Abstract
Background:
Nonfermenting gram-negative bacilli have emerged as important healthcare-associated pathogens. It is important to correctly identify all clinically significant nonfermenting gram-negative bacilli considering the intrinsic multidrug resistance exhibited by these bacteria.
Materials and Methods:
A retrospective study was undertaken to identify the various nonfermenting gram-negative bacilli other than Pseudomonas aeruginosa and Acinetobacter spp. isolated from respiratory samples (n = 9363), to understand their clinical relevance and to analyze their antibiotic susceptibility pattern.
Results:
Nonfermenting gram-negative bacilli were isolated from 830 (16.4%) samples showing significant growth. Thirty-three (4%) isolates constituted nonfermenting gram-negative bacilli other than P. aeruginosa and Acinetobacter spp. Stenotrophomonas maltophilia (15, 45.5%) was the most common isolate followed by Burkholderia cepacia (4, 12.1%), Sphingomonas paucimobilis (3, 9.1%), and Achromobacter xylosoxidans (3, 9.1%). On the basis of clinicomicrobiological correlation, pathogenicity was observed in 69.7% (n = 23) isolates. Timely and correct treatment resulted in clinical improvement in 87.9% cases.
Conclusion:
Any nonfermenting gram-negative bacilli isolated from respiratory tract infection should not be ignored as mere contaminant, but correlated clinically for its pathogenic potential and identified using standard methods so as to institute appropriate and timely antibiotic coverage.
Keywords: Intrinsic resistance, Nonfermenting gram-negative bacilli, S. maltophilia
INTRODUCTION
Injudicious and empirical use of antibiotics has enabled non-fermenting gram-negative bacilli (NFGNB) emerge as important healthcare-associated pathogens.[1] These organisms are ubiquitous in nature particularly in soil and water. In the hospital environment, they may be isolated from instruments such as ventilator machine humidifiers, mattresses, and other equipments as well as from the skin of healthcare workers.[2] All these organisms have the potential to spread horizontally on fomites or the hands of healthcare workers.[1,2,3] The identification of these non-fermenters is important because of the fact that most of them show multidrug-resistant pattern and inherent resistance to many antibiotics.[3] Majority of the earlier studies have only focused on the identification of Pseudomonas spp. and Acinetobacter spp. because of their higher isolation rates. However, in immunocompromised patients especially, NFGNB other than Pseudomonas spp. and Acinetobacter spp. can also cause the disease. So, it becomes imperative to know about the occurrence and susceptibility pattern of these non-fermenters.
This study was undertaken to identify the various NFGNB other than P. aeruginosa and Acinetobacter spp. isolated from the respiratory samples, to understand their clinical relevance, and to analyze their antibiotic susceptibility pattern.
MATERIALS AND METHODS
A retrospective study was conducted in the Clinical Microbiology Laboratory of a tertiary care center in Coastal Karnataka from December 2009 to November 2011. All the respiratory samples (sputum or bronchoalveolar lavage fluid) received during this time period were included. Sputum gram stains were read at ×100 magnification and evaluated according to the Bartlett criteria.[4] Specimens were scored 0, +1, or +2 according to the number of leukocytes seen per field and 0, –1, and –2 according to the number of squamous epithelial cells seen per field. Specimens with total scores of 0 or less were considered inadequate and heavily contaminated with oropharyngeal flora. Those containing greater than 25 leucocytes and fewer than 10 squamous epithelial cells per field were optimal specimens and further processed for culture.[4] All samples were inoculated on sheep blood agar, MacConkey agar and chocolate agar, and incubated at 37°C for 18-24 hours. Bronchoalveolar lavage fluid was processed by quantitative culture with positive threshold of 104 CFU/mL.[5] The NFGNB other than P. aeruginosa and Acinetobacter spp. isolated as the predominant organism from sputum (at least two samples per patient) or ≥104 CFU/mL of bronchoalveolar lavage fluid and correlating with microscopic findings of the sample were considered significant and included in the study. The organisms isolated were subjected to routine biochemical reactions like oxidase test, growth on Triple Sugar Iron agar, mannitol motility test medium, indole production, hydrogen sulfide production, urea hydrolysis, and citrate utilization. The non-fermenters which could not be identified on the basis of above-mentioned biochemical reactions were subjected to Vitek 2 system for identification. The susceptibility testing was performed using Kirby-Bauer disc diffusion method using commercially available discs according to Clinical Laboratory Standards Institute guidelines.[6]
Details of patients such as the demographic parameters, clinical presentation, radiological findings, location of stay in the hospital, duration of hospital stay, and prognostic outcomes were noted in a structured proforma retrospectively from the medical record department. The clinical significance of the NFGNB isolates was assessed by correlating with clinical and radiological findings of the patient. Culture isolates obtained as scanty growth or mixed growth with more than two types of organisms from sputum samples; or <104 CFU/mL from bronchoalveolar lavages and not correlating with gram stain findings and clinical criterion of nosocomial pneumonia were considered as colonizers and not included in the analysis. Descriptive analysis of results was performed using statistical software, SPSS version 16.0. Frequencies and percentages were derived for categorical data. For continuous data, mean was obtained.
Nosocomial pneumonia was defined as a new or progressive and persistent infiltrate or consolidation or cavitation on two serial chest radiographs that occurred at least 48 h after hospital admission in the presence of at least one of the following clinical signs: Fever (temperature >38°C) with no other recognized cause, leukopenia (<4.0 × 109 cells/L) or leukocytosis (>12.0 × 109 cells/L), altered mental status with no other recognized cause and any two of the following: New onset of purulent sputum, change in character of sputum, increased respiratory secretions, or increased suctioning requirements, new-onset or worsening cough, or dyspnea, or tachypnea, rales or bronchial breath sounds, worsening gas exchange (e.g., oxygen desaturation ratio [PaO2-FiO2] ≤240, increased oxygen requirement, or increased ventilation demand).[7]
RESULTS
