assays were then performed as described in A. (C) Effect of wortmannin (Wm) on CREB–KIX interactions. HEK 293T cells were exposed to 20 μM Wm or vehicle for 30 min and then exposed to IR (20 Gy) or mock-irradiated. After 1 h, cell extracts were incubated with GST–KIX and the levels of bound CREB determined by immunoblotting. (D) CREB3A shows enhanced coimmunoprecipitation with full-length CBP. HEK 293T cells were cotransfected with FLAG-CBP and either CREBWT or CREB3A expression vectors. At 24 h later, cells were exposed to IR or mock irradiated, and cell extracts were immunoprecipitated with α-FLAG. Levels of the coimmunoprecipitated CREB proteins were determined by blotting with α-CREB and expression levels of CREB and FLAG-CBP were assessed by blotting with α-CREB and α-FLAG, respectively.
The effects of IR on the CREB–KIX interaction were examined also. IR exposure inhibited the interaction between CREBWT and GST–KIX, but it had no effect on the binding of the CREB3A mutant (Fig. 3A). During these experiments, we noted that, in comparison with CREBWT, the binding of CREB133A to GST–KIX was more strongly inhibited by IR (Fig. 3A). This finding suggested that ATM-mediated phosphorylation of CREB133A disrupted a low affinity CREB133A–KIX interaction. Consistent with this possibility, the KIX-binding activity of a CREB3A/133A mutant, containing Ala substitutions at Ser-133 and all three ATM sites, was resistant to IR (Fig. 3B). IR also inhibited the interaction between endogenous CREB and GST–KIX, and the observed inhibition was reversed by the ATM inhibitor, wortmannin (Fig. 3C). The combined findings imply that ATM phosphorylation sites antagonize CREB–KIX interactions and that phosphorylation of Ser-133 counteracts the inhibitory effects of these residues.
Further experiments were performed to confirm that ATM phosphorylation sites modulate CREB–CBP interactions in intact cells. HEK 293T cells were cotransfected with CREBWT or CREB3A and full-length FLAG-CBP expression plasmids. Cells were exposed to 20 Gy of IR or mock irradiated, and cell extracts were immunoprecipitated with α-FLAG. Immunoblot analysis using α-CREB revealed that IR inhibited the coimmunoprecipitation of CREBWT with FLAG-CBP. In contrast, CREB3A demonstrated enhanced coimmunoprecipitation with CBP that was resistant to inhibition by IR (Fig. 3D). These results confirm the GST–KIX binding data and suggest that ATM phosphorylation sites antagonize CREB–CBP complex formation in vivo.
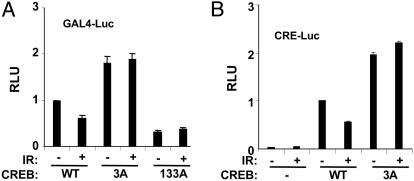
Effects of ATM Phosphorylation Site Mutations on CREB Transactivation Potential. The CBP-binding results described above suggested that ATM phosphorylation sites may impact CREB transactivation potential. To test this possibility, we used a reporter gene assay in which the DNA binding domain of yeast Gal4 is fused to the N-terminal 280 aa of CREB (see Materials and Methods). HEK 293T cells were cotransfected with a luciferase construct containing tandem GAL4 binding sites (GAL4-Luc) and plasmids encoding Gal4-CREBWT, Gal4-CREB3A, or Gal4-CREB133A. The transfected cells were then exposed to IR or mock irradiated, and luciferase assays were performed 1 h later. This assay revealed that the transcriptional activity of Gal4-CREB3A was consistently 2-fold higher than the activity of Gal4-CREBWT in the absence of stimulus (Fig. 4A). Exposure to IR inhibited the basal activity of Gal4-CREBWT but affected the activity of Gal4-CREB3A only marginally. The inhibition of Gal4-CREBWT was modest, ranging from 30% to 50% in multiple experiments. The activating effects of ATM phosphorylation site mutations on CREB transactivation potential were also observed by using a 5X-CRE-luciferase reporter and full-length CREB constructs, and they could be recapitulated by single amino acid substitutions at Ser-111 or Ser-121 (Figs. 4B and 8B, which is published as supporting information on the PNAS web site). In addition, cellular exposure to the ATM inhibitors wortmannin (20 μM) or caffeine (5 mM) had a slight enhancing effect on CREB transactivation potential (Fig. 8A). These data suggest that ATM-mediated phosphorylation antagonizes CREB transactivation potential.
