Abstract
Recent studies from a number of laboratories have revealed a surprising number of connections between RNA polymerase II transcription and the ubiquitin/proteasome pathway. We now find yet another intersection of these pathways by showing that the 26S proteasome associates with regions of the GAL1, GAL10, and HSP82 genes, including the 3′ ends, in a transcription-dependent fashion. The appearance of the proteasome on these inducible genes correlates with both the accumulation of transcripts and the buildup of RNA polymerase II complexes in the same region. Furthermore, the 26S proteasome and RNA polymerase II coimmunoprecipitate, and inhibition of 26S proteolytic activity leads to increased read through of a transcription termination site. We suggest that the proteasome is generally recruited to the DNA at sites of stalled RNA polymerase and may act to resolve these complexes.
Recent research has demonstrated several connections between the 26S proteasome, or its subunits, and RNA polymerase II (pol II) transcription. We demonstrated that several proteins from the 19S regulatory particle, but not the 20S proteolytic unit (1), of the 26S proteasome (2) associate directly with the yeast Gal4 activator and are recruited to activated GAL promoters (3). The 19S has also been shown to be critical for efficient elongation by RNA pol II in both yeast and mammalian cells, and this activity does not require the proteolytic activity of the proteasome (4, 5). In addition to this role in elongation, we have also found that this proteasome subcomplex, which we have termed APIS (3), negatively regulates Gal4–promoter interactions in vivo, again in a nonproteolytic fashion (A.D., F.G., A. Ferdous, T.K., and S.A.J., unpublished observations). In contrast to the evidence for nonproteolytic activities of the proteasome in transcription, several laboratories noted an inverse correlation between the potency of many transcriptional activators and the protein level in the cell (6–9), suggesting an intimate mechanistic link between the process of transcriptional activation and the turnover of the activator via the ubiquitin–proteasome pathway. This idea has been further supported by the recent finding that proteasome inhibitors strongly inhibit estrogen receptor-mediated transcription and specifically block cycling of the estrogen receptor and other transcription factors on and off estrogen-responsive promoters (10). These results suggest that the 20S as well as the 19S subunit might be directly involved in the transcriptional process. Here, we examine this possibility and report another intersection between the transcription and ubiquitin–proteasome pathways.
In our previous study of the recruitment of the APIS complex to the GAL1/10 promoter, we showed that promoter DNA was coprecipitated with antibodies raised against several of the proteasomal ATPases in a chromatin immunoprecipitation (ChIP) assay, but that little or no promoter DNA was brought down by using antibodies raised against the 20S core subunit of the proteasome or proteins in the lid (a subunit of the 19S regulatory particle; ref. 3). In addition, the use of PCR primers that support the amplification of different regions of the GAL1 gene provided evidence that the APIS complex, but again not the 20S core or lid complexes, was broadly distributed along the gene soon after induction, which is consistent with a role in elongation. We report here the results of a more extensive ChIP analysis designed to ask whether the proteasome might associate with an active gene sometime after induction of transcription. Indeed, we find evidence for 26S proteasome association with regions of induced genes that correlate with locations of RNA pol II build-up, including the 3′ end of the gene, sites of UV damage, and other regions that may represent pause sites. Furthermore, we demonstrate that an inhibitor of the 26S proteasome increases read through of a transcriptional termination site. Finally, coimmunoprecipitation experiments reveal a physical association between pol II and the proteasome. Thus, the evidence clearly supports a physical and functional interaction between these two complexes. Because the promoter regions of the genes that we analyzed seem to be the only sites where pol II is detected, but the 20S core particle is not, we hypothesize that the 26S proteasome associates with stalled elongation complexes, including those at termination sites, and that its activity may be important in resolving these stalled complexes.
Materials and Methods
Yeast Strains. Strain SC691 was described previously, as were the Pre1-Flag and Cim5-Flag containing strains (3, 11). The Rpb3-Flag strain was made by first transforming yeast with an HpaI-linearized pRS306 RPB3 plasmid containing ≈500 bp of upstream sequence and an amino-terminal flag sequence fused in frame with the RPB3 coding region. After identifying integrated clones, 5-fluoroorotic acid selection was used to screen for a second recombination event. The gal1::HIS3 strain was made by cloning the 3′ half of the GAL1 gene and inserting the 2.6-kb ScaI/SmaI fragment from pRS 303 into the HindIII site.
ChIP Assays. ChIP assays for Figs. 1 and 5 were performed as described (3). ChIP assays for the Flag-epitope containing proteins were performed essentially the same; however, the chromatin was diluted 1/100 with chromatin dilution buffer (1% Triton X-100/1.2 mM EDTA/16.7 mM Tris·HCl, pH 8.1/167 mM NaCl/0.5 mM PMSF) before immunoprecipitation with 20 μl (50% slurry) of anti-flag agarose beads (Sigma), resulting in 1/10 the starting material after immunoprecipitation as compared with the original protocol.
