Abstract
Damaged and misfolded proteins accumulate during the aging process, impairing cell function and tissue homeostasis. These perturbations to protein homeostasis (proteostasis) are hallmarks of age-related neurodegenerative disorders such as Alzheimer’s, Parkinson’s or Huntington’s disease. Damaged proteins are degraded by cellular clearance mechanisms such as the proteasome, a key component of the proteostasis network. Proteasome activity declines during aging, and proteasomal dysfunction is associated with late-onset disorders. Modulation of proteasome activity extends lifespan and protects organisms from symptoms associated with proteostasis disorders. Here we review the links between proteasome activity, aging and neurodegeneration. Additionally, strategies to modulate proteasome activity and delay the onset of diseases associated to proteasomal dysfunction are discussed herein.
Keywords: Aging, Alzheimer’s disease, Huntington’s disease, Parkinson’s disease, Proteasome, Proteostasis, Stem cells.
INTRODUCTION
Protein homeostasis, or proteostasis, is regulated by a complex network of cellular mechanisms that monitors the concentration, folding, cellular localization, and interactions of proteins from their synthesis through their degradation [1-3]. The accumulation of reactive oxygen species (ROS) during aging challenges the structure of proteins and the quality of the proteome. In addition, the ability to maintain a functional proteome declines during the aging process [1, 4-6]. This decline in proteostasis is considered one of the hallmarks of aging [7]. The accumulation of damaged proteins contributes to multiple age-related diseases such as Alzheimer’s (AD) [8], Parkinson’s (PD) [9] or Huntington’s disease (HD) [10]. Misfolded, damaged, aggregated or unnecessary proteins are degraded by the proteasome or through autophagy-lysosome [11-14]. Proteasome-mediated degradation is a complex mechanism where proteins are first targeted for degradation by the ubiquitination machinery and then recognized, unfolded and proteolyzed by the proteasome (Fig. 1). The impact of a failure in the ubiquitination pathway on neurodegenerative diseases has been previously reviewed in [15]. This review focuses on the proteasomal machinery and its demise in aging and neurodegenerative diseases. Proteasome functionality declines during the aging process and triggers the onset of age-related diseases. Mechanisms that promote proteasome activity can slow aging and decrease the incidence of age-related diseases [16, 17]. Likewise, mechanisms that promote autophagy can extend healthspan [13, 14, 18-22]. Several longevity-promotingpathways, such as reduced insulin/IGF-1 signaling pathway (IIS) or dietary restriction (DR), ameliorate proteome dysfunction in age-related diseases [3]. Notably, these longevity pathways modulate the proteostasis network (i.e., proteasome activity) providing increased stability of the proteome and protecting from the symptoms associated to diseases such as AD, PD or HD [3, 17, 23, 24]. Here, we discuss the mechanistic links between proteasome activity, aging and neurodegenerative diseases.
Fig. (1).
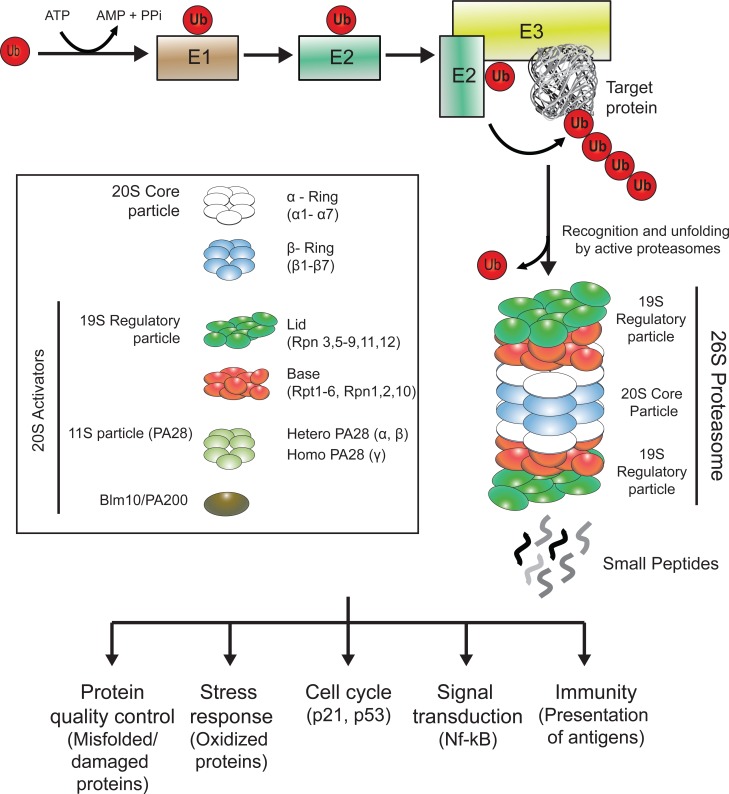
The Ubiquitin-Proteasome System (UPS). Ubiquitin is activated through the ubiquitin-activating enzyme (E1) and next transferred to the ubiquitin-conjugating enzyme (E2). Both E2 and the target protein bind to the ubiquitin-ligase (E3) which links the activated ubiquitin from E2 to the target protein. The sequential conjugation of ubiquitins to the target protein generates a poly-ubiquitin chain, which acts as a signal for the protein to be degraded through the proteasome. Active proteasomes are formed by the interaction of proteasomal regulatory particles with a core particle (20S), which contains the proteolytic active sites. Free intracellular 20S are normally found in an inactive/closed state and require the binding of 20S activators to degrade proteins. The major assembly of the 20S proteasome is with the 19S regulatory protein, which recognizes the polyubiquitylated substrate, removes the ubiquitin moieties and unfolds the substrate to translocate it into the 20S proteolytic chamber. The 20S can also be activated by the PA28 complex (also known as 11S) or by the Blm10/PA200 protein. Finally, the protein is cleaved into short peptides. The proteasome is involved in a variety of cellular functions, such as quality control of the proteome, stress response or cell cycle regulation.
THE UBIQUITIN-PROTEASOME SYSTEM (UPS)
The UPS is the primary selective degradation system in eukaryotic cells [16]. The UPS is a precise and carefully timed mechanism that is critical for maintaining the proper concentration of many regulatory proteins involved in cell cycle, stem cell function, development, signal transduction, metabolism, gene transcription, DNA repair, senescence, apoptosis, inflammation and other biological processes [11, 14, 25-28] (Fig. 1). In addition, the UPS is a key component of the protein quality control system needed to terminate damaged or misfolded proteins [11, 13, 14]. The first step of the UPS-mediated degradation is the conjugation of ubiquitin, a highly conserved 76 amino acid residue polypeptide, to the substrate protein [11, 29-31] (Fig. 1). The attachment of one ubiquitin molecule is achieved through an enzymatic sequential mechanism involving three distinct classes of enzymes [11, 32, 33]. First, the ubiquitin-activating enzyme (E1) activates the C-terminal glycine residue of a ubiquitin in an ATP-dependent manner. Activated ubiquitin is next transferred to a cysteine site of a ubiquitin-conjugating enzyme (E2). In the third step, a ubiquitin ligase (E3) links ubiquitin from the E2 enzyme by its C-terminus to the ε-amino group of a lysine residue of the target protein. While apparently there are only two E1, there are several E2 enzymes and many E3 ubiquitin ligases, each of which recognizes one or several of specific protein motifs [34]. Therefore, E3 ubiquitin ligases are responsible for the selectivity of the process targeting specific proteins to degradation by the proteasome. The same cascade links additional molecules to the primary ubiquitin via internal ubiquitin lysines, forming a ubiquitin chain. Ubiquitin has seven lysine residues, all of which can form polymer chains [35]. A chain of at least four lysine 48-linked ubiquitins is the primary signal for degradation [36], while other linear ubiquitin chains participate in different processes such as signal transduction [27, 37]. After the ubiquitination sequential mechanism, the polyubiquitylated protein is now recognized by the machinery responsible for its degradation, the proteasome.
The proteasome is a complex proteolytic machine formed by the assembly of several subunits [11]. The core particle (20S) of the proteasome exhibits a barrel-like structure in which the 28 subunits are assembled into four seven membered rings [11]. The two outer rings are composed by seven α subunits (named α1 to α7), while the two inner rings are composed by seven β subunits (named β1 to β7) [38]. Three of the β subunits contain proteolytic active sites: β1, β2 and β5 present caspase-like, trypsin-like and chymotrypsin-like activities, respectively [39, 40]. A specialized form of the 20S proteasome, known as the immunoproteasome because of its prevalence in antigen-presenting cells, is generated by replacement of the catalytic subunits by β1i, β2i and β5i [41, 42]. The function of the α-rings is to control the substrate entry into the catalytic cavity. Although 20S particles can exist in a free form, its default status is closed and requires the binding of proteasome activators to degrade polyubiquitylated proteins. Therefore, 20S particles are considered to be inactive, unable to degrade polyubiquitylated proteins [43]. However, it is important to remark that free 20S particles have a detectable activity independent of ubiquitination and ATP towards small proteins [44].
Active proteasome exists in several forms, but its major assembly is formed through the assembly of the 20S and the 19S, a subunit that also imparts regulation on the activity of the holo-complex (26S, single capped or 30S, double capped) [45-47]. The 19S recognizes polyubiquitylated proteins and unfolds and translocates these proteins to the 20S for degradation in an ATP dependent process [48]. The 19S is composed of two subcomplexes: a base adjacent to the 20S and a lid that sits on top of the base [48]. The base contains six ATPases (Rpt1-Rpt6), which are members of the AAA family of ATPases and three non ATPases subunits (Rpn1, Rpn2 and Rpn10) [11, 49]. The lid complex forms the distal mass of the 19S and is critical for substrate recognition and deubiquitination [11]. The 19S lid is formed by eight subunits (Rpn3, Rpn5, Rpn6, Rpn7, Rpn8, Rpn9, Rpn11, and Rpn12) [50].
In addition to regulation by 19S, the core particle can be activated by other regulatory particles. For example, PA28 (also known as 11S) [51] is formed by hetero-heptameric rings of 28-kDa proteins (PA28α and PA28β) or homo-heptameric rings of PA28γ [52]. PA28 binds to the cylinder end of the core particle, thus opening the channel [53, 54]. In contrast to the 19S regulatory particle, PA28 lacks ATPase activity and the ability to bind to ubiquitin conjugates [51, 55]. PA28αβ is inducible by interferon-γ [56] and modulates the presentation and generation of specific viral antigens [57]. PA28γ is involved in cell cycle regulation promoting the degradation of small proteins such as p21 [58]. The 20S proteasome can also be activated by Blm10/PA200, a monomeric protein of 250 kDa [59]. Blm10/PA200 forms hybrid complexes in which this protein binds to one end of the 20S proteasome and the 19S to the opposite end [60, 61]. Taken together, proteasome activity is tightly modulated by different proteasome activators.
PROTEASOME ACTIVITY DECLINES DURING CELLULAR SENESCENCE AND AGING
A hallmark of aging is the progressive decline in cellular proteostasis and the accumulation of misfolded and damaged proteins [7]. This failure of proteostasis with age involves mechanisms such a decline in stabilization of correctly folded proteins and protein clearance systems. Chaperones assure the proper cellular localization and folding of proteins throughout their life cycle [4, 62]. When the stability of the proteome is challenged, a series of cellular responses are activated to maintain the proper folding of proteins such as the heat shock response (HSR) or the unfolded protein response (UPR) [1, 63]. Induction of chaperones is highly impaired in aging [64]. In addition, an overexpression of chaperones in either D. melanogaster or C. elegans promotes longevity [65-68]. In mice, long-lived mutant strains express high levels of specific heat shock proteins while mice lacking a heat-shock-family chaperone exhibit an accelerated aging phenotype [69, 70]. Similar to the chaperone network, protein clearance mechanisms such as autophagy also decline during aging [12, 71]. Studies on the aged human brain have shown decreased markers of autophagy [72]. Mice overexpressing LAMP2a, a receptor of the chaperone-mediated autophagy, avoid aging-associated autophagy impairment and exhibit improved liver functions when aged [73]. Interestingly, administration of rapamycin, an autophagy inducer, considerably delays aging in mice and other models [74-76]. In flies, induction of specific autophagy genes promotes lifespan and confers protection upon different stress factors [77]. In yeast, worms and flies, autophagy is required for the rapamycin-induced lifespan extension [12, 78].