Fig. 4.
Ala substitutions at ATM phosphorylation sites enhance CREB transcriptional activity. (A) HEK 293T cells were cotransfected with GAL4-Luc, and Gal4-CREBWT, Gal4-CREB3A, or Gal4-CREB133A. Transfected cells were exposed to IR (20 Gy) or mock irradiated and harvested 1 h later for luciferase assays. The data were normalized for cotransfected β-Gal expression and are plotted as relative luciferase activity. The data represent the averaged results of at least three experiments (Gal4-CREBWT and Gal4-CREB3A, n = 5). (B) Transcriptional activity of full-length CREB toward a CRE-Luc promoter construct. HEK 293T cells were cotransfected with a 5X-CRE-Luc reporter construct and expression plasmids encoding CREBWT or CREB3A. Transfected cells were exposed to IR or mock irradiated and processed for luciferase assays as described in A.
Mutation of the three ATM phosphorylation sites activated CREB, whereas mutation of the PKA site at Ser-133 inhibited CREB activity. Therefore, we tested whether the CREB3A mutation could rescue the activation defect of the CREB133A mutant. The transactivation potential of Gal4-CREB3A/133A mutant containing Ala substitutions at Thr-100, Ser-111, Ser-121, and Ser-133 was comparable with that of Gal4-CREB133A, indicating that Ala substitutions at ATM sites are insufficient to activate Gal4-CREB independently of the Ser-133 phosphorylation site (Fig. 8B). In addition, IR preexposure did not attenuate forskolin-induced CREB activation, suggesting that the canonical phospho-Ser-133-dependent activation pathway is dominant to the ATM pathway in this assay (data not shown). In summary, the combined data suggest that ATM phosphorylation sites negatively regulate CREB transactivation potential and are congruent with the CBP-binding results.
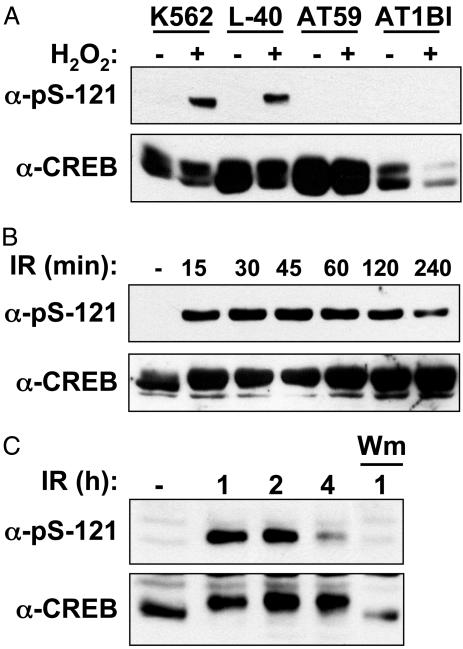
Ser-121 Is a Highly Inducible ATM Phosphorylation Site. The results described above strongly suggested that ATM phosphorylates CREB in response to DNA damage and that the putative ATM phosphorylation sites at Thr-100, Ser-111, and Ser-121 negatively regulate CREB transactivation potential and binding to CBP. To prove unequivocally that Thr-100, Ser-111, and Ser-121 are bona fide ATM phosphorylation sites, we attempted to generate phospho-specific Abs that recognize each residue. We successfully generated an Ab that recognizes phosphorylated Ser-121 (α-pS-121). α-pS-121 demonstrated robust and phosphatase-sensitive reactivity with CREB after cellular exposure to H2O2 (Figs. 5A and 9A, which is published as supporting information on the PNAS web site). As a specificity control, we demonstrated that α-pS-121 recognized ectopically expressed CREBWT but not CREB121A (Fig. 9B). Phosphorylation of CREB on Ser-121 was defective in the ATM-deficient cell lines AT59 and AT1, indicating that ATM is responsible for phosphorylation of this site in vivo (Fig. 5A). As expected, IR strongly induced the phosphorylation of Ser-121 (Fig. 5B). UV light, aphidicolin, and the hypoxia-mimetic drug deferoxamine mesylate also induced Ser-121 phosphorylation but with delayed kinetics and lower magnitude in comparison with IR or H2O2 (see Fig. 10, which is published as supporting information on the PNAS web site, and data not shown). From these findings, we conclude that the phosphorylation of CREB on Ser-121 represents a general response to cellular stress.