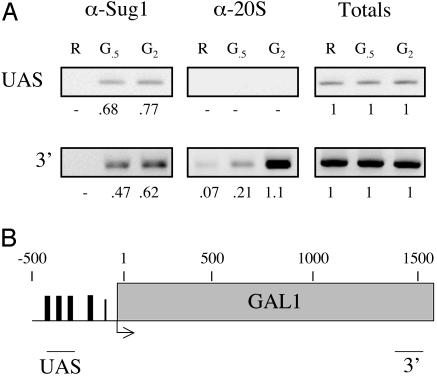
Fig. 1.
Association of the proteasome with the 3′ end of the GAL1 gene revealed by ChIP analysis. (A) A map of the GAL1 gene showing the regions amplified by PCR. (B) GAL1 PCR products from a ChIP assay. The sample points are in raffinose before GAL gene induction (R), 30 min after galactose addition (G.5), or 2 h after galactose addition (G2) are shown. The numbers correspond to the amount of specific DNA precipitated as a percentage of the input DNA. Totals are 1% of the input DNA.
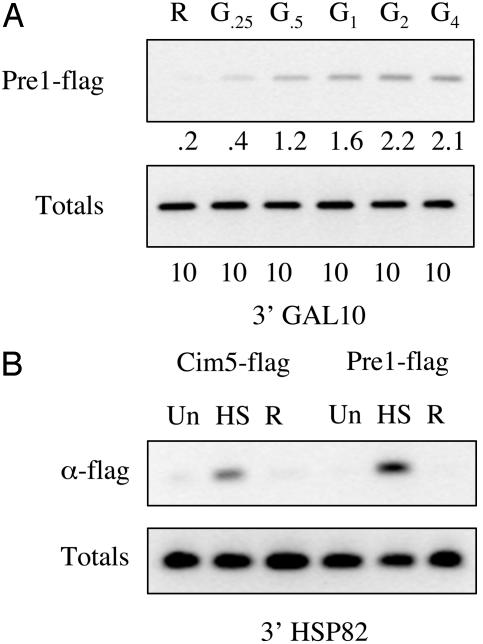
Fig. 5.
Association of the proteasome with the 3′ ends of the GAL10 and HSP82 genes. (A) GAL10 3′ region PCR products from a ChIP assay. Primers are located in a region at the very 3′ end of the GAL10 coding region. The sample points are in raffinose before GAL gene induction (R), 15 min (G.25), 30 min (G.5), 1 h (G1), 2 h (G2), or 4 h (G4) after galactose addition. Totals are 10% of the input DNA (B) HSP82 PCR products from a ChIP assay. The sample points are before heat shock (Un), after heat shock for 15 min (HS), and after removing from heat shock for 20 min (R). Totals are 10% of the input DNA.
Conditions for Galactose Induction. Yeast cells were grown at 30°C to midlog phase with 2% raffinose as the carbon source. The cells were induced by addition of galactose to 2%. For ChIP experiments, formaldehyde was added at the time points indicated. Aliquots were taken before cross-linking for Northern analysis. For the UV treatment, the strains were irradiated in dishes such that the depth of the cell suspension was <3 mm. Each strain was exposed to UV light at a dose of 40 J from a germicidal lamp at a fluence of 1 J per s per m2 with constant agitation. Formaldehyde cross-linking was done immediately after irradiation.
Conditions for Heat Shock Induction. Yeast cells were grown at 30°C to midlog phase, and the heat shock genes were induced for 15 min by the addition of an equal volume of media prewarmed to 53°C. Heat shock induction was reversed by the addition of 0.8 volumes of media at 4°C.
Conditions for Immunoprecipitation. Either the RPB3-flag strain or the untagged control were grown to midlog phase, and the cells were pelleted. Cells were washed with water, resuspended in MTB buffer (500 mM Hepes, pH 7.5/100 mM K-acetate/5 mM Mg-acetate/1 mM EGTA/0.1 mM DTT/10% glycerol/protease inhibitors) with 5 mm ATP and an ATP regenerating system (15 mM creatine phosphate and 50 μg/ml creatine phosphokinase), and extracts were made by bead beating. After measuring protein concentration, equal amounts of protein were mixed with anti-Flag agarose beads equilibrated with the extract buffer. Immunoprecipitates were carried out at 4°C for 4 h, and beads were pelleted and washed extensively with the binding buffer. The samples were run out on SDS/10% PAGE and Western blotted by using standard methods.