As a key component of the cellular proteostasis network, proteasome function declines during aging. This dysfunction can occur at different levels such as decreased expression of proteasome subunits [79, 80], alteration and/or replacement of proteasome subunits [81], disassembly of proteasomes [82] or inactivation by interacting with protein aggregates [83]. This later mechanism could induce a catastrophic proteostasis feedback during aging, since inhibition of proteostasis itself can induce the accumulation of protein inclusions, which in turn can further inhibit proteasome activity. In yeast, the failure of the UPS during aging induces the accumulation of protein inclusions [84]. These protein aggregates obstruct UPS function. By increasing disaggregase activity, the accumulation of proteins inclusions during aging diminishes and degradation by the 26S proteasome is restored [84].
A decline in proteasome function during aging and senescence has been observed in several mammalian tissues and cells such as human skin, epidermal cells, fibroblasts and lymphocytes [85-88], bovine eye lens [89] and rat liver, lung, heart, kidney, spinal cord, hippocampus, cerebral cortex and muscle tissues [90-95]. Human fibroblasts are a well-established model to define senescence mechanisms. In comparison to cells that have undergone fewer passages, human senescent fibroblasts exhibit a reduction in the levels of all three proteasome activities and proteasome subunit levels [87]. In addition, fibroblasts treated with proteasome inhibitors exhibit a shortened replicative lifespan and a senescent-like phenotype [96]. Notably, a transgenic mouse with decreased proteasomal chymotrypsin-like activity exhibits a shortened lifespan, premature age-related phenotypes and aggravation of age-related metabolic disorders such as obesity or excessive accumulation of triglycerides in hepatic cells [97].
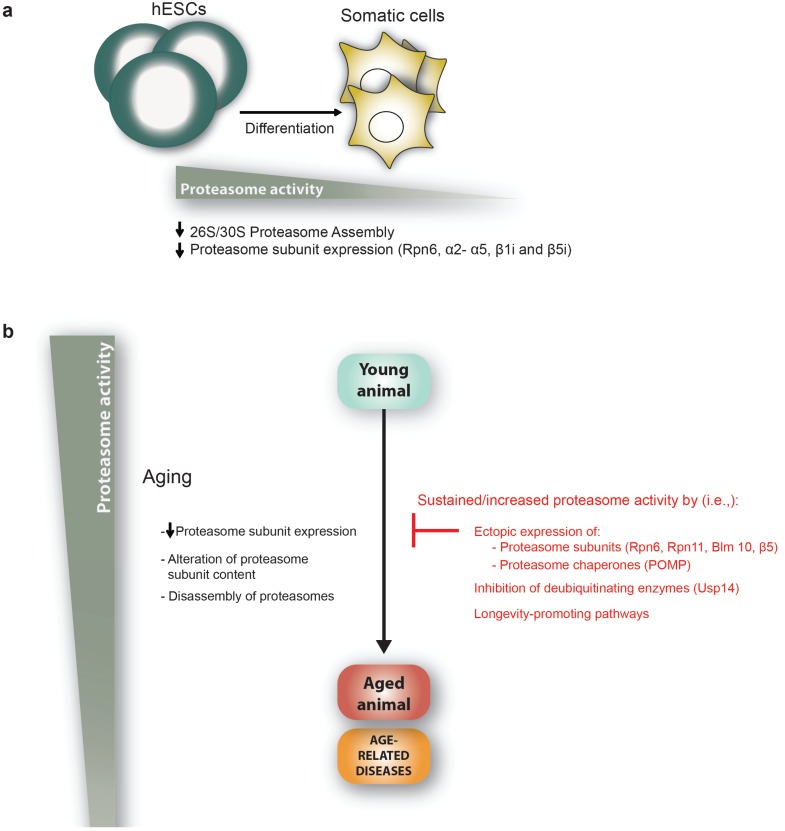
These observations indicate that proteasome activity declines during the aging process. It has been shown that different tissues exhibit different proteasome activity [98, 99]. Taken together, these findings raise an intriguing question: Is the proteasome equally affected in different cell types during aging? This question can be addressed by using cell-specific photoconvertible reporters to measure proteasome activity in real time in living animals [100]. For instance, a study using this methodology in C. elegans has shown that dorsorectal neurons exhibit a more severe decline in proteasome activity during aging than body-wall muscle cells [100]. The specific biological purpose of every cell type may define cellular differences in proteasome activity levels and how aging impacts on their proteasome machinery. Indeed, recent findings in embryonic stem cells (ESC) support this hypothesis. ESCs do not undergo replicative senescence and are considered to be immortal in culture [101, 102]. The immortality of ESCs suggests that these cells could have increased mechanisms to protect their proteome and, therefore, avoid senescence. Accordingly, human ESCs (hESCs) exhibit high proteasome activity compared to their differentiated counterparts such as neurons, fibroblasts or trophoblasts [28] (Fig. 2A). This increased proteasome activity is correlated in human and mouse with increased levels of the 19S proteasome subunit PSMD11/Rpn6 [25, 28, 103], which is an essential subunit for the activity of the 26S/30S proteasome that stabilizes the otherwise weak interaction between the 20S core and the 19S cap [28, 104]. Moreover, hESCs also exhibit increased levels of other proteasome subunits such as α2- α5 [103] and the immunoproteasome subunits β1i (PSMB9) and β5i (PSMB8) [105]. Besides Psmd11/Rpn6, mouse ESCs also express high levels of the deubiquitinating enzyme Psmd14/Rpn11 [25]. These findings raise the intriguing question of why ESCs cells need enhanced activity of the proteasome. Increased proteasome activity in these cells could be required to avoid senescence and maintain an intact proteome for self-renewal. In contrast to fibroblasts, proteasome activity does not differ in hESCs depending on the passage number [28]. Since the passage of damaged proteins to daughter cells could compromise the aging process of an organism, increased proteasome activity in ESCs could also be required to generate a cell lineage with an intact proteome. Notably, degradation of damaged proteins is triggered upon the first signs of mouse ESC differentiation [106, 107]. Induction of the proteasome activator PA28αβ is required for degradation of these damaged proteins during the first signs of cell fate specification [107].
Fig. (2).
The links between proteasome activity, aging and age-related diseases. a, Human embryonic stem cells (hESCs) exhibit higher levels of proteasome activity than their differentiated counterparts (i.e. fibroblasts, trophoblasts or neurons). This high activity is required for the maintenance of stem cell function and is mediated, among others, through increased expression of the 19S proteasomal subunit Rpn6. b, Proteasome activity declines with aging as a consequence of a reduction in the expression of proteasomal subunits, alteration or replacement of several proteasomal subunits, disassembly of the proteasome or inactivation of the proteasome through direct interaction with age-induced protein-aggregates. Consequently, interventions that increase and sustain proteasome activity such as overexpression of proteasomal subunits and chaperones can extend lifespan and delay the onset of age-related diseases. Several longevity-promoting pathways (i.e., dietary restriction (DR) or reduced insulin/IGF-1 signaling pathway (IIS)) increase proteasome activity. Likewise, long-lived organisms (i.e., the naked mole rat and the giant clam) and centenarians exhibit increased proteasome activity.
Since adult stem cell exhaustion is considered one of the tentative hallmarks of aging in organisms [7], it will be fascinating to examine whether proteasome activity is increased in these cells. Adult organisms have two types of stem cells: 1) germ line stem cells (GSCs) [108] and 2) adult somatic stem cells which are found in several tissues and rejuvenate them. Adult somatic stem cells persist throughout the lifespan of organism. However, adult somatic stem cell function declines during the aging process in tissues such as the brain, skin, blood, bone and skeletal muscle and this failure may contribute to age-related diseases [7, 109]. Whether adult somatic stem cells also have an enhanced proteasome activity remains to be elucidated, but the maintenance of this activity may critically impact organismal aging. In contrast, GSCs are designed to maintain an unlimited proliferative capacity in order to fulfill their biological purpose: to be passed from one generation to the next. GSCs can acquire in vitro properties similar to those of ESCs such as pluripotency [110]. GSCs generate the gametes that will produce the embryos after reproduction. ESCs and oocytes share a common transcriptome signature [103, 111]. Similar to hESC, human oocytes have an increased expression in the levels of PSMD11 [103]. In D. melanogaster, gonads (ovaries and spermathecae) and maturating oocytes have an increased 26S proteasome activity and accumulate less damaged proteins compared to aging somatic tissues [98, 99]. Proteasome activity is already down-regulated in middle-aged flies, when signs of aging first appeared [98, 99]. Although proteasome activity declines in somatic tissues during the aging process, maturating oocytes and gonads maintain their high activity compared to age-matched somatic tissues [98]. Taken together, these results show that proteasome activity and its decline during aging are differentially regulated depending on the cell type. In addition, these results support the disposable soma theory of aging formalized by Thomas Kirkwood in 1977 [112]. In this hypothesis, resources are allocated from the soma to the germline under stress conditions and aging in order to prevent and repair damage to the germline. By this mechanism, the germline would ensure the fitness of the next generation. Therefore, increased proteasome activity in maturating oocytes and gonads may contribute to safeguard reproduction and ensure the generation a pristine proteome in the ensuing generation. In yeast, aggregated and damaged proteins can be sequestered and retained in mother cells during division [113]. This asymmetric division enables the generation of a rejuvenated, germ-like, daughter cell lineage [114, 115]. The aforementioned results in flies suggest that an evolutionarily conserved mechanism may have evolved to avoid replicative senescence by establishing an aging (somatic tissues) and rejuvenated/immortal (germ cells) lineage, the later with an increased proteasome activity that reduces the accumulation of damaged proteins.
PROTEASOME DYSFUNCTION IN AGE-RELATED DISEASES
Gain-of-toxic-function diseases, in which aggregation-mediated proteotoxicity exceeds the cellular clearance machinery, have been associated with aging [116, 117]. Among these diseases are neurodegenerative disorders such as AD, HD, PD and amyotrophic lateral sclerosis (ALS) [3, 8-10, 14, 16]. Protein aggregation and deposition are common features of these disorders [8-10] that share emergence patterns and are more frequent late in life, even though different toxic proteins are involved in their onset [116]. The pathophysiological significance of protein aggregates is vaguely known. The accumulation of abnormal proteins may lead to perturbed cellular functions and eventually to neuronal death, ultimately manifesting as neurodegenerative disease. Changes in the proteasome machinery may explain why aging is a risk factor for AD, PD and HD [117]. Furthermore, loss-of-function experiments have shown that decreased proteasomal activity can enhance the neurodegenerative phenotype [17, 118]. In addition, the inclusion bodies of AD, PD, HD and ALS contain abnormal amounts of ubiquitin, suggesting a link between proteasome dysfunction and neurodegeneration [15, 119].