Fig. 5.
Stress-induced phosphorylation of CREB on Ser-121. (A) H2O2-induced phosphorylation of CREB on Ser-121 is ATM-dependent. Cells expressing WT ATM(K562, L40) or ATM-deficient cells (AT59, AT1BI) were left untreated or exposed to 200 μM H2O2 and harvested 1 h later. Cell extracts were analyzed by SDS/PAGE and immunoblotting with α-pS-121 (Upper) and α-CREB (Lower). (B) Time course of IR-induced Ser-121 phosphorylation. HEK 293T cell extracts were prepared at the indicated time points after irradiation with 10 Gy and analyzed by immunoblotting with α-pS121 and α-CREB. (C) ATM-mediated phosphorylation of CREB in CGNs. Mouse CGNs were exposed to 10 Gy of IR or mock irradiated. Cell extracts were prepared at the indicated times and analyzed by immunoblotting with α-pS121 and α-CREB. Where indicated, the samples were treated with wortmannin (20 μM) 15 min before irradiation.
The phosphorylation of endogenous CREB on Ser-121 was highly inducible by IR, yet a fraction of CREB was phosphorylated on Ser-111 in the absence of DNA damage, as evidenced by the presence of the slow mobility form on SDS/PAGE gels (Figs. 1 A and 2C). Therefore, we postulated that Ser-111 phosphorylation is required for phosphorylation of Ser-121. Consistent with this possibility, a Ser-111→Ala substitution completely blocked the H2O2-induced phosphorylation of Ser-121 (see Fig. 10C). These data are consistent with a model in which the phosphorylation of Ser-111 primes the inducible phosphorylation of Ser-121 in response to stress stimuli.
CREB has been implicated as a positive regulator of neuron survival, whereas ATM promotes apoptosis of neurons in response to DNA damage (7, 15). Therefore, the neuronal functions of ATM and CREB may be linked. As a first step toward establishing this possibility, we tested whether CREB was phosphorylated on Ser-121 in cerebellar granule neurons (CGNs) isolated from 7-day-old mice. Exposure of CGNs to IR resulted in Ser-121 phosphorylation that was maximal 1–2 hours after IR and was suppressed by wortmannin (Fig. 5C). From this result, we conclude that the ATM–CREB pathway is activated by DNA damage in primary neurons.
Discussion
We have delineated an ATM–CREB signaling pathway linking the genome surveillance apparatus to a key regulator of gene expression and cell survival. Our findings indicate that ATM regulates CREB by means of a noncanonical, Ser-133 phosphorylation-independent mechanism involving the direct phosphorylation of three closely spaced amino acids (Thr-100, Ser-111, and Ser-121) located within the KID of CREB. Given that the ATM phosphorylation sites are clustered, and that phosphorylation of at least two of the sites (Ser-111 and Ser-121) is induced by a wide variety of stress stimuli, we collectively refer to Thr-100/Ser-111/Ser-121 as a S/TQ domain. ATM is required for both IR and H2O2-induced CREB phosphorylation. However, CREB was also phosphorylated on Ser-121 in response to UV light, aphidicolin, and hypoxia, which are stimuli that activate the ATM-related kinase, ATR (36). It is, therefore, likely that ATM and ATR regulate CREB phosphorylation collectively in response to stress stimuli. Our findings further suggest that the phosphorylation of Ser-111 is required for the DNA damage-inducible phosphorylation of Ser-121 (see Fig. 10C). Interestingly, Ser-111, which is a preferred ATM phosphorylation site in vitro, also conforms to a consensus casein kinase 2 site. Casein kinase 2 displays high basal activity in vivo and has been implicated as a CREB kinase (37, 38). It is, therefore, conceivable that casein kinase 2 plays a role in priming the damage-inducible phosphorylation of Ser-121.