β-Galactosidase (β-gal) Reporter Assays. The ise1 strain (12) was transformed with pHZ18Δ2 or pL101 and maintained on Ura drop out media for selection. Cells were grown to midlog phase and induced with galactose for 4 h (in the presence or absence of 100 μM MG132). Cells were pelleted and washed with dH2O. Extracts were made by bead beating in buffer A (25 mM Tris·HCl, pH 7.5/150 mM NaCl/5 mM MgCl2/10% glycerol/protease inhibitors). Protein levels were measured by Bradford and normalized, and β-gal activity was measured as the rate of Chlorophenol red β-d-galactopyranoside hydrolysis per microgram of extract by using standard methods.
Results
The Proteasome Associates with the 3′ End of GAL1. ChIP analysis was used to determine whether the full 26S proteasome associated with the GAL1 gene at any point after induction. Fig. 1 A shows that a signal representing association of the 20S core particle with the 3′ end of the GAL1 gene (see Fig. 1B) was observed within 30 min after induction of transcription. No 20S-dependent signal was observed by using primers that amplified a site in the GAL1 promote that included the upstream activation sequences. However, the Sug1 protein, one of the proteasomal ATPases and a component of the APIS complex, is clearly present at this site 30 min postinduction. First, this argues that the lack of a signal for the 20S core at the upstream promoter site is not a result of poor cross-linking of the proteasome to the DNA or some other trivial reason and validates our previous conclusion that Gal4 specifically recruits APIS (3), not the entire 26S proteasome (13), to the activated promoter. More importantly for this study, this surprising result suggests that the entire proteasome is eventually recruited to the GAL1 gene.
The 20S-specific signal in the ChIP assay was almost completely dependent upon induction of GAL gene expression with galactose and increased in intensity in a time-dependent fashion, reaching a peak 2 h after induction (Fig. 1 A). Even at 2 h postinduction, however, no 20S-specific signal was observed on the 5′ end of the gene. As reported previously, the 3′ end of the gene also coimmunoprecipitated with anti-Sug1 antibodies. In this case, however, a strong signal was observed 30 min after induction, which increased in intensity only modestly, if at all, by 2 h postinduction (Fig. 1 A). The different kinetics and spatial distribution of Sug1 and 20S association with the DNA are consistent with the idea that the APIS complex is required early in the transcription process, presumably to drive elongation, and functions independent of the proteasome core, as proposed previously (3, 4). They also support the suggestion that the 20S core complex, presumably in the context of the intact 26S proteasome, is recruited to the 3′ end of the gene to participate in some distinct process.
To further test this result, the assay was repeated by using a yeast strain carrying a Flag epitope-tagged Pre1 protein (11), a 20S component. The results are shown in Fig. 2A Lower. In this case, a small amount of promoter DNA coimmunoprecipitated with the anti-Flag antibody. The intensity of the band increased slightly after induction. It should be noted that the levels of anti-Flag precipitable DNA at the highest point were ≈3- to 10-fold more than was precipitated by using the anti-20S or anti-Sug1 antibody, respectively (Figs. 1 A and 2B). Thus, the weak signal observed in this more efficient immunoprecipitation might represent a low level of 20S association with the promoter region that was undetectable by using the anti-20S antibodies. Alternatively, the signal might be due to a small amount of sheared DNA fragments long enough to include both the upstream activation sequence region and sequences well 3′ of the start site where the 20S complex clearly associates (vide infra), which was also undetectable with the anti-20S antibodies. In any case, this signal was much weaker than that observed on the 3′ end of the gene, which again increased 30 min after addition of galactose and reached a peak between 1 and 2 h postinduction (Fig. 2 and data not shown).
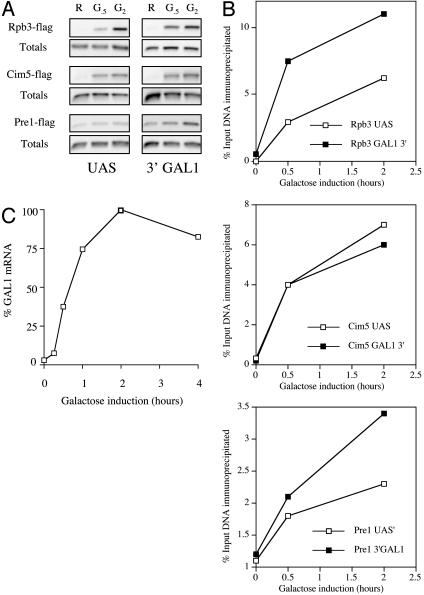
Fig. 2.
Detailed analysis of the association of Rpb3 and proteasomal proteins with GAL1 by using the ChIP assay. (A) GAL1 PCR products from a ChIP assay. Upstream activation sequence refers to the GAL1–10 promoter region and 3′ to the 3′ region of GAL1 (see Fig. 1A). The sample points are in raffinose before GAL gene induction (R), 30 min (G.5), and 2 h (G2) after galactose addition. Totals are 10% of the input DNA. (B) Graph of the ChIP results. The numbers correspond to the amount of specific DNA precipitated as a percentage of the input DNA. (C) Time course of GAL1 gene induction. GAL1 mRNA levels were normalized to ACT1 as measured by Northern analysis and plotted as a percentage of the peak level over time.