AD is the most common of the neurodegenerative disorders marked by protein misfolding and aggregation. AD results in progressive dementia and loss of memory, reasoning and language qualities [8]. Proteasome activity is decreased in brains from AD patients compared with age-matched controls [120]. AD is characterized by the deposition of two different protein aggregates: (i) extracellular senile (amyloid) plaques and (ii) intracellular neurofibrillary tangles [8]. The latter are mostly composed by hyperphosphorylated tau, a microtubule-associated protein [8]. An in vitro study has shown that tau protein can be degraded by the 20S proteasome in an ATP/ubiquitin independent manner [121]. However, other studies showed that tau degradation by the proteasome is at least partially ubiquitin-dependent in cells [122, 123]. The ubiquitin ligase CHIP promotes tau degradation [122, 123]. When tau is not bound to microtubules, it associates with the chaperone Hsp70. CHIP interacts directly with Hsp70 and promotes tau ubiquitination and degradation [122, 123]. Furthermore, inhibition of the proteasome-associated deubiquitinating enzyme Usp14 promotes tau degradation [124]. In addition, the proteasome activator Blm10 accelerates tau degradation in vitro [125]. Tau clearance is blocked by FK506 binding protein 51 kDa (FKBP51), which forms a mature chaperone complex with Hsp90 that prevents tau degradation [126]. In mice, FKBP51 and HSP90 synergize to block tau clearance by the proteasome, resulting in tau oligomerization [127]. Notably, in humans, FKBP51 levels increase relative to age and higher FKBP51 levels are associated with AD progression [127]. These results suggest a model in which age-associated increases in FKBP51 and its interaction with Hsp90 promote neurotoxic tau accumulation. Strikingly, aggregated tau can inhibit proteasome activity [120], which in turn could contribute to the pathology of AD. Senile plaques are mostly formed by aggregates of amyloid-β (Aβ), which is generated via proteolytic cleavage of the amyloid precursor protein (APP) [8]. Incorrect cleavage of APP leads to Aβ aggregation. Interestingly, intracellular Aβ oligomers can inhibit proteasome activity [128].
PD is a neurodegenerative movement disorder characterized by the accumulation of Lewy Bodies in dopaminergic neurons [9]. The major components of Lewy bodies are α-synuclein, ubiquitin, the E3 ligase parkin and other UPS-related proteins. Different approaches have been undertaken to uncover the role of the proteasome in the pathology of PD. A rat model for this disease was established by treating animals with proteasome inhibitors [129]. These rats exhibited characteristics of PD, such as bradykinesia and tremor, which was accompanied at a molecular level by α-synuclein and ubiquitin-containing inclusions in dopaminergic neurons. However, other studies could not obtain a similar output [119]. As an alternative approach, proteasome subunits knock-out mouse models were generated. Since the constitutive deletion of most of the proteasome genes is embryonically lethal, conditional knock-out models had to be developed [13]. Deletion of the ATPase subunit Psmc1/Rpt2 specifically in dopaminergic neurons lead to ubiquitin and α-synuclein positive inclusions which resulted in neuronal death, thus resembling the hallmarks of PD [130]. In addition, variations in the gene PSMC4/Rpt3 correlate with the age of PD onset in patients [131]. These results suggest a role for the 26S/30S in α-synuclein degradation. However, α-synuclein monomers can also be degraded by free 20S in a ubiquitin-independent process [132]. ALS is another progressive neurodegenerative movement disorder. ALS affects motor neurons [9]. Impairment of the proteasome in these neurons by conditional knock-out of Psmc4/Rpt3 revealed loss of motor neurons, locomotor dysfunction and accumulation of aggregates of TDP-43 and FUS proteins, features typical of ALS [133].
HD is the most studied disorder of polyglutamine (polyQ) diseases. HD is an autosomal dominant disease that affects muscle coordination and leads to progressive cognitive decline and psychosis [10]. HD displays selective neurodegeneration that occurs preferentially in the brain striatum [10, 134, 135]. The disorder is caused by the expansion of a CAG triplet repeat region in the huntingtin gene, which translates into a polyQ stretch in the N-terminal domain of the protein and results in fibril formation and aggregation [10, 136, 137]. An expansion of glutamines of over 40 can trigger the development of HD. In turn, the length of the CAG stretches correlates with the disease progression and types of symptoms [137, 138]; unusually long CAG expansions (>50) cause an early onset of the disease, known as juvenile HD [139]. Huntingtin inclusions contain ubiquitinated proteins, chaperones and components of the proteasome [140]. Enhancement of the proteasome machinery has been proven beneficial in HD models. Increased expression of PA28γ in a cellular model of HD improves cell survival [141]. Furthermore, ectopic expression of PSMD11/Rpn6 in a C. elegans model of HD ameliorates the symptoms associated to this disease [17]. In contrast to the soluble form of huntingtin, the aggregated form has been found to be ubiquitinated itself [142, 143], suggesting an impairment of the capacity of the proteasome to degrade aberrant huntingtin [144-146]. In addition, proteasome activity is reduced in brains from HD patients and mice models [144]. Nevertheless, whether the formation of inclusion bodies in neurons is protective or toxic is still under debate. Considering the correlation between behavioral deficits and the load of misfolded huntingtin in HD mouse models, the inclusions themselves have been proposed to be on the base of the toxicity leading to neuronal death [140, 147, 148]. The exact mechanisms of this toxicity, however, remain unsolved but a general impairment of the proteasomal function resulting in proteostasis collapse and cellular death has been proposed [149, 150]. An in vitro study suggested that the proteasome is not able to cleave within PolyQ stretches and the occasional failure of these long undegradable sequences to exit the proteasome may interfere with its function [151]. Accordingly, incubation of proteasomes with mutant huntingtin exerts an inhibitory effect on the proteasome by directly impeding the entrance of other substrates [152]. However, other studies found that huntingtin aggregates do not affect proteasome activity suggesting that proteasomal dysfunction may be a consequence of a general proteostasis collapse [142, 153].
Aging is also a primary risk factor for cancer [7, 154]. Cancer is the consequence of an aberrant gain of cellular fitness whereas aging is characterized by a loss of fitness. Although aging and cancer may seem to be opposite processes, both can share common origins: the cellular damage accumulated during aging triggers cellular senescence, but this damage occasionally provides aberrant advantages to specific cells triggering cancer [7, 154]. Several studies have suggested that proteasomal activity itself is elevated in human cancers [155-157]. This enhancement is consistent with the special requirements of cancerous cells, such as the elimination of aberrant proteins. The high degree of cell division and high mutation rates in these cells lead to an accumulation of misfolded proteins, which activate stress responses and/or apoptosis. Upregulation of proteasomal activity eliminates aberrant proteins that otherwise would be toxic for the cell [158]. Therefore, immortal cells such as cancer cells and hESCs exhibit a high proteasome activity.
Proteasome activity could have an important relevance in theories that suggest a relationship between aging somatic stem cell health and the onset of cancer [154]. Stem cells and progenitor cells persist through the lifespan of an organism and accumulate DNA damage from stresses such as intracellular oxidants. Typically, these damaged cells undergo cell arrest, apoptosis or senescence. Therefore, this process decreases the number and functionality of adult somatic cells and progenitor cells contributing to the aging process and age-related pathologies. However, some rare cells can escape from the default pathway, acquiring additional mutations that allow them to continue to proliferate and increase the likelihood of cancer [154]. As mentioned above, whether adult somatic stem cells have an increased proteasome activity is unknown. Therefore, a better knowledge of how adult somatic stem cells regulate their proteasome activity may help to understand how cancer stem cells are generated in an organism and to find specific treatment against these cells. Since both stem cells and cancer stem cells rely on similar protective mechanisms, specifically targeting these pathways in cancer cells may be difficult. Deciphering the differences in proteasome activity regulation between stem cells and cancer stem cells might be needed for efficient treatment implementation.
It is not clear whether inhibition of the proteasome would result in selective inhibition of malignant cell growth, but a number of identified compounds that inhibit proteasomal degradation have been shown to induce apoptosis [159, 160], kill tumor cells [161], overcome drug resistance [162, 163] and enhance radiation sensitivity [161]. The underlying mechanisms for this selectivity might be that malignant cells show greater sensitivity to the cytotoxic effects of proteasome inhibition than non-cancer cells [164-166]. Likewise, hESCs are more sensitive to proteasome inhibition than their differentiated counterparts [28]. Accordingly, proteasome inhibition has become a valuable and powerful tool in the treatment of certain types of cancer [167-170]. Bortezomib, a proteasome inhibitor that reversibly inhibits the chymotrypsin site in the 20S particle, was approved in 2003 for treatment of multiple myeloma (MM) [171, 172]. MM cells produce high amounts of aberrant immunoglobins and, consequently, strongly rely on proteasome function for the continual clearance of abnormal proteins. Bortezomib-mediated programmed cell death is accompanied by generation of ROS, transmembrane mitochondrial potential gradient dissipation, release of propaptotitc mitochondrial proteins, such as cytochrome c, and activation of intrinsic, caspase-9-mediated and extrinsic, caspase-8-mediated apoptosis. Other mechanisms include induction of aggregosome formation, endoplasmic reticulum stress and the unfolded protein response [173, 174]. Bortezomib has also shown great efficiency against related hematological malignancies, namely Waldenström's macroglobulinemia [175, 176] and mantle cell lymphoma [177, 178]. Furthermore, two second-generation compounds have entered phase I trials: NPI-0052 [179] and carfilzomib [180], which also inhibit the chymotrypsin-like activity of the 20S particle but show improved pharmacological properties. Unlike bortezomib, which binds to the proteasome in a slowly reversible manner, NPI-0052 and carfilzomib bind irreversibly, abrogating a possible recovery from proteasome inhibition by the release of the drug.
PROTEASOME ACTIVITY AS A DETERMINANT OF LONGEVITY AND STRESS RESISTANCE
Several reports suggest that sustained proteasome activity correlates with longevity. Notably, proteasome activity is increased in long-lived humans (centenarians) [181]. In this study, the levels of several proteasome subunits and proteasome activity were analyzed in fibroblasts derived from healthy centenarians and compared with fibroblast from young and old control donors. Strikingly, proteasome activity and subunit levels in healthy centenarian-derived fibroblasts are more similar to the younger than the older control donors-derived fibroblasts [181]. Furthermore, increased proteasome activity was also found in extremely long-lived animals such as the naked mole rat [182] and the giant clam [183]. As mentioned before, increased proteasome activity is increased in immortal cells such as hESCs, which can proliferate continuously in the absence of senescence [28].
This link between increased proteasome activity and longevity has been further supported by genetic approaches (Fig. 2B). For instance, ectopic expression of 19S proteasome subunits extends lifespan in organismal models such as D. melanogaster and C. elegans. A genetic gain-of-function screen in D. melanogaster characterized Rpn11 as a determinant of aging [118]. Rpn11 overexpression suppresses the age-dependent reduction of 26S/30S proteasome activity and extends lifespan of flies [118]. In addition, increased levels of Rpn11 suppress expanded PolyQ-induced progressive neurodegeneration [118]. In C. elegans, overexpression of Rpn6 is sufficient to extend longevity under proteotoxic stress conditions and reduces toxic aggregates in PolyQ-disease models [17]. Increased assembly of active proteasomes induced by overexpression of the 20S proteasome subunit β5 confers resistance to oxidative stress and delays senescence in human fibroblasts [184]. In addition, overexpression of the proteasome chaperone POMP, a protein involved in proteasome assembly, increases proteasome function and protects from oxidative stress in human fibroblasts [185]. In Saccharomyces cerevisae, overexpression of the yeast homolog of POMP (UMP1) induces an enhanced preservation of proteasome-mediated protein degradation and increased viability during stationary-phase, a model for post-mitotic aging [186]. Modulation of transcription factors that regulate the expression of proteasome subunits is also a valid approach to increased proteasome activity. In yeast, a transcription factor named Rpn4 regulates the levels of proteasome subunits [187]. Rpn4 degradation is promoted by the E3 ubiquitin ligase Ubr2 [188]. In addition, Rpn4 expression is regulated by stress inducible transcription factors such as Hsf1 [189], the primary regulator of heat-stress response [190]. In yeast, loss of UBR2 results in elevated levels of Rpn4 and increased replicative lifespan and resistance to proteotoxic stress [191].