Although the functional implications of the ATM–CREB pathway remain to be fully elucidated, our findings suggest that one outcome of ATM-mediated CREB phosphorylation is the inhibition of CBP binding. Combination or single ATM phosphorylation-site mutations enhanced the CBP-binding affinity of CREB 4- to 5-fold, whereas IR inhibited the binding of both endogenous CREB and transiently transfected CREB to CBP. Relative to endogenous CREB, the CBP-binding of overexpressed CREBWT was generally less sensitive to IR. We suspect that the difference is caused by the strong constitutive phosphorylation of transiently transfected CREB on Ser-111 (Fig. 2C), which may blunt the IR-inducible component of the response. The enhanced CBP-binding activity of CREB3A relative to CREBWT, even in the absence of IR, supports this hypothesis (Fig. 3A). Notably, the 4- to 5-fold increased CBP-binding capacity of CREB3A translated into an ≈2-fold enhancement of CREB transactivation potential. The modest activating effects of ATM phosphorylation site mutations on CREB activity are reminiscent of findings that examined the impact of Ala substitutions at the inhibitory CaMKII phosphorylation site at Ser-142 (23, 24). However, the nonlinearity between the CBP binding and transactivation parameters suggests that the impact of the ATM phosphorylation sites on CREB transcriptional functions may not be modeled adequately by generic CREB reporter assays but instead may be gene and promoter context-dependent.
Precisely how ATM phosphorylation sites modulate the CREB–KIX interaction is not known. The Ser-111 and Ser-121 phosphorylation sites lie within α-helix A of the KID, which does not contact the KIX domain directly (39). We propose that ATM-mediated phosphorylation induces a conformational change in the KID that attenuates binding to KIX. Consistent with this possibility, an Ala substitution at Ser-111 grossly altered CREB electrophoretic mobility (Fig. 2C), suggesting that phosphorylation of this residue may impact CREB conformation. Our results further suggest that the phospho-Ser-133-dependent binding of CREB to the KIX domain is dominant over the inhibitory effects of the ATM phosphorylation sites. Specifically, the CREB133A mutant, which displays weak CBP-binding affinity, unmasked a strong inhibitory effect of IR on the CREB–KIX interaction that was reversed when the ATM phosphorylation sites were mutated (Fig. 3B). This finding suggests that the phosphorylation of CREBWT on Ser-133 opposes the antagonistic effects of ATM phosphorylation sites on CREB–KIX complex formation.
The broad implications of the ATM pathway for CREB-dependent gene expression are presently unclear. It is conceivable that, by modulating CBP binding affinity, ATM alters the repertoire of CREB-dependent gene expression. Given that the PKA pathway preferentially activates CRE-containing promoters that possess a TATA box (40), it is possible that ATM principally affects the expression of TATA-less CREB targets, which include DNA repair factors and cell cycle regulators (40). Further studies are needed to examine comprehensively the impact of the ATM–CREB pathway on basal and stress-induced expression of CREB target genes. Finally, given that CREB is a neuron survival factor, it is attractive to speculate that ATM–CREB pathway regulates neuron homeostasis and/or apoptosis. Consistent with a neuronal function for the ATM–CREB pathway, we found that ATM phosphorylated CREB in CGNs exposed to IR (Fig. 5C). Future studies will be needed to determine the function of the ATM–CREB pathway in neurons and to ascertain whether dysregulation of the ATM–CREB pathway contributes to neuropathogenesis in A–T.
Supplementary Material
Acknowledgments
We thank Dr. Jennifer Nyborg for providing GST–KIX; Dr. Pang Yao for providing FLAG-CBP; Drs. Yosef Shiloh and Martin Lavin for supplying ATM-deficient cell lines; and Rebecca Kirkland and Dr. Jim Franklin for assistance with CGN cultures. This work was supported by National Institutes of Health Grant GM67868-01 and by a Shaw Scientist Award from the Greater Milwaukee Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: A–T, ataxia–telangiectasia; CBP, CREB-binding protein; CGN, cerebellar granule neuron; HEK, human embryonic kidney; IR, ionizing radiation; KID, kinase-inducible domain.