The same experiment was repeated by using a strain that expressed a Flag-tagged Cim5/Rpt1 protein, another of the proteasomal ATPases (Fig. 2 A). The results were similar to those observed by using the anti-Sug1 antibody. Flag-tagged Cim5/Rpt1 protein was found to associate with both the promoter region and the 3′ end of the GAL1 gene. The fact that the ChIP assays using a monoclonal antibody in combination with epitope-tagged proteins provided results similar to those which used antibodies raised against the native factors argues strongly that the 26S proteasome indeed associates with the 3′ end of the GAL1 gene, but weakly or not at all with the promoter region, and that the bands observed in the ChIP assays are not the result of cross-reactivity of the antibodies with some other protein.
Is the Proteasome Associated with Pol II and Involved in Termination? Why would the proteasome be present at the 3′ end of a transcriptionally active gene? One possibility is that some proteasome-mediated event might be involved in the termination of transcription. Termination is not well understood mechanistically (14, 15), and there is no evidence for or against a role for the proteasome in this process. If the proteasome were involved in termination, one would predict that the kinetics of proteasome association with the GAL1 gene would correlate with the accumulation of RNA pol II complexes in this region. To test this theory, another set of ChIP assays was conducted by using a strain expressing a Flag-tagged Rpb3 protein. As shown in Fig. 2 A (and graphed in Fig. 2B), the signal corresponding to Rpb3 association with the 3′end of GAL1 increases in intensity 30 min after galactose addition, finally peaking 2 h postinduction. pol II–promoter associations show similar kinetics.
We also examined the time course of GAL1 mRNA synthesis by Northern blot analysis. The GAL1 mRNA levels were normalized to ACT1 mRNA. As shown in Fig. 2C, the level of GAL1 transcript increased steadily over a 1-h period, reached a maximum 2 h after induction, and then settled at a somewhat reduced steady state 4 h postinduction (later time points not shown). Thus, the accumulation of the 26S proteasome and pol II at the 3′ end of the GAL1 gene correlate well with each other and with the appearance of full-length transcript.
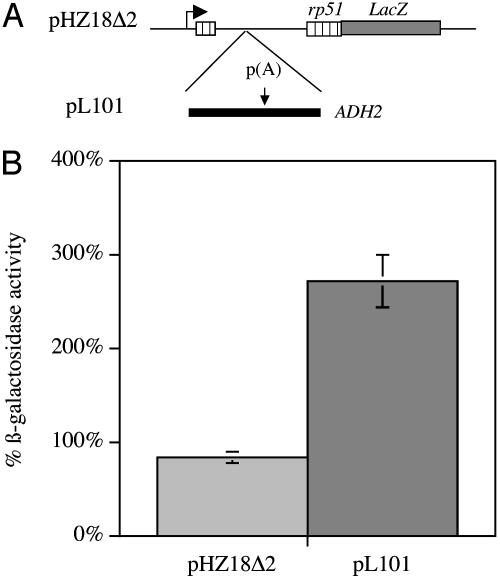
If the proteasome does play a role in termination, then one would predict that chemical inhibitors of its activity would have an effect on the efficiency of this process. To test this possibility, we used a β-gal reporter assay developed by Hyman and Moore (16). This assay uses a fusion gene controlled by the GAL1 upstream activation sequence that consists of the ribosomal protein 51 gene (5′ exon, intron and part of the 3′ exon) fused in frame to the lacZ gene. In the control construct (pHZ18Δ2) after galactose induction, the pre-mRNA is spliced and β-gal is produced. The test construct (pL101) contains the termination region of the ADH2 gene inserted into the intron, so any β-gal produced is a result of polymerase reading through that site.
The constructs were transformed into a yeast strain sensitive to proteasome inhibitors (12) and grown under appropriate selection. Three separate colonies from each transformation were isolated and grown to midlog phase in Raffinose. Induction was carried out by the addition of galactose in the presence or absence of the 26S proteasome inhibitor MG132. Extracts were made from the samples 4 h after induction and tested for β-gal activity. Fig. 3 shows that the addition of proteasome inhibitor to the strains carrying the plasmid lacking a termination site in front of lacZ (pHZ18Δ2) resulted in a decrease in β-gal activity to a level 83.3 ± 6.6% compared with the untreated sample. In contrast, addition of the proteasome inhibitor to the strains carrying the plasmid containing a termination site in front of lacZ (pL101) resulted in an increase in β-gal activity to a level 271.3 ± 28.7% compared with the untreated sample. This effect of the proteasome inhibitor on the frequency of termination site read through is consistent with the idea that the 26S plays a functional role in this process. It should be noted, however, that the effect of the proteasome inhibitor on termination efficiency is modest. Whereas pL101 normally gives a β-gal activity level of ≈1% of pHZ18Δ2, in the presence of MG132, this increases to ≈3% in the presence of the inhibitor. This result is not surprising, because termination seems to be a remarkably plastic process that is notoriously tolerant of various mutations and pharmacologic agents (14, 17).