A series of signaling pathways promote longevity and provide increased stability of the proteome delaying the onset of age-related diseases [3, 23, 24, 192]. Reduced food-intake without malnutrition (DR) extends lifespan in multiple species [193, 194]. DR decreases protein synthesis by modulating translational rates [195, 196] which can improve proteostasis maintenance. The decrease in the load of nascent polypeptides to the proteostasis machinery may allow more efficient protein folding and degradation and, therefore, decrease the accumulation of misfolded and damaged proteins. In mice, DR induces the expression of the 19S proteasome subunit Psmc3/Rpt5 and the proteasome activator PA28 [79]. In C. elegans, the HECT E3 ubiquitin ligase WWP-1 is a positive regulator of lifespan in response to DR [197]. This lifespan extension induced by WWP-1 is dependent on the FOXA transcription factor pha-4 [197]. In addition, the
oxidative-stress-responsive transcription factor skn-1 is also required for DR-induced longevity in C. elegans [198]. skn-1 promotes the expression of aip-1/AIRAP (arsenic-inducible RNA-associated protein) under metabolic stress [199]. aip-1/AIRAP binds to the 19S increasing proteasome activity and clearance of damaged proteins [200]. Notably, aip-1/AIRAP ameliorates β-amyloid peptide and PolyQ toxicity [201, 202]. In addition, skn-1 and its mammalian orthologues Nrf1 and Nrf2 upregulate the expression of proteasomal genes in response to proteasome inhibition and oxidative stress [203-210]. Inducible Nrf2 activation in D. melanogaster promotes youthful expression of proteasome subunits in aged somatic tissues [99]. This result suggests that age-dependent Nrf2 dysfunction may contribute to the decrease in proteasome subunit expression with age. Interestingly, Nrf2-mediated proteasome activation delays senescence in human fibroblasts [204].
Reduced insulin/IGF-1 signaling pathway (IIS) extends lifespan in both invertebrates and vertebrates [211-214]. IIS reduction correlates with increased longevity of humans [215-217]. The insulin/IGF-1 receptor activates a conserved PI3-kinase/PDK/AKT signaling cascade that phosphorylates FOXO transcription factors, thereby preventing their nuclear localization. When IIS signaling is reduced, FOXO accumulates in the nucleus and regulates downstream genes that extend lifespan and increase stress resistance in worms, flies and mice [212, 214, 218-222]. In C. elegans, the CUL-1 E3 ubiquitin ligase complex (Skp1-Cul1-F-Box) is required for the extended lifespan of IIS mutant worms. This regulation is achieved, at least in part, by promoting the transcriptional activity of the worm FOXO transcription factor DAF-16 [223]. In worms, decreased IIS induces proteasome activity [224]. This regulation is achieved through DAF-16 transcriptional repression of the proteasome-associated deubiquitinating enzyme ubh-4 [224]. Uchl5, the human ortholog of ubh-4, increases degradation of toxic proteins in mammalian cells [224]. Delayed aging, by IIS reduction, protects worms and mice from protein aggregation toxicity [23, 24, 219, 225]. Different invertebrate models have been established to study proteostasis-associated human diseases. Studies in these disease models have shown that expression of human disease proteins in invertebrates can be toxic and results in physiological and behavioral changes [23, 225, 226]. Experiments using C. elegans expressing polyQ in body wall muscle showed that toxic protein aggregation is age-dependent [225]. IIS reduction is able to delay PolyQ aggregation and toxicity, suggesting that this pathway modulates the proteostasis network to enable cells to deal with toxic proteins to a much later age [225]. Whether reduced IIS ameliorates PolyQ aggregation in body well muscle by modulating, at least partially, proteasome activity has not been examined. Interestingly, overexpression of rpn-6, a target of DAF-16, is sufficient to reduce toxic aggregates in neuronal PolyQ-disease models [17]. In a C. elegans model of AD, expression of Aβ1-42 peptide in the body wall muscle cells produces a progressive paralysis phenotype [227]. In a DAF-16 dependent manner, reduced function of the IIS can protect from the toxicity of Aβ1-42 expression [23].Interestingly, reduced IIS induces the aggregation of small toxic Aβ1-42 oligomers into larger, less toxic structures suggesting the activation of an aggregation mechanism [23]. These results have been substantiated in mice, in which a heterozygous mutation in the IGF-1 receptor is protective in a mouse model of AD [24]. However, it has been also suggested that autophagic degradation of the β-amyloid peptide is required for the protective effect of reduced IIS in Aβ1-42-expressing worms [228]. Whether activation of DAF-16 also ameliorates the toxicity of Aβ1-42 through modulation of proteasome activity has not been examined.
Besides IIS, other growth factors can regulate aging [229]. Epidermal Growth Factor (EGF) signalling promotes longevity in C. elegans [230]. Upregulation of both proteasome activity and polyubiquitination, and a consequent decrease in protein aggregation are required for the lifespan extension induced by EGF signalling [231].
Among invertebrates, birds and mammals, experimental paradigms that limit reproductive investment also cause lifespan extension [232]. Hypothetically, the need for repairing and preventing damage to the germline dominates resource allocation strategies, while the somatic tissues age and deteriorate [112]. In support of such theories, modulations of reproduction that eliminate germ cells in C. elegans and D. melanogaster provide effective mechanisms for extending lifespan [232-234], phenotypes that may be caused by heightened resource availability and proteome stability within the post-mitotic soma [17, 235]. Inhibiting germline proliferation delays the onset of PolyQ-dependent aggregation and toxicity [235]. Proteasome activity and RPN-6 protein levels are increased in germline-lacking worms [17]. In these long-lived animals, increased proteasome activity, rpn-6 expression and longevity are modulated by DAF-16 [17]. Similar to these long-lived worms, FOXO4 is necessary for increased proteasome activity and PSMD11/Rpn6 levels in immortal hESCs [28, 236]. Interestingly, it has been recently reported that DNA damage in germ cells of C. elegans induce a systemic response that protects somatic tissues by increasing their proteasome activity [237].
Reduction of the activity of the mitochondrial electron transport chain (ETC) is another evolutionary conserved pathway that delays aging in invertebrates and mice [238-242]. However, proteasome activity is not increased in ETC long-lived C. elegans [17]. The activity of the proteasome in other longevity-promoting mechanisms has not been established yet.
CONCLUDING REMARKS
Increasing evidences point to the decline of a functional proteome and accumulation of misfolded and damaged proteins as determinants of the aging process. In turn, damaged proteins are associated to late-onset neurodegenerative diseases such as AD, PD and HD. The proteasome constitutes one of the main cellular protein clearance mechanisms and is involved in key biological aspects such as cell cycle, signal transduction or apoptosis, among others. Immortal cells, long-lived laboratory models and centenarians, positively correlate with increased proteasome activity. Furthermore, genetic enhancement of proteasome function not only extends lifespan, but also increases stress resistance and ameliorates the symptoms and incidence of age-related disorders. Taken together, these findings indicate that the age-related decline of proteasome functionality is an important determinant of aging and age-related neurodegenerative diseases. Recently, nine cellular and molecular hallmarks have been categorized and proposed to lie on the base of the aging process [7]: genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, and altered intercellular communication. The UPS is involved in the regulation of the maintenance of a stable and functional proteome, the clearance of ROS-induced damaged proteins caused by mitochondrial dysfunction and the regulation of tumour suppressor proteins involved in cellular senescence. In addition, proteostasis and cellular stress responses regulate somatic stem cell function [243]. This suggests that the proteasome has a role in several of the hallmarks of aging and, therefore, is a key component of the aging process. This could be translated into a valuable therapeutic approach for the treatment of progressive, age-related neurodegenerative diseases. However, considering the links between enhanced proteasomal activity and malignant cells, some detrimental effects of this approach should not be discarded.
ACKNOWLEDGEMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) (CECAD) and the European Commission (FP7-PEOPLE-2013-CIG). We thank Suzanne Wolff and Peter M. Douglas for providing a critical review of the manuscript.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–9. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 2.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009;78:959–91. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 3.Taylor RC, Dillin A. Aging as an event of proteostasis col-lapse.old Spring Harb. Perspect. Biol. 2011;3(5) doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22(11):1427–38. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morimoto RI, Cuervo AM. Protein homeostasis and aging: taking care of proteins from the cradle to the grave. J. Gerontol. A Biol. Sci. Med. Sci. 2009;64(2):167–70. doi: 10.1093/gerona/gln071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vabulas R M, Raychaudhuri S, Hayer-Hartl M, Hartl F U. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb. Perspect. Biol. 2010;2(12):a004390. doi: 10.1101/cshperspect.a004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Otin C, Blasco M A, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selkoe D J. Alzheimer's disease.Cold Spring Harb. Perspect. Biol. 2011;3(7):a004457. doi: 10.1101/cshperspect.a004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosco D A, LaVoie M J, Petsko G A, Ringe D. Proteostasis and movement disorders: Parkinson's disease and amyotrophic lateral sclerosis.old Spring Harb. Perspect. Biol. 2011;3(10):a007500. doi: 10.1101/cshperspect.a007500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkbeiner S. Huntington's Disease. Cold Spring Harb. Perspect. Biol. 2011;3(6):a007476. doi: 10.1101/cshperspect.a007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. nnu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubinsztein D C, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146(5):682–95. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 13. Tanaka K, Matsuda N. Proteostasis and neurodegeneration: The roles of proteasomal degradation and autophagy. Biochim. Biophys. Acta. 2013;1843(1):197–204. doi: 10.1016/j.bbamcr.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Wong E, Cuervo A. M.Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb. Perspect. Biol. 2010;2(12):a006734. doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Q, Figueiredo-Pereira M E. Ubiquitin/proteasome pathway impairment in neurodegeneration: therapeutic implications. Apoptosis. 2010;15(11):1292–311. doi: 10.1007/s10495-010-0466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt M, Finley D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta. 2013;1843(1):13–25. doi: 10.1016/j.bbamcr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilchez D, Morantte I, Liu Z, Douglas P M, Merkwirth C, Rodrigues A P, Manning G, Dillin A. RPN-6 determines C.elegans longevity under proteotoxic stress conditions. Nature. 2012;489(7415):263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 18.Hansen M, Chandra A, Mitic L L, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C.elegans. PLoS Genet. 2008;4(2):e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hars E S, Qi H, Ryazanov A G, Jin S, Cai L, Hu C, Liu L F. Autophagy regulates ageing in C.elegans. Autophagy. 2007;3(2):93–5. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- 20.Lapierre L R, Hansen M. Lessons from C.elegans: signaling pathways for longevity. Trends Endocrinol. Metab. 2012;23(12):637–44. doi: 10.1016/j.tem.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lapierre L R, Melendez A, Hansen M. Autophagy links lipid metabolism to longevity in C.elegans. Autophagy. 2012;8(1):144–6. doi: 10.4161/auto.8.1.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melendez A, Talloczy Z, Seaman M, Eskelinen E L, Hall D H, Levine B. Autophagy genes are essential for dauer development and life-span extension in C.elegans. Science. 2003;301(5638):1387–91. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 23.Cohen E, Bieschke J, Perciavalle R M, Kelly J W, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313(5793):1604–10. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 24.Cohen E, Paulsson J F, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham H M, Holzenberger M, Kelly J W, Masliah E, Dillin A. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139(6):1157–69. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buckley S M, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth C L, Koller A A, Dasgupta R, Silva J C, Stadtfeld M, Hochedlinger K, Chen E I, Aifantis I. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11(6):783–98. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okita Y, Nakayama K I. UPS delivers pluripotency. Cell Stem Cell. 2012;11(6):728–30. doi: 10.1016/j.stem.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka K. The proteasome: from basic mechanisms to emerging roles. Keio J. Med. 2013;62(1):1–12. doi: 10.2302/kjm.2012-0006-re. [DOI] [PubMed] [Google Scholar]