References
- 1.Becker-Catania, S. G. & Gatti, R. A. (2001) Adv. Exp. Med. Biol. 495, 191–198. [DOI] [PubMed] [Google Scholar]
- 2.Savitsky, K., Bar-Shira, A., Gilad, S., Rotman, G., Ziv, Y., Vanagaite, L., Tagle, D. A., Smith, S., Uziel, T. & Sfez, S. (1995) Science 268, 1749–1753. [DOI] [PubMed] [Google Scholar]
- 3.Shiloh, Y. (2003) Nat. Rev. Cancer 3, 155–168. [DOI] [PubMed] [Google Scholar]
- 4.Barlow, C., Hirotsune, S., Paylor, R., Liyanage, M., Eckhaus, M., Collins, F., Shiloh, Y., Crawley, J. N., Ried, T., Tagle, D., et al. (1996) Cell 86, 159–171. [DOI] [PubMed] [Google Scholar]
- 5.Xu, Y., Ashley, T., Brainerd, E. E., Bronson, R. T., Meyn, M. S. & Baltimore, D. (1996) Genes Dev. 10, 2411–2422. [DOI] [PubMed] [Google Scholar]
- 6.Herzog, K. H., Chong, M. J., Kapsetaki, M., Morgan, J. I. & McKinnon, P. J. (1998) Science 280, 1089–1091. [DOI] [PubMed] [Google Scholar]
- 7.Chong, M. J., Murray, M. R., Gosink, E. C., Russell, H. R., Srinivasan, A., Kapsetaki, M., Korsmeyer, S. J. & McKinnon, P. J. (2000) Proc. Natl. Acad. Sci. USA 97, 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rotman, G. & Shiloh, Y. (1997) BioEssays 19, 911–917. [DOI] [PubMed] [Google Scholar]
- 9.Barzilai, A., Rotman, G. & Shiloh, Y. (2002) DNA Repair 1, 3–25. [DOI] [PubMed] [Google Scholar]
- 10.Kamsler, A., Daily, D., Hochman, A., Stern, N., Shiloh, Y., Rotman, G. & Barzilai, A. (2001) Cancer Res. 61, 1849–1854. [PubMed] [Google Scholar]
- 11.Stern, N., Hochman, A., Zemach, N., Weizman, N., Hammel, I., Shiloh, Y., Rotman, G. & Barzilai, A. (2002) J. Biol. Chem. 277, 602–608. [DOI] [PubMed] [Google Scholar]
- 12.Barlow, C., Dennery, P. A., Shigenaga, M. K., Smith, M. A., Morrow, J. D., Roberts, L. J., Wynshaw-Boris, A. & Levine, R. L. (1999) Proc. Natl. Acad. Sci. USA 96, 9915–9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quick, K. L. & Dugan, L. L. (2001) Ann. Neurol. 49, 627–635. [PubMed] [Google Scholar]
- 14.Takao, N., Li, Y. & Yamamoto, K. (2000) FEBS Lett. 472, 133–136. [DOI] [PubMed] [Google Scholar]
- 15.Lonze, B. E. & Ginty, D. D. (2002) Neuron 35, 605–623. [DOI] [PubMed] [Google Scholar]
- 16.Shaywitz, A. J. & Greenberg, M. E. (1999) Ann. Rev. Biochem. 68, 821–861. [DOI] [PubMed] [Google Scholar]
- 17.Mayr, B. & Montminy, M. (2001) Nat. Rev. Mol. Cell Biol. 2, 599–609. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, G. A. & Montminy, M. R. (1989) Cell 59, 675–680. [DOI] [PubMed] [Google Scholar]
- 19.Chrivia, J. C., Kwok, R. P., Lamb, N., Hagiwara, M., Montminy, M. R. & Goodman, R. H. (1993) Nature 365, 855–859. [DOI] [PubMed] [Google Scholar]
- 20.Arias, J., Alberts, A. S., Brindle, P., Claret, F. X., Smeal, T., Karin, M., Feramisco, J. & Montminy, M. (1994) Nature 370, 226–229. [DOI] [PubMed] [Google Scholar]
- 21.Brindle, P., Linke, S. & Montminy, M. (1993) Nature 364, 821–824. [DOI] [PubMed] [Google Scholar]