Fig. 3.
Inhibition of the 26S proteasome results in increased read through of a termination site. (A) Schematic representation of the reporter constructs. (B) Effect of MG132 on β-gal activity from the reporter constructs. Results of three independent experiments in which strains were induced by galactose for 4 h in the presence or absence of 100 μM MG132, and extracts were obtained. Results are shown as the activity in the untreated sample over the activity of the treated sample.
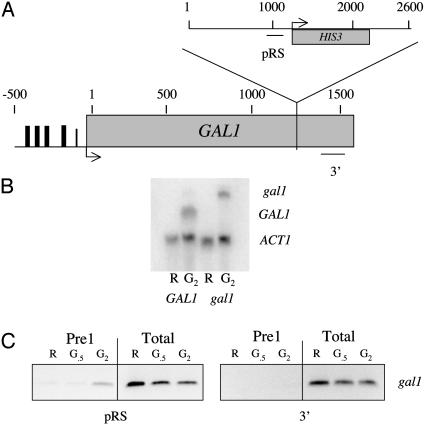
Altering the Site of Transcription Termination Results in a Corresponding Alteration in the Site of Proteasome Association. The data presented above argue that the proteasome is enriched on the 3′ end of several transcriptionally active genes and plays a functional role in termination. If so, then if the site of termination were altered, one would predict that there would be a corresponding alteration in the association of the proteasome with the gene. To test this hypothesis, a gal1::HIS3 strain was constructed by inserting a 2.6-kbp fragment into the coding region of GAL1 (Fig. 4A). Northern analysis showed that this insertion resulted in an altered GAL1 transcript that was ≈800 base pairs longer than that observed in the wild-type strain (Fig. 4B). Thus, termination occurred within the insert, and the polymerase did not reach the native termination site of GAL1 on the other end of the insert. ChIP assays were conducted by using a PCR primer pair that amplified the original 3′ termination site of the GAL1 gene and another pair that amplified an area within the inserted region. The mutant gal1::HIS3 strain showed no transcription-dependent accumulation of Pre1 at the original 3′ end of GAL1 (Fig. 4C). In contrast, a transcription-dependent accumulation of Pre1 was observed within the inserted DNA in the region which Northern analysis suggests transcription occurs (Fig. 4C). These data show clearly that by moving the termination site, the location of 26S proteasome association with the DNA is also altered accordingly.
Fig. 4.
Altering the site of termination alters the association of the proteasome with the DNA. (A) Map showing the site of insertion within the GAL1 coding region and location of the primers used in the assay. (B) Northern blot showing the shift in mobility of the GAL1 transcript in the insertion mutant. Sample points are raffinose (R) and 2 h after addition of galactose (G2). Blot was probed with both ACT1 and GAL1 fragments. (C) Association of proteasomal proteins with mutant gal1 strain by using the ChIP assay. Amplified regions are as indicated. The sample points are in raffinose before GAL gene induction (R), 30 min (G.5), and 2 h (G2) after galactose addition. Totals are 10% of the input DNA.
The Proteasome Is Observed at the 3′ End of Other Inducible Genes. To begin to address the generality of this observation, ChIP assays were conducted by using PCR primers that amplify the 3′ ends of other inducible genes. GAL1 and GAL10 are positioned next to each other and share the same promoter but have opposite orientations. ChIP assays were carried out in the Flag-tagged Pre1 strain to assess whether the 26S proteasome could be detected on the 3′ end of GAL10 after induction. The results of the PCR show a similar time- and transcription-dependent occupancy by the 26S proteasome on the 3′ region of GAL10 to that observed on the 3′ region of GAL1 (Fig. 5A).
To extend this analysis beyond the GAL genes, we examined HSP82, a heat shock gene. Transcription of HSP82 can be induced rapidly by the addition of prewarmed media and then silenced through the addition of chilled media (18). Cells expressing either Flag-tagged Cim5/Rpt1 or Pre1 were grown to midlog phase and treated with formaldehyde before heat shock (uninduced), 15 min after raising the temperature to 39°C (induced), or 20 min after reversing the heat shock by returning the temperature to 30°C (reversed). As shown in Fig. 5B, little or no DNA from the 3′ end of HSP82 was precipitated by the anti-Flag antibody in either strain in the absence of induction or when the cells were heat-shocked and were then returned to normal temperature before cross-linking. However, a strong signal was observed when cross-linking was carried out at the elevated temperature. No signal was observed on the GAL1/10 promoter under these conditions, nor was a signal observed when formaldehyde was added at the same time as the warmed media (data not shown). The data obtained from the GAL10 and HSP82 genes argue that transcription-dependent association of the 26S proteasome with the 3′ end of the gene is not restricted to GAL1.
Colocalization of Pol II and the Proteasome at Sites Outside Termination Regions. The data shown above reveal an association of the proteasome with the 3′ end of several transcriptionally active genes and also argue that the complex plays a functional role in termination. However, are proteasome–gene associations restricted entirely to the 3′ end of active genes? In the process of conducting the ChIP experiments shown in Fig. 4, we detected clear association of the Flag-tagged Pre1 protein with a region of the altered GAL1 gene that was well separated from the alternative termination site (data not shown). This result, combined with the well known fact that transcriptional elongation occurs in fits and starts and that there are many pause sites in any given gene (19), led us to consider a more general idea. This hypothesis is that the proteasome associates with genes in several places that correspond to sites of pol II buildup, of which termination sites are simply a special case (20, 21). If true, then one should observe strongly enhanced association of the 20S core particle with a gene locus when a novel RNA pol II pause site is inserted at that site.
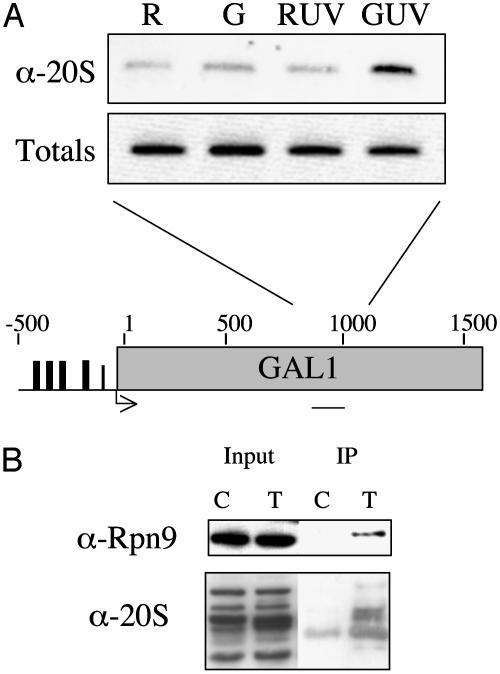
To test this idea, we examined the effect of UV irradiation on the association of the 20S core particle with the middle region of the GAL1 gene (Fig. 6A). UV light-induced lesions are known to act as severe blocks to transcriptional elongation (22, 23). When the cells were grown in raffinose (noninducing conditions), only the background level of signal (see also Fig. 1) was observed in the ChIP assay (Fig. 6A, first lane) by using anti-20S antibodies. The same result was obtained when the uninduced cells were UV-irradiated before cross-linking (third lane). Induction of GAL1 transcription before cross-linking resulted in a slight signal above background in nonirradiated cells (second lane). This signal was clearly real, as determined by similar experiments by using the more efficient anti-FLAG antibody and Flag-Pre1 strains (not shown), arguing for a low level of 20S association with this region of GAL1 under normal conditions. However, when the induced cells were UV-irradiated before cross-linking, a pronounced increase in the coimmunoprecipitated DNA was observed (Fig. 6A, last lane). This was not the case when uninduced cells were irradiated. Thus, proteasome recruitment depends on both DNA damage and active transcription, consistent with the model that association of the 26S complex with the gene is in response to polymerase stalling. The observation of UV-induced proteasome recruitment is consistent with some previous observations, as it has been shown that blocking elongation by creating lesions in the template or by the addition of α-amanitin leads to polyubiquitylation and subsequent 26S proteasome-dependent degradation of RNA pol II (23). However, the data shown in Fig. 6A indicate that these events may be initiated at the site of the stalled complex and suggest a specific model that proteasome action acts to clear the blocked complex from the template.
Fig. 6.
The proteasome interacts with RNA pol II and associates with transcriptionally active GAL1 after UV irradiation. (A) GAL1 PCR products representing the middle region of the gene from a ChIP assay by using anti-20S antibodies. The sample points are in raffinose (R), raffinose after UV treatment (RUV), 30 min after galactose induction (G), or 30 min after galactose induction and UV treatment (GUV). Totals are 1% of the input DNA. A map of the GAL1 gene shows the region amplified by PCR. (B) Western analysis of immunoprecipitated Rpb3 samples. Extracts from a strain containing a Flag-tagged Rpb3 (T) or an untagged control (C) were immunoprecipitated with anti-Flag antibody and probed for 26S subunits as indicated. Input represents 1% of the total protein added.
The 26S Proteasome and RNA Pol II Associate Physically. All of the data reported above were obtained by using ChIP assays, which report both direct and indirect interactions of protein with the DNA. Therefore, it was of interest to test more directly for a pol II–proteasome association, so a coimmunoprecipitation experiment was carried out. By using the Rpb3-Flag-carrying strain or an untagged strain as a negative control, we immunoprecipitated the RNA pol II complex with anti-Flag antibodies. A Western blot using anti-20S antibodies showed that the 20S complex was highly enriched in the immunoprecipitated material by using extract from the Rpb3-Flag strain, but not from the untagged strain. These data are consistent with the idea that some fraction of the 26S and RNA pol II are physically associated in the cell.
Discussion
Why Is the Proteasome Recruited to Stalled Polymerase Complexes? The data presented above show that the proteasome is recruited to sites in genes that correspond to regions in which RNA pol II complexes build up, including 3′ regions proximal to termination sites, sites of UV damage, and certain other sequences that may represent elongation pause sites. We suggest that these observations reflect association of the 26S proteasome with the transcription machinery in response to sequences or structures in the template that result in pronounced stalling. These observations further suggest the possibility that events that occur at stalled complexes in front of DNA lesions may be mechanistically similar to those that occur when the elongation becomes stalled in termination regions. There is evidence that the same proteins involved in resolving RNA pol II at sites of damage are also involved in transcriptional elongation. The Saccharomyces cerevisiae protein Def1 forms a complex with Rad26 (the CSB homolog in yeast) and is involved in the ubiquitylation of RNA pol II after UV irradiation (24). def1 deletion strains are also sensitive to the transcription elongation inhibitor 6-azauracil (24). Additionally, a genetic interaction has been established with ESS1, which encodes a prolyl-isomerase required for RNA pol II termination and the E3 ubiquitin ligase gene RSP5 (25). This interaction seems to occur through Rpb1 (25).
The Yin and Yang of Transcriptional Elongation. The mechanism by which the proteasome may act on stalled polymerase complexes, whether at sites of DNA damage or in termination regions, is unclear. Obviously, one possibility is that polymerase and/or some associated proteins are degraded, resulting in release of the blocked complex. Our lab has previously shown that 19S subunits of the proteasome are critical for efficient elongation by RNA pol II in a manner independent of the 20S and proteolysis (4, 5). Here, we show that inhibition of the 26S proteasome results in the increased read through of a transcriptional termination site. In other words, inhibition of the 19S decreases elongation whereas inhibition of the 20S decreases termination. This result suggests a balance of the two activities is important for efficient transcription. One possible model is that the chaperone-like activity of the 19S would remodel a stalled polymerase complex resulting in increased elongation, where as a terminally paused polymerase would require the more drastic function of the whole 26S proteasome and presumably degradation. During the normal course of transcription, this degradation pathway would account for only a small percentage of pausing events. However events such as DNA damage, which increase the number of terminally stalled polymerases, would then result in a subsequent increase in stalled complexes being resolved by this pathway.
In striking contrast to the coincidence of pol II and the 20S core particle on the 3′ end and certain internal regions of the genes studies here, little or no 20S was observed on the promoter of GAL1. This is consistent with the idea that the full proteasome associates with elongating polymerase complexes, either constitutively or specifically at strong pause sites, but that it is not part of the preinitiation complex. The proteasomal ATPases, such as Sug1, Sug2, and Cim5, are clearly present at the promoter, however, which we have proposed is a reflection of Gal4-mediated recruitment of the APIS complex (3). A recent report by Morris et al. (13) has shown that components of the 26S proteasome are found at the CDC20 promoter. In their hands, hemagglutinin-tagged Pre1 could also be found associated with the GAL1 promoter, in contrast to our results (Figs. 1 and 2). However, we do observe the 26S proteasome in the coding region of actively transcribing genes. One possible explanation for this discrepancy could be different degrees of shearing during the chromatin preparation. In our hands, the average size of DNA fragments is ≈400–500 base pairs after sonication. Given the high expression levels of GAL1 (and high levels of Pre1 associated with the gene outside of the promoter), incomplete shearing could account for the results observed by those authors (i.e., amplification of a minor component of long DNA by using 5′-directed primers). Consistent with that observation is that the promoter regions that showed no Pre1 association in the work of Morris et al. (CLB2, CLN1, and CLN2) are not proximal to the 3′ end of any genes. With that in mind, it is important to note that only the 19S mutant strain sug1–25 and not the 20S double-mutant strain pre1-1 pre4-1 showed any defect in CDC20 transcriptional activation, suggesting a nonproteolytic role for the proteasome in this process (13).
Conclusion
In summary, we have shown that the 26S proteasome associates with transcriptionally active genes at sites of DNA damage and the 3′ ends of active genes. Furthermore, we show that the 26S proteasome and RNA pol II can interact physically and functionally. These results suggest that the proteasome plays important roles in transcriptional termination and the clearance of dead complexes blocked by DNA damage and suggests that these processes may be related mechanistically. These observations have revealed yet another intimate connection between the proteasome and RNA pol II transcription and should stimulate other avenues of investigation in this area.
Acknowledgments
We thank Frank Amini for providing the yeast strain that produces Flag-tagged Rpb3; Claire Moore for the plasmids used in the Fig. 3 experiment; and John Nitiss for the ise1 yeast strain. This work was supported by National Institutes of Health Grant GM-066380 and by unrestricted funds to the Center for Biomedical Inventions.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ChIP, chromatin immunoprecipitation; pol II, polymerase II; β-gal, β-galactosidase.
References
- 1.Glickman, M. H., Rubin, D. M., Fried, V. A. & Finley, D. (1998) Mol. Cell. Biol. 18, 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumeister, W., Walz, J., Zuhl, F. & Seemuller, E. (1998) Cell 92, 367–380. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez, F., Delahodde, A., Kodadek, T. & Johnston, S. A. (2002) Science 296, 548–550. [DOI] [PubMed] [Google Scholar]
- 4.Ferdous, A., Gonzalez, F., Sun, L., Kodadek, T. & Johnston, S. A. (2001) Mol. Cell 7, 981–991. [DOI] [PubMed] [Google Scholar]
- 5.Ferdous, A., Kodadek, T. & Johnston, S. A. (2002) Biochemistry 41, 12798–12805. [DOI] [PubMed] [Google Scholar]
- 6.Molinari, E., Gilman, M. & Natesan, S. (1999) EMBO J. 18, 6439–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salghetti, S. E., Kim, S. Y. & Tansey, W. P. (1999) EMBO J. 18, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salghetti, S. E., Muratani, M., Wijnen, H., Futcher, B. & Tansey, W. P. (2000) Proc. Natl. Acad. Sci. USA 97, 3118–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salghetti, S. E., Caudy, A. A., Chenoweth, J. G. & Tansey, W. P. (2001) Science 293, 1651–1653. [DOI] [PubMed] [Google Scholar]
- 10.Reid, G., Hübner, M. R., Metivier, R., Brand, H., Denger, S., Manu, D., Beaudouin, J., Ellenberg, J. & Gannon, F. (2003) Mol. Cell 11, 695–707. [DOI] [PubMed] [Google Scholar]
- 11.Verma, R., Chen, S., Feldman, R., Schieltz, D., Yates, J., Dohmen, J. & Deshaies, R. J. (2000) Mol. Biol. Cell 11, 3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nitiss, J. & Wang, J. C. (1988) Proc. Natl. Acad. Sci. USA 85, 7501–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris, M. C., Kaiser, P., Rudyak, S., Baskerville, C., Watson, M. H. & Reed, S. I. (2003) Nature 423, 1009–1013. [DOI] [PubMed] [Google Scholar]
- 14.Zhao, J., Hyman, L. & Moore, C. (1999) Microbiol. Mol. Biol. Rev. 63, 404–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Proudfoot, N. J., Furger, A. & Dye, M. J. (2002) Cell 108, 501–512. [DOI] [PubMed] [Google Scholar]
- 16.Hyman, L. E. & Moore, C. (1993) Mol. Cell. Biol. 13, 5159–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Proudfoot, N. J. & O'Sullivan, J. (2002) Curr. Biol. 12, R855–R857. [DOI] [PubMed] [Google Scholar]
- 18.Borkovich, K. A., Farrelly, F. W., Finkelstein, D. B., Taulien, J. & Lindquist, S. (1989) Mol. Cell. Biol. 9, 3919–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uptain, S. M., Kane, C. M. & Chamberlin, M. J. (1997) Annu. Rev. Biochem. 66, 117–172. [DOI] [PubMed] [Google Scholar]
- 20.Yonaha, M. & Proudfoot, N. J. (1999) Mol. Cell 3, 593–600. [DOI] [PubMed] [Google Scholar]
- 21.Enriquez-Harris, P., Levitt, N., Briggs, D. & Proudfoot, N. J. (1991) EMBO J. 10, 1833–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratner, J. N., Balasubramanian, B., Corden, J., Warren, S. L. & Bregman, D. B. (1998) J. Biol. Chem. 273, 5184–5189. [DOI] [PubMed] [Google Scholar]
- 23.Lee, K. B., Wang, D., Lippard, S. J. & Sharp, P. A. (2002) Proc. Natl. Acad. Sci. USA 99, 4239–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woudstra, E. C., Gilbert, C., Fellows, J., Jansen, L., Brouwer, J., Erdjument-Bromage, H., Tempst, P. & Svejstrup, J. Q. (2002) Nature 415, 929–933. [DOI] [PubMed] [Google Scholar]
- 25.Wu, X., Chang, A., Sudol, M. & Hanes, S. D. (2001) Curr. Genet. 40, 234–242. [DOI] [PubMed] [Google Scholar]