- 28.Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren T W, Gage F H, Dillin A. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489(7415):304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciehanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem. Biophys. Res. Commun. 1978;81(4):1100–5. doi: 10.1016/0006-291x(78)91249-4. [DOI] [PubMed] [Google Scholar]
- 30.Hershko A, Ciechanover A, Heller H, Haas A L, Rose I A. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc. Natl. Acad. Sci. USA. 1980;77(4):1783–6. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkinson KD, Urban MK, Haas AL. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J. Biol. Chem. 1980;255(16):7529–32. [PubMed] [Google Scholar]
- 32.Ciechanover A, Elias S, Heller H, Hershko A. "Covalent affinity" purification of ubiquitin-activating enzyme. J. Biol. Chem. 1982;257(5):2537–42. [PubMed] [Google Scholar]
- 33.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system.Resoltion affinity purification, and role in protein breakdown. . J. Biol. Chem. . 1983;258(13):8206–14. [PubMed] [Google Scholar]
- 34.Pickart C M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 35.Iwai K. iverse ubiquitin signaling in NF-kappaB activation. Trends Cell. Biol. 2012;22(7):355–64. doi: 10.1016/j.tcb.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19(1):94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals.'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 2008;9(6):536–42. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. tructure of 20S proteasome from yeast at 2. A resolution. Nature. 1997;386(6624):463–71. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 39.Orlowski M. The multicatalytic proteinase complex, a major extralysosomal proteolytic system. Biochem. 1990;29(45):10289–97. doi: 10.1021/bi00497a001. [DOI] [PubMed] [Google Scholar]
- 40.Orlowski M, Cardozo C, Michaud C. Evidence for the presence of five distinct proteolytic components in the pituitary multicatalytic proteinase complex.Properties of two components cleaving bonds on the carboxyl side of branched chain and small neutral amino acids. Biochem. 1993;32(6):1563–72. doi: 10.1021/bi00057a022. [DOI] [PubMed] [Google Scholar]
- 41.Gaczynska M, Rock KL, Goldberg AL. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature. 1993;365(6443):264–7. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 42.Monaco JJ, McDevitt HO. H-2-linked low-molecular weight polypeptide antigens assemble into an unusual macromolecular complex. Nature. 1984;309(5971):797–9. doi: 10.1038/309797a0. [DOI] [PubMed] [Google Scholar]
- 43.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 2005;398:364–78. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- 44.Baugh JM, Viktorova EG, Pilipenko EV. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J. Mol. Biol. 2009;386(3):814–27. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffman L, Pratt G, Rechsteiner M. Multiple forms of the 20 S multicatalytic and the 26 S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. J. Biol. Chem. 1992;267(31):22362–8. [PubMed] [Google Scholar]
- 46.Hough R, Pratt G, Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J. Biol. Chem. 1987;262(17):8303–13. [PubMed] [Google Scholar]
- 47.Waxman L, Fagan JM, Goldberg AL. Demonstration of two distinct high molecular weight proteases in rabbit reticulocytes, one of which degrades ubiquitin conjugates. J. Biol. Chem. 1987;262(6):2451–7. [PubMed] [Google Scholar]
- 48.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 1994;269(10):7059–61. [PubMed] [Google Scholar]
- 49.Beyer A. Sequence analysis of the AAA protein family. Protein Sci. 1997;6(10):2043–58. doi: 10.1002/pro.5560061001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Udvardy A. Purification and characterization of a multiprotein component of the Drosophila 26 S (1500 kDa) proteolytic complex. J. Biol. Chem. 1993;268(12):9055–62. [PubMed] [Google Scholar]
- 51.Ma CP, Slaughter CA, DeMartino GN. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain). J. Biol. Chem. 1992;267(15):10515–23. [PubMed] [Google Scholar]
- 52.Ahn K, Erlander M, Leturcq D, Peterson PA, Fruh K, Yang Y. In vivo characterization of the proteasome regulator PA28. J. Biol. Chem. 1996;271(30):18237–42. doi: 10.1074/jbc.271.30.18237. [DOI] [PubMed] [Google Scholar]
- 53.Demartino GN, Gillette TG. Proteasomes: machines for all reasons. Cell. 2007;129(4):659–62. doi: 10.1016/j.cell.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 54.Forster A, Masters E I, Whitby F G, Robinson H, Hill C P. The 1. A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell. 2005;18(5):589–99. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 55.Dubiel W, Pratt G, Ferrell K, Rechsteiner M. Purification of an 11 S regulator of the multicatalytic protease. J. Biol. Chem. 1992;267(31):22369–77. [PubMed] [Google Scholar]
- 56.Realini C, Jensen C C, Zhang Z, Johnston S C, Knowlton J R, Hill C P, Rechsteiner M. Characterization of recombinant REGalpha, REGbeta, and REGgamma proteasome activators. J. Biol. Chem. 1997;272(41):25483–92. doi: 10.1074/jbc.272.41.25483. [DOI] [PubMed] [Google Scholar]
- 57.Sijts A, Sun Y, Janek K, Kral S, Paschen A, Schadendorf D, Kloetzel P M. The role of the proteasome activator PA28 in MHC class I antigen processing. Mol. Immunol. 2002;39(3-4):165–9. doi: 10.1016/s0161-5890(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 58.Li X, Amazit L, Long W, Lonard DM, Monaco JJ, O'Malley BW. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol. Cell. 2007;26(6):831–42. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 59.Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21(13):3516–25. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blickwedehl J, Agarwal M, Seong C, Pandita R K, Melendy T, Sung P, Pandita T K, Bangia N. Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability. Proc. Natl. Acad. Sci. USA. 2008;105(42):16165–70. doi: 10.1073/pnas.0803145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt M, Haas W, Crosas B, Santamaria P G, Gygi S P, Walz T, Finley D. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat. Struct. Mol.iol. . 2005; 12(4):294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- 62.Hartl F U, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–32. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 63.Gidalevitz T, Prahlad V, Morimoto R I. The stress of protein misfolding: from single cells to multicellular organisms. Cold Spring Harb. Perspect. Biol. 2011;3(6) doi: 10.1101/cshperspect.a009704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Calderwood S K, Murshid A, Prince T. The shock of aging molecular chaperones and the heat shock response in longevity and aging--a mini-review. Gerontology. 2009;55(5):550–8. doi: 10.1159/000225957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu A L, Murphy C T, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300(5622):1142–5. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 66.Morley J F, Morimoto R I. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell. 2004;15(2):657–64. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morrow G, Samson M, Michaud S, Tanguay R M. verexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 2004;18(3):598–9. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- 68.Walker G A, Lithgow G J. Lifespan extension in C.elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell. 2003;2(2):131–9. doi: 10.1046/j.1474-9728.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- 69.Min J N, Whaley R A, Sharpless N E, Lockyer P, Portbury A L, Patterson C. CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol. Cell Biol. 2008;28(12):4018–25. doi: 10.1128/MCB.00296-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Swindell WR, Masternak MM, Kopchick JJ, Conover CA, Bartke A, Miller RA. Endocrine regulation of heat shock protein mRNA levels in long-lived dwarf mice. Mech. Ageing Dev. 2009;130(6):393–400. doi: 10.1016/j.mad.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 2000;275(40):31505–13. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 72.Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J. Biol. Chem. 2006;281(20):14474–85. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 73.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat. Med. 2008;14(9):959–65. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blagosklonny M V. Rapamycin-induced glucose intolerance: hunger or starvation diabetes. Cell Cycle. 2011;10(24):4217–24. doi: 10.4161/cc.10.24.18595. [DOI] [PubMed] [Google Scholar]
- 75.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–5. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilkinson JE, Burmeister L, Brooks SV, Chan CC, Friedline S, Harrison DE, Hejtmancik JF, Nadon N, Strong R, Wood LK, Woodward MA, Miller RA. Rapamycin slows aging in mice. Aging Cell. 2012;11(4):675–82. doi: 10.1111/j.1474-9726.2012.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simonsen A, Cumming RC, Finley KD. Linking lysosomal trafficking defects with changes in aging and stress response in Drosophila. Autophagy. 2007;3(5):499–501. doi: 10.4161/auto.4604. [DOI] [PubMed] [Google Scholar]
- 78.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11(1):35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee C K, Klopp R G, Weindruch R, Prolla T A. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285(5432):1390–3. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 80.Ly D H, Lockhart D J, Lerner R A, Schultz P G. Mitotic misregulation and human aging. Science. 2000;287(5462):2486–92. doi: 10.1126/science.287.5462.2486. [DOI] [PubMed] [Google Scholar]
- 81.Ferrington D A, Husom A D, Thompson L V. Altered proteasome structure, function, and oxidation in aged muscle. FASEB J. 2005;19(6):644–6. doi: 10.1096/fj.04-2578fje. [DOI] [PubMed] [Google Scholar]
- 82.Vernace V A, Arnaud L, Schmidt-Glenewinkel T, Figueiredo-Pereira M E. Aging perturbs 26S proteasome assembly in Drosophila melanogaster. FASEB J. 2007;21(11):2672–82. doi: 10.1096/fj.06-6751com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grune T, Jung T, Merker K, Davies KJ. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and 'aggresomes' during oxidative stress, aging, and disease. Int. J. Biochem. Cell Bio.l. 2004;36(12):2519–30. doi: 10.1016/j.biocel.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 84.Andersson V, Hanzen S, Liu B, Molin M, Nystrom T. Enhancing protein disaggregation restores proteasome activity in aged cells. Aging (Albany NY) 2013;5(11):802–812. doi: 10.18632/aging.100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bulteau AL, Petropoulos I, Friguet B. Age-related alterations of proteasome structure and function in aging epidermis. Exp. Gerontol. 2000;35(6-7):767–77. doi: 10.1016/s0531-5565(00)00136-4. [DOI] [PubMed] [Google Scholar]
- 86.Carrard G, Dieu M, Raes M, Toussaint O, Friguet B. Impact of ageing on proteasome structure and function in human lymphocytes. Int. J. Biochem. Cell Biol. 2003;35(5):728–39. doi: 10.1016/s1357-2725(02)00356-4. [DOI] [PubMed] [Google Scholar]
- 87.Chondrogianni N, Stratford F L, Trougakos I P, Friguet B, Rivett A J, Gonos E S. Central role of the proteasome in senescence and survival of human fibroblasts: induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J. Biol. Chem. 2003;278(30):28026–37. doi: 10.1074/jbc.M301048200. [DOI] [PubMed] [Google Scholar]
- 88.Petropoulos I, Conconi M, Wang X, Hoenel B, Bregegere F, Milner Y, Friguet B. Increase of oxidatively modified protein is associated with a decrease of proteasome activity and content in aging epidermal cells. J. Gerontol. A Biol. Sci. Med. Sci. 2000;55(5):B220–7. doi: 10.1093/gerona/55.5.b220. [DOI] [PubMed] [Google Scholar]
- 89.Wagner B J, Margolis J W. Age-dependent association of isolated bovine lens multicatalytic proteinase complex (proteasome) with heat-shock protein 90, an endogenous inhibitor. Arch. Biochem. Biophys. 1995;323(2):455–62. doi: 10.1006/abbi.1995.0067. [DOI] [PubMed] [Google Scholar]
- 90.Bardag-Gorce F, Farout L, Veyrat-Durebex C, Briand Y, Briand M. Changes in 20S proteasome activity during ageing of the LOU rat. Mol. Biol. Rep. 1999;26(1-2):89–93. doi: 10.1023/a:1006968208077. [DOI] [PubMed] [Google Scholar]
- 91.Bulteau A L, Szweda L I, Friguet B. Age-dependent declines in proteasome activity in the heart. Arch. Biochem. Biophys. 2002;397(2):298–304. doi: 10.1006/abbi.2001.2663. [DOI] [PubMed] [Google Scholar]
- 92.Conconi M, Szweda L I, Levine R L, Stadtman E R, Friguet B. Age-related decline of rat liver multicatalytic proteinase activity and protection from oxidative inactivation by heat-shock protein 90. Arch. Biochem. Biophys. 1996;331(2):232–40. doi: 10.1006/abbi.1996.0303. [DOI] [PubMed] [Google Scholar]
- 93.Husom A D, Peters E A, Kolling E A, Fugere N A, Thompson L V, Ferrington D A. Altered proteasome function and subunit composition in aged muscle. Arch. Biochem. Biophys. 2004;421(1):67–76. doi: 10.1016/j.abb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 94.Keller J N, Hanni K B, Markesbery W R. Possible involvement of proteasome inhibition in aging: implications for oxidative stress. Mech. Ageing Dev. 2000;113(1):61–70. doi: 10.1016/s0047-6374(99)00101-3. [DOI] [PubMed] [Google Scholar]
- 95.Shibatani T, Nazir M, Ward W F. Alteration of rat liver 20S proteasome activities by age and food restriction. J. Gerontol. A Biol. Sci. Med. Sci. 1996;51(5):B316–22. doi: 10.1093/gerona/51a.5.b316. [DOI] [PubMed] [Google Scholar]
- 96.Torres C, Lewis L, Cristofalo V J. Proteasome inhibitors shorten replicative life span and induce a senescent-like phenotype of human fibroblasts. J. Cell Physiol. 2006;207(3):845–53. doi: 10.1002/jcp.20630. [DOI] [PubMed] [Google Scholar]
- 97.Tomaru U, Takahashi S, Ishizu A, Miyatake Y, Gohda A, Suzuki S, Ono A, Ohara J, Baba T, Murata S, Tanaka K, Kasahara M. Decreased proteasomal activity causes age-related phenotypes and promotes the development of metabolic abnormalities. Am. J. Pathol. 2012;180(3):963–72. doi: 10.1016/j.ajpath.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 98.Fredriksson A, Johansson Krogh E, Hernebring M, Pettersson E, Javadi A, Almstedt A, Nystrom T. Effects of aging and reproduction on protein quality control in soma and gametes of Drosophila melanogaster. Aging Cell. 2012;11(4):634–43. doi: 10.1111/j.1474-9726.2012.00823.x. [DOI] [PubMed] [Google Scholar]
- 99.Tsakiri E N, Sykiotis G P, Papassideri I S, Gorgoulis V G, Bohmann D, Trougakos I P. Differential regulation of proteasome functionality in reproductive vs.somatic tissues of Drosophila during aging or oxidative stress. FASEB J. 2013;27(6):2407–20. doi: 10.1096/fj.12-221408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hamer G, Matilainen O, Holmberg CI. A photoconvertible reporter of the ubiquitin-proteasome system in vivo. Nat. Methods. 2010;7(6):473–8. doi: 10.1038/nmeth.1460. [DOI] [PubMed] [Google Scholar]
- 101.Evans M J, Kaufman M H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–6. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 102.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 103.Assou S, Cerecedo D, Tondeur S, Pantesco V, Hovatta O, Klein B, Hamamah S, De Vos J. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics. 2009;10:10. doi: 10.1186/1471-2164-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pathare GR, Nagy I, Bohn S, Unverdorben P, Hubert A, Korner R, Nickell S, Lasker K, Sali A, Tamura T, Nishioka T, Forster F, Baumeister W, Bracher A. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc. Natl. Acad. Sci. USA. 2012;109(1):149–54. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Atkinson S P, Collin J, Irina N, Anyfantis G, Kyung B K, Lako M, Armstrong L. A putative role for the immunoproteasome in the maintenance of pluripotency in human embryonic stem cells. Stem Cells. 2012;30(7):1373–84. doi: 10.1002/stem.1113. [DOI] [PubMed] [Google Scholar]
- 106.Hernebring M, Brolen G, Aguilaniu H, Semb H, Nystrom T. Elimination of damaged proteins during differentiation of embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2006;103(20):7700–5. doi: 10.1073/pnas.0510944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hernebring M, Fredriksson A, Liljevald M, Cvijovic M, Norrman K, Wiseman J, Semb H, Nystrom T. Removal of damaged proteins during ES cell fate specification requires the proteasome activator PA28. Sci. Rep. 2013;3:1381. doi: 10.1038/srep01381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li L, Xie T. Stem cell niche: structure and function. Annu. Rev. Cell Dev. Biol. 2005;21:605–31. doi: 10.1146/annurev.cellbio.21.012704.131525. [DOI] [PubMed] [Google Scholar]
- 109.Signer R A, Morrison S J. Pluripotency of spermatogonial stem cells from adult mouse testis.Mechanisms that regulate stem cell aging and life span. Cell Stem Cell. 2013;12(2):152–65. doi: 10.1016/j.stem.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guan K, Nayernia K, Maier LS, Wagner S, Dressel R, Lee JH, Nolte J, Wolf F, Li M, Engel W, Hasenfuss G. Reference Titile. Nature. 2006;440(7088):1199–203. doi: 10.1038/nature04697. [DOI] [PubMed] [Google Scholar]
- 111.Nicholas C R, Haston K M, Grewall A K, Longacre T A, Reijo Pera R A. Transplantation directs oocyte maturation from embryonic stem cells and provides a therapeutic strategy for female infertility. Hum. Mol. Genet. 2009;18(22):4376–89. doi: 10.1093/hmg/ddp393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kirkwood T B. Evolution of ageing. Nature. 1977;270(5635):301–4. doi: 10.1038/270301a0. [DOI] [PubMed] [Google Scholar]
- 113.Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299(5613):1751–3. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 114.Erjavec N, Cvijovic M, Klipp E, Nystrom T. Selective benefits of damage partitioning in unicellular systems and its effects on aging. Proc. Natl. Acad. Sci. USA. 2008;05(48):18764–9. doi: 10.1073/pnas.0804550105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Erjavec N, Nystrom T. Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2007;104(26):10877–81. doi: 10.1073/pnas.0701634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cohen E, Dillin A. The insulin paradox: aging, proteotoxicity and neurodegeneration. Nat. Rev. Neurosci. 2008;9(10):759–67. doi: 10.1038/nrn2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zabel C, Nguyen HP, Hin SC, Hartl D, Mao L, Klose J. Proteasome and oxidative phoshorylation changes may explain why aging is a risk factor for neurodegenerative disorders. J. Proteomics. 2010;73(11):2230–8. doi: 10.1016/j.jprot.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 118.Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol. Cell Biol. 2009;29(4):1095–106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Matsuda N, Tanaka K. Does impairment of the ubiquitin-proteasome system or the autophagy-lysosome pathway predispose individuals to neurodegenerative disorders such as Parkinson's disease? J. Alzheimers Dis. 2010;19(1):1–9. doi: 10.3233/JAD-2010-1231. [DOI] [PubMed] [Google Scholar]
- 120.Keck S, Nitsch R, Grune T, Ullrich O. Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer's disease. J. Neurochem. 2003;85(1):115–22. doi: 10.1046/j.1471-4159.2003.01642.x. [DOI] [PubMed] [Google Scholar]
- 121.Grune T, Botzen D, Engels M, Voss P, Kaiser B, Jung T, Grimm S, Ermak G, Davies K J. Tau protein degradation is catalyzed by the ATP/ubiquitin-independent 20S proteasome under normal cell conditions. Arch. Biochem. Biophys. 2010;500(2):181–8. doi: 10.1016/j.abb.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dickey CA, Koren J, Zhang YJ, Xu YF, Jinwal UK, Birnbaum M J, Monks B, Sun M, Cheng J Q, Patterson C, Bailey R M, Dunmore J, Soresh S, Leon C, Morgan D, Petrucelli L. Akt and CHIP coregulate tau degradation through coordinated interactions. Proc. Natl. Acad. Sci. USA. 2008;105(9):3622–7. doi: 10.1073/pnas.0709180105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G, Kim J, Dillmann W H, Browne S E, Hall A, Voellmy R, Tsuboi Y, Dawson T M, Wolozin B, Hardy J, Hutton M. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum. Mol. Genet. 2004;13(7):703–14. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 124.Lee M J, Lee J H, Rubinsztein D C. Tau degradation: the ubiquitin-proteasome system versus the autophagy-lysosome system. Prog. Neurobiol. 2013;105:49–59. doi: 10.1016/j.pneurobio.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 125.Dange T, Smith D, Noy T, Rommel P C, Jurzitza L, Cordero R J, Legendre A, Finley D, Goldberg A L, Schmidt M. Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism. J. Biol. Chem. 2011;286(50):42830–9. doi: 10.1074/jbc.M111.300178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jinwal U K, Koren J, Borysov S I, Schmid A B, Abisambra J F, Blair L J, Johnson A G, Jones J R, Shults C L, O'Leary J C, Jin Y, Buchner J, Cox M B, Dickey C A. The Hsp90 cochaperone, FKBP51, increases Tau stability and polymerizes microtubules. J. Neurosci. 2010;30(2):591–9. doi: 10.1523/JNEUROSCI.4815-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Blair L J, Nordhues B A, Hill S E, Scaglione K M, O'Leary J C, Fontaine S N, Breydo L, Zhang B, Li P, Wang L, Cotman C, Paulson H L, Muschol M, Uversky V N, Klengel T, Binder E B, Kayed R, Golde T E, Berchtold N, Dickey C A. Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. J. Clin. Invest. 2013;123(10):4158–69. doi: 10.1172/JCI69003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tseng B P, Green K N, Chan J L, Blurton-Jones M, LaFerla F M. Abeta inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiol. Aging. 2008;29(11):1607–18. doi: 10.1016/j.neurobiolaging.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McNaught K S, Perl D P, Brownell A L, Olanow C W. Systemic exposure to proteasome inhibitors causes a progressive model of Parkinson's disease. Ann. Neurol. 2004;56(1):149–62. doi: 10.1002/ana.20186. [DOI] [PubMed] [Google Scholar]
- 130.Bedford L, Hay D, Devoy A, Paine S, Powe D G, Seth R, Gray T, Topham I, Fone K, Rezvani N, Mee M. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J. Neurosci. 2008; 28(33):8189–98. doi: 10.1523/JNEUROSCI.2218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wahl C, Kautzmann S, Krebiehl G, Strauss K, Woitalla D, Muller T, Bauer P, Riess O, Kruger R. A comprehensive genetic study of the proteasomal subunit S6 ATPase in German Parkinson's disease patients. J. Neural Transm. 2008;115(8):1141–8. doi: 10.1007/s00702-008-0054-3. [DOI] [PubMed] [Google Scholar]
- 132.Tofaris GK, Layfield R, Spillantini MG. alpha-synuclein metabolism and aggregation is linked to ubiquitin-independent degradation by the proteasome. FEBS Lett. 2001;509(1):22–6. doi: 10.1016/s0014-5793(01)03115-5. [DOI] [PubMed] [Google Scholar]
- 133.Tashiro Y, Urushitani M, Inoue H, Koike M, Uchiyama Y, Komatsu M, Tanaka K, Yamazaki M, Abe M, Misawa H, Sakimura K, Ito H, Takahashi R. Motor neuron-specific disruption of proteasomes, but not autophagy, replicates amyotrophic lateral sclerosis. J. Biol. Chem. 2012;287(51):42984–94. doi: 10.1074/jbc.M112.417600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gusella J F, MacDonald M E, Ambrose C M, Duyao MP. Molecular genetics of Huntington's disease. Arch. Neurol. 1993;50(11):1157–63. doi: 10.1001/archneur.1993.00540110037003. [DOI] [PubMed] [Google Scholar]
- 135.Vonsattel J P, DiFiglia M. Huntington disease. J. Neuropathol. Exp. Neurol. 1998;57(5):369–84. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 136.Kumail N. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes.The Huntington's Disease Collaborative Research Group. Cell. 1993;72(6):971–83. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 137. Sanchez I, Mahlke C, Yuan J. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 2003;421(6921):373–9. doi: 10.1038/nature01301. [DOI] [PubMed] [Google Scholar]
- 138.Snell RG, MacMillan JC, Cheadle JP, Fenton I, Lazarou LP, Davies P, MacDonald M E, Gusella JF, Harper P S, Shaw D J. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat. Genet. 1993;4(4):393–7. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 139.Nance M A, Myers R H. Juvenile onset Huntington's disease--clinical and research perspectives. Ment. Retard. Dev. Disabil. Res. Rev. 2001;7(3):153–7. doi: 10.1002/mrdd.1022. [DOI] [PubMed] [Google Scholar]
- 140.Holmberg C I, Staniszewski K E, Mensah K N, Matouschek A, Morimoto R I. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 2004;23(21):4307–18. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Seo H, Sonntag K C, Kim W, Cattaneo E, Isacson O. Proteasome activator enhances survival of Huntington's disease neuronal model cells. PLOS One. 2007;2(2):e238. doi: 10.1371/journal.pone.0000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hipp MS, Patel CN, Bersuker K, Riley BE, Kaiser SE, Shaler TA, Brandeis M, Kopito R R. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington's disease. J. Cell Biol. 2012;196(5):573–87. doi: 10.1083/jcb.201110093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kalchman MA, Graham RK, Xia G, Koide HB, Hodgson JG, Graham KC, Goldberg YP, Gietz RD, Pickart CM, Hayden MR. Huntingtin is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J. Biol. Chem. 1996;271(32):19385–94. doi: 10.1074/jbc.271.32.19385. [DOI] [PubMed] [Google Scholar]
- 144.Finkbeiner S, Mitra S. The ubiquitin-proteasome pathway in Huntington's disease. ScientificWorldJournal. 2008;8:421–33. doi: 10.1100/tsw.2008.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sieradzan K A, Mechan A O, Jones L, Wanker E E, Nukina N, Mann D M. Huntington's disease intranuclear inclusions contain truncated, ubiquitinated huntingtin protein. Exp. Neurol. 1999;156(1):92–9. doi: 10.1006/exnr.1998.7005. [DOI] [PubMed] [Google Scholar]
- 146.Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker E E. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell. 2001;12(5):1393–407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bence N F, Sampat R M, Kopito R R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292(5521):1552–5. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 148.Diaz-Hernandez M, Valera AG, Moran MA, Gomez-Ramos P, Alvarez-Castelao B, Castano JG, Hernandez F, Lucas JJ. Inhibition of 26S proteasome activity by huntingtin filaments but not inclusion bodies isolated from mouse and human brain. J. Neuro hem. 2006;98(5):1585–96. doi: 10.1111/j.1471-4159.2006.03968.x. [DOI] [PubMed] [Google Scholar]
- 149.Ardley HC, Hung CC, Robinson PA. The aggravating role of the ubiquitin-proteasome system in neuro-degeneration. FEBS Lett. 2005;579(3):571–6. doi: 10.1016/j.febslet.2004.12.058. [DOI] [PubMed] [Google Scholar]
- 150.Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40(2):427–46. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 151.Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg A L. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol. Cell. 2004;14(1):95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- 152.Bennett EJ, Bence NF, Jayakumar R, Kopito RR. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol. Cell. 2005;17(3):351–65. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 153.Pratt G, Rechsteiner M. Proteasomes cleave at multiple sites within polyglutamine tracts: activation by PA28gamma(K188E). J. Biol. Chem. 2008;283(19):12919–25. doi: 10.1074/jbc.M709347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448: (7155)767–74. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 155. Arlt A, Bauer I, Schafmayer C, Tepel J, Muerkoster SS, Brosch M, Roder C, Kalthoff H, Hampe J, Moyer MP, Folsch UR, Schafer H. Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2). Oncogene. 2009;28(45):3983–96. doi: 10.1038/onc.2009.264. [DOI] [PubMed] [Google Scholar]
- 156.Chen L, Madura K. Increased proteasome activity, ubiquitin-conjugating enzymes, and eEF1A translation factor detected in breast cancer tissue. Cancer Res. 2005;65(13):5599–606. doi: 10.1158/0008-5472.CAN-05-0201. [DOI] [PubMed] [Google Scholar]
- 157.Zhang W G, Yu J P, Wu Q M, Tong Q, Li S B, Wang X H, Xie GJ. Inhibitory effect of ubiquitin-proteasome pathway on proliferation of esophageal carcinoma cells. World J. Gastroenterol. 2004;10(19):2779–84. doi: 10.3748/wjg.v10.i19.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Whitesell L, Lindquist S L. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer. 2005;5(10):761–72. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 159.Ling Y H, Liebes L, Ng B, Buckley M, Elliott P J, Adams J, Jiang J D, Muggia F M, Perez-Soler R. PS-341, a novel proteasome inhibitor, induces Bcl-2 phosphorylation and cleavage in association with G2-M phase arrest and apoptosis. Mol. Cancer Ther. 2002;1(10):841–9. [PubMed] [Google Scholar]
- 160.Pei X Y, Dai Y, Grant S. The proteasome inhibitor bortezomib promotes mitochondrial injury and apoptosis induced by the small molecule Bcl-2 inhibitor HA14-1 in multiple myeloma cells. Leukemia. 2003;17(10):2036–45. doi: 10.1038/sj.leu.2403109. [DOI] [PubMed] [Google Scholar]
- 161.Teicher BA, Ara G, Herbst R, Palombella V J, Adams J. The proteasome inhibitor PS-341 in cancer therapy. Clin. Cancer Res. 1999;5(9):2638–45. [PubMed] [Google Scholar]
- 162.Frankel A, Man S, Elliott P, Adams J, Kerbel R S. Lack of multicellular drug resistance observed in human ovarian and prostate carcinoma treated with the proteasome inhibitor PS-341. Clin. Cancer Res.. 2000;6(9):3719–28. [PubMed] [Google Scholar]
- 163.Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliott PJ, Adams J, Anderson K C. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001;61(7):3071–6. [PubMed] [Google Scholar]
- 164.Delic J, Masdehors P, Omura S, Cosset JM, Dumont J, Binet JL, Magdelenat H. The proteasome inhibitor lactacystin induces apoptosis and sensitizes chemo- and radioresistant human chronic lymphocytic leukaemia lymphocytes to TNF-alpha-initiated apoptosis. Br. J. Cancer. 1998;77(7):1103–7. doi: 10.1038/bjc.1998.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Orlowski RZ, Eswara JR, Lafond-Walker A, Grever MR, Orlowski M, Dang CV. Tumor growth inhibition induced in a murine model of human Burkitt's lymphoma by a proteasome inhibitor. Cancer Res. 1998;58(19):4342–8. [PubMed] [Google Scholar]
- 166.Soligo D, Servida F, Delia D, Fontanella E, Lamorte G, Caneva L, Fumiatti R, Lambertenghi Deliliers G. The apoptogenic response of human myeloid leukaemia cell lines and of normal and malignant haematopoietic progenitor cells to the proteasome inhibitor PSI. Br. J. Haematol. 2001;113(1):126–35. doi: 10.1046/j.1365-2141.2001.02683.x. [DOI] [PubMed] [Google Scholar]
- 167.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5(5):417–21. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 168.Almond J B, Cohen G M. The proteasome: a novel target for cancer chemotherapy. Leukemia. 2002;16(4):433–43. doi: 10.1038/sj.leu.2402417. [DOI] [PubMed] [Google Scholar]
- 169.Chen W, Hu X T, Shi Q L, Zhang F B, He C. Knockdown of the novel proteasome subunit Adrm1 located on the 20q13 amplicon inhibits colorectal Cancer Cell migration, survival and tumorigenicity. Oncol. Rep. 2009;21(2):531–7. [PubMed] [Google Scholar]
- 170.Goy A, Younes A, McLaughlin P, Pro B, Romaguera J E, Hagemeister F, Fayad L, Dang N H, Samaniego F, Wang M, Broglio K, Samuels B, Gilles F, Sarris A H, Hart S, Trehu E, Schenkein D, Cabanillas F, Rodriguez A M. Phase II study of proteasome inhibitor bortezomib in relapsed or refractory B-cell non-Hodgkin's lymphoma. J. Clin. Oncol. 2005;23(4):667–75. doi: 10.1200/JCO.2005.03.108. [DOI] [PubMed] [Google Scholar]
- 171.Orlowski R Z, Kuhn D J. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin. Cancer Res. 2008;14(6):1649–57. doi: 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- 172.Richardson P G, Mitsiades C, Schlossman R, Ghobrial I, Hideshima T, Munshi N, Anderson K C. Bortezomib in the front-line treatment of multiple myeloma. Expert Rev. Anticancer Ther. 2008;8(7):1053–72. doi: 10.1586/14737140.8.7.1053. [DOI] [PubMed] [Google Scholar]
- 173.Nencioni A, Grunebach F, Patrone F, Ballestrero A, Brossart P. Proteasome inhibitors: antitumor effects and beyond. Leukemia. 2007;21(1):30–6. doi: 10.1038/sj.leu.2404444. [DOI] [PubMed] [Google Scholar]
- 174.Richardson P G, Mitsiades C, Hideshima T, Anderson K C. Bortezomib: proteasome inhibition as an effective anticancer therapy. Annu. Rev. Med. 2006;57:33–47. doi: 10.1146/annurev.med.57.042905.122625. [DOI] [PubMed] [Google Scholar]
- 175.Chen C I, Kouroukis C T, White D, Voralia M, Stadtmauer E, Stewart A K, Wright J J, Powers J, Walsh W, Eisenhauer E. Bortezomib is active in patients with untreated or relapsed Waldenstrom's macroglobulinemia: a phase II study of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 2007;25(12):1570–5. doi: 10.1200/JCO.2006.07.8659. [DOI] [PubMed] [Google Scholar]
- 176.Treon S P, Hunter Z R, Matous J, Joyce R M, Mannion B, Advani R, Cook D, Songer J, Hill J, Kaden B R, Sharon D, Steiss R, Leleu X, Branagan A R, Badros A. Multicenter clinical trial of bortezomib in relapsed/refractory Waldenstrom's macroglobulinemia: results of WMCTG Trial 03-248. Clin. Cancer Res. 2007;13(11):3320–5. doi: 10.1158/1078-0432.CCR-06-2511. [DOI] [PubMed] [Google Scholar]
- 177.Belch A, Kouroukis C T, Crump M, Sehn L, Gascoyne R D, Klasa R, Powers J, Wright J, Eisenhauer E A. A phase II study of bortezomib in mantle cell lymphoma: the National Cancer Institute of Canada Clinical Trials Group trial IND. 50. Ann. Oncol. 2007;18(1):116–21. doi: 10.1093/annonc/mdl316. [DOI] [PubMed] [Google Scholar]
- 178.Fisher RI, Bernstein SH, Kahl BS, Djulbegovic B, Robertson MJ, de Vos S, Epner E, Krishnan A, Leonard JP, Lonial S, Stadtmauer EA, O'Connor OA, Shi H, Boral A L, Goy A. Multicenter phase II study of bortezomib in patients with relapsed or refractory mantle cell lymphoma. J. Clin. Oncol. 2006;24(30):4867–74. doi: 10.1200/JCO.2006.07.9665. [DOI] [PubMed] [Google Scholar]
- 179.Chauhan D, Catley L, Li G, Podar K, Hideshima T, Velankar M, Mitsiades C, Mitsiades N, Yasui H, Letai A, Ovaa H, Berkers C, Nicholson B, Chao TH, Neuteboom ST, Richardson P, Palladino M A, Anderson KC. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Cancer Cell. 2005;8(5):407–19. doi: 10.1016/j.ccr.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 180.Kuhn DJ, Chen Q, Voorhees PM, Strader JS, Shenk KD, Sun CM, Demo SD, Bennett MK, van Leeuwen FW, Chanan-Khan AA, Orlowski RZ. otent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood. 2007;110(9):3281–90. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chondrogianni N, Petropoulos I, Franceschi C, Friguet B, Gonos E S. Fibroblast cultures from healthy centenarians have an active proteasome. Exp. Gerontol. 2000;35(6-7):721–8. doi: 10.1016/s0531-5565(00)00137-6. [DOI] [PubMed] [Google Scholar]
- 182.Perez V I, Buffenstein R, Masamsetti V, Leonard S, Salmon A B, Mele J, Andziak B, Yang T, Edrey Y, Friguet B, Ward W, Richardson A, Chaudhuri A. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc. Natl. Acad. Sci. USA. 2009;106(9):3059–64. doi: 10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ungvari Z, Csiszar A, Sosnowska D, Philipp E E, Campbell C M, McQuary P R, Chow T T, Coelho M, Didier ES, Gelino S, Holmbeck MA, Kim I, Levy E, Sonntag W E, Whitby P W, Austad S N, Ridgway I. Testing predictions of the oxidative stress hypothesis of aging using a novel invertebrate model of longevity: the giant clam (Tridacna derasa). J. Gerontol. A Biol. Sci. Med. Sci. 2013;68(4):359–67. doi: 10.1093/gerona/gls159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chondrogianni N, Tzavelas C, Pemberton A J, Nezis I P, Rivett A J, Gonos E S. Overexpression of proteasome beta5 assembled subunit increases the amount of proteasome and confers ameliorated response to oxidative stress and higher survival rates. J. Biol. Chem. 2005;280(12):11840–50. doi: 10.1074/jbc.M413007200. [DOI] [PubMed] [Google Scholar]
- 185.Chondrogianni N, Gonos E S. Overexpression of hUMP1/POMP proteasome accessory protein enhances proteasome-mediated antioxidant defence. Exp. Gerontol. 2007;42(9):899–903. doi: 10.1016/j.exger.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 186.Chen Q, Thorpe J, Dohmen J R, Li F, Keller J N. Ump1 extends yeast lifespan and enhances viability during oxidative stress: central role for the proteasome? Free Radic. Biol. Med. 2006;40(1):120–6. doi: 10.1016/j.freeradbiomed.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 187. Dohmen R J, Willers I, Marques A J. Biting the hand that feeds: Rpn4-dependent feedback regulation of proteasome function. Biochim. Biophys. Acta. 2007;1773(11):1599–604. doi: 10.1016/j.bbamcr.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 188.Wang L, Mao X, Ju D, Xie Y. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J. Biol. Chem. 2004;279(53):55218–23. doi: 10.1074/jbc.M410085200. [DOI] [PubMed] [Google Scholar]
- 189.Ma M, Liu Z L. Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genomics. 2010;11:660. doi: 10.1186/1471-2164-11-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Morimoto R I. The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 2011;76:91–9. doi: 10.1101/sqb.2012.76.010637. [DOI] [PubMed] [Google Scholar]
- 191.Kruegel U, Robison B, Dange T, Kahlert G, Delaney J R, Kotireddy S, Tsuchiya M, Tsuchiyama S, Murakami C J, Schleit J, Sutphin G, Carr D, Tar K, Dittmar G, Kaeberlein M, Kennedy B K, Schmidt M. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet. 2011;7(9):e1002253. doi: 10.1371/journal.pgen.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wolff S, Dillin A. The trifecta of aging in Caenorhabditis elegans. Exp. Gerontol. 2006;41(10):894–903. doi: 10.1016/j.exger.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 193.Fontana L, Partridge L, Longo V D. Extending healthy life span--from yeast to humans. Science. 2010;328(5976):321–6. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 2008;77:727–54. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- 195.Wullschleger S, Loewith R, Hall M N. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 196.Zid B M, Rogers A N, Katewa S D, Vargas M A, Kolipinski M C, Lu T A, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139(1):149–60. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Carrano AC, Liu Z, Dillin A, Hunter T. A conserved ubiquitination pathway determines longevity in response to diet restriction. Nature. 2009;460(7253):396–9. doi: 10.1038/nature08130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bishop N A, Guarente L. Two neurons mediate diet-restriction-induced longevity in C.elegans. Nature. 2007;447(7144):545–9. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 199.Ferguson AA, Springer MG, Fisher AL. skn-1-Dependent and -independent regulation of aip-1 expression following metabolic stress in Caenorhabditis elegans. Mol. Cell Biol. 2010;30(11):2651–67. doi: 10.1128/MCB.01340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Stanhill A, Haynes CM, Zhang Y, Min G, Steele MC, Kalinina J, Martinez E, Pickart C M, Kong X P, Ron D. An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol. Cell. 2006;23(6):875–85. doi: 10.1016/j.molcel.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 201.Hassan WM, Merin DA, Fonte V, Link CD. AIP-1 ameliorates beta-amyloid peptide toxicity in a Caenorhabditis elegans Alzheimer's disease model. Hum. Mol. Genet. 2009;18(15):2739–47. doi: 10.1093/hmg/ddp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yun C, Stanhill A, Yang Y, Zhang Y, Haynes C M, Xu C F, Neubert T A, Mor A, Philips M R, Ron D. Proteasomal adaptation to environmental stress links resistance to proteotoxicity with longevity in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2008;105(19):7094–9. doi: 10.1073/pnas.0707025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kahn N W, Rea S L, Moyle S, Kell A, Johnson T E. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem. J. 2008;409(1):205–13. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- 204.Kapeta S, Chondrogianni N, Gonos E S. Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J. Biol. Chem. 2010;285(11):8171–84. doi: 10.1074/jbc.M109.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kraft D C, Deocaris C C, Wadhwa R, Rattan S I. Preincubation with the proteasome inhibitor MG-132 enhances proteasome activity via the Nrf2 transcription factor in aging human skin fibroblasts. Ann. N. Y. Acad. Sci. 2006;1067:420–4. doi: 10.1196/annals.1354.060. [DOI] [PubMed] [Google Scholar]
- 206.Kwak M K, Wakabayashi N, Greenlaw J L, Yamamoto M, Kensler T W. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol. Cell Biol. 2003;23(23):8786–94. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Li X, Matilainen O, Jin C, Glover-Cutter K M, Holmberg C I, Blackwell T K. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 2011;7(6):e1002119. doi: 10.1371/journal.pgen.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pickering A M, Linder R A, Zhang H, Forman H J, Davies K J. Nrf2-dependent induction of proteasome and Pa28alphabeta regulator are required for adaptation to oxidative stress. J. Biol. Chem. 2012;287(13):10021–31. doi: 10.1074/jbc.M111.277145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Pickering A M, Staab T A, Tower J, Sieburth D, Davies K J. A conserved role for the 20S proteasome and Nrf2 transcription factor in oxidative stress adaptation in mammals, Caenorhabditis elegans and Drosophila melanogaster. J. Exp. Biol. 2013;216(4):543–53. doi: 10.1242/jeb.074757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Radhakrishnan S K, Lee C S, Young P, Beskow A, Chan J Y, Deshaies R J. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell. 2010;38(1):17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bartke A. Insulin and aging. Cell Cycle. 2008;7(21):3338–43. doi: 10.4161/cc.7.21.7012. [DOI] [PubMed] [Google Scholar]
- 212.Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even P C, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421(6919):182–7. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 213.Panowski S H, Dillin A. Signals of youth: endocrine regulation of aging in Caenorhabditis elegans. Trends Endocrinol. Metab. 2009;20(6):259–64. doi: 10.1016/j.tem.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 214.Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299(5611):1346–51. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 215.Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc. Natl. Acad. Sci. USA. 2009;106(8):2700–5. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Suh Y, Atzmon G, Cho M O, Hwang D, Liu B, Leahy D J, Barzilai N, Cohen P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl. Acad. Sci. USA. 2008;105(9):3438–42. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Willcox B J, Donlon T A, He Q, Chen R, Grove J S, Yano K, Masaki K H, Willcox D C, Rodriguez B, Curb J D. FOXO3A genotype is strongly associated with human longevity. Proc. Natl. Acad. Sci. USA. 2008;105(37):13987–92. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bluher M, Kahn B B, Kahn C R. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299(5606):572–4. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 219.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120(4):449–60. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 220.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C.elegans mutant that lives twice as long as wild type. . Nature. 1993;366(6454):461–4. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 221.Panowski S H, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C.elegans. Nature. 2007;447(7144):550–5. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 222.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292(5514):107–10. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 223.Ghazi A, Henis-Korenblit S, Kenyon C. Regulation of Caenorhabditis elegans lifespan by a proteasomal E3 ligase complex. Proc. Natl. Acad. Sci. USA. 2007;104(14):5947–52. doi: 10.1073/pnas.0700638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Matilainen O, Arpalahti L, Rantanen V, Hautaniemi S, Holmberg C I. Insulin/IGF-1 signaling regulates proteasome activity through the deubiquitinating enzyme UBH-4. Cell Rep. 2013;3(6):1980–95. doi: 10.1016/j.celrep.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 225.Morley J F, Brignull H R, Weyers J J, Morimoto R I. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2002;99(16):10417–22. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kerr F, Augustin H, Piper MD, Gandy C, Allen MJ, Lovestone S, Partridge L. Dietary restriction delays aging, but not neuronal dysfunction, in Drosophila models of Alzheimer's disease. Neurobiol. Aging. 2011;32(11):1977–89. doi: 10.1016/j.neurobiolaging.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Link C D. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 1995;92(20):9368–72. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Florez-McClure M L, Hohsfield LA, Fonte G, Bealor MT, Link CD. Decreased insulin-receptor signaling promotes the autophagic degradation of beta-amyloid peptide in C.elegans. Autophagy. 2007;3(6):569–80. doi: 10.4161/auto.4776. [DOI] [PubMed] [Google Scholar]
- 229.Bartke A. Growth hormone, insulin and aging: the benefits of endocrine defects. Exp. Gerontol. 2011;46(2-3):108–11. doi: 10.1016/j.exger.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Iwasa H, Yu S, Xue J, Driscoll M. Novel EGF pathway regulators modulate C.elegans healthspan and lifespan via EGF recptor PLC-gamma, and IP3R activation. . Aging Cell. 2010; 9(4):490–505. doi: 10.1111/j.1474-9726.2010.00575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Liu G, Rogers J, Murphy C T, Rongo C. EGF signalling activates the ubiquitin proteasome system to modulate C.elegans lifespan. EMBO J. 2011;30(15):2990–3003. doi: 10.1038/emboj.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Partridge L, Gems D, Withers DJ. Sex and death what is the connection. Cell. 2005;120(4):461–72. doi: 10.1016/j.cell.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 233.Flatt T, Min K J, D'Alterio C, Villa-Cuesta E, Cumbers J, Lehmann R, Jones D L, Tatar M. Drosophila germ-line modulation of insulin signaling and lifespan. Proc. Natl. Acad. Sci. USA. 2008;105(17):6368–73. doi: 10.1073/pnas.0709128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kenyon C. A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann. N. Y. Acad. Sci. 2010;1204:156–62. doi: 10.1111/j.1749-6632.2010.05640.x. [DOI] [PubMed] [Google Scholar]
- 235.Shemesh N, Shai N, Ben-Zvi A. Germline stem cell arrest inhibits the collapse of somatic proteostasis early in Caenorhabditis elegans adulthood. Aging Cell. 2013;12(5):814–22. doi: 10.1111/acel.12110. [DOI] [PubMed] [Google Scholar]
- 236.Vilchez D, Boyer L, Lutz M, Merkwirth C, Morantte I, Tse C, Spencer B, Page L, Masliah E, Berggren WT, Gage FH, Dillin A. FOXO4 is necessary for neural differentiation of human embryonic stem cells. Aging Cell. 2013;12(3):518–22. doi: 10.1111/acel.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ermolaeva MA, Segref A, Dakhovnik A, Ou HL, Schneider JI, Utermohlen O, Hoppe T, Schumacher B. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature. 2013;501(7467):416–20. doi: 10.1038/nature12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Copeland JM, Cho J, Lo T, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr.Biol. 2009;19(19):1591–8. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 239.Dillin A, Hsu A L, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser A G, Kamath R S, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298(5602):2398–401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 240.Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell. 2001;1(5):633–44. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 241.Houtkooper R H, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams R W, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497(7450):451–7. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19(20):2424–34. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Vilchez D, Simic M S, Dillin A. Proteostasis and aging of stem cells. Trends Cell. Biol. 2013 doi: 10.1016/j.tcb.2013.09.002. [DOI] [PubMed] [Google Scholar]