- 22.Kwok, R. P., Lundblad, J. R., Chrivia, J. C., Richards, J. P., Bachinger, H. P., Brennan, R. G., Roberts, S. G., Green, M. R. & Goodman, R. H. (1994) Nature 370, 223–226. [DOI] [PubMed] [Google Scholar]
- 23.Kornhauser, J. M., Cowan, C. W., Shaywitz, A. J., Dolmetsch, R. E., Griffith, E. C., Hu, L. S., Haddad, C., Xia, Z. & Greenberg, M. E. (2002) Neuron 34, 221–233. [DOI] [PubMed] [Google Scholar]
- 24.Sun, P., Enslen, H., Myung, P. S. & Maurer, R. A. (1994) Genes Dev. 8, 2527–2539. [DOI] [PubMed] [Google Scholar]
- 25.Parker, D., Ferreri, K., Nakajima, T., LaMorte, V. J., Evans, R., Koerber, S. C., Hoeger, C. & Montminy, M. R. (1996) Mol. Cell. Biol. 16, 694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riccio, A., Ahn, S., Davenport, C. M., Blendy, J. A. & Ginty, D. D. (1999) Science 286, 2358–2361. [DOI] [PubMed] [Google Scholar]
- 27.Rudolph, D., Tafuri, A., Gass, P., Hammerling, G. J., Arnold, B. & Schutz, G. (1998) Proc. Natl. Acad. Sci. USA 95, 4481–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lonze, B. E., Riccio, A., Cohen, S. & Ginty, D. D. (2002) Neuron 34, 371–385. [DOI] [PubMed] [Google Scholar]
- 29.Mantamadiotis, T., Lemberger, T., Bleckmann, S. C., Kern, H., Kretz, O., Martin, V., Tronche, F., Kellendonk, C., Gau, D., Kapfhammer, J., et al. (2002) Nat. Genet. 31, 47–54. [DOI] [PubMed] [Google Scholar]
- 30.Shieh, P. B., Hu, S. C., Bobb, K., Timmusk, T. & Ghosh, A. (1998) Neuron 20, 727–740. [DOI] [PubMed] [Google Scholar]
- 31.Tao, X., Finkbeiner, S., Arnold, D. B., Shaywitz, A. J. & Greenberg, M. E. (1998) Neuron 20, 709–726. [DOI] [PubMed] [Google Scholar]
- 32.Bao, S., Tibbetts, R. S., Brumbaugh, K. M., Fang, Y., Richardson, D. A., Ali, A., Chen, S. M., Abraham, R. T. & Wang, X. F. (2001) Nature 411, 969–974. [DOI] [PubMed] [Google Scholar]
- 33.Van Orden, K. Giebler, H. A., Lemasson, I., Gonzales, M. & Nyborg, J. K. (1999) J. Biol. Chem. 274, 26321–26328. [DOI] [PubMed] [Google Scholar]
- 34.Sarkaria, J. N., Tibbetts, R. S., Busby, E. C., Kennedy, A. P., Hill, D. E. & Abraham, R. T. (1998) Cancer Res. 58, 4375–4382. [PubMed] [Google Scholar]
- 35.Tibbetts, R. S., Brumbaugh, K. M., Williams, J. M., Sarkaria, J. N., Cliby, W. A., Shieh, S. Y., Taya, Y., Prives, C. & Abraham, R. T. (1999) Genes Dev. 13, 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abraham, R. T. (2001) Genes Dev. 15, 2177–2196. [DOI] [PubMed] [Google Scholar]
- 37.Meggio, F. & Pinna, L. A. (2003) FASEB J. 17, 349–368. [DOI] [PubMed] [Google Scholar]
- 38.Saeki, K., Yuo, A. & Takaku, F. (1999) Biochem. J. 338, 49–54. [PMC free article] [PubMed] [Google Scholar]
- 39.Radhakrishnan, I., Perez-Alvarado, G. C., Parker, D., Dyson, H. J., Montminy, M. R. & Wright, P. E. (1999) J. Mol. Biol. 287, 859–865. [DOI] [PubMed] [Google Scholar]
- 40.Conkright, M. D., Guzman, E., Flechner, L., Su, A. I., Hogenesch, J. B. & Montminy, M. (2003) Mol. Cell 11, 1101–1108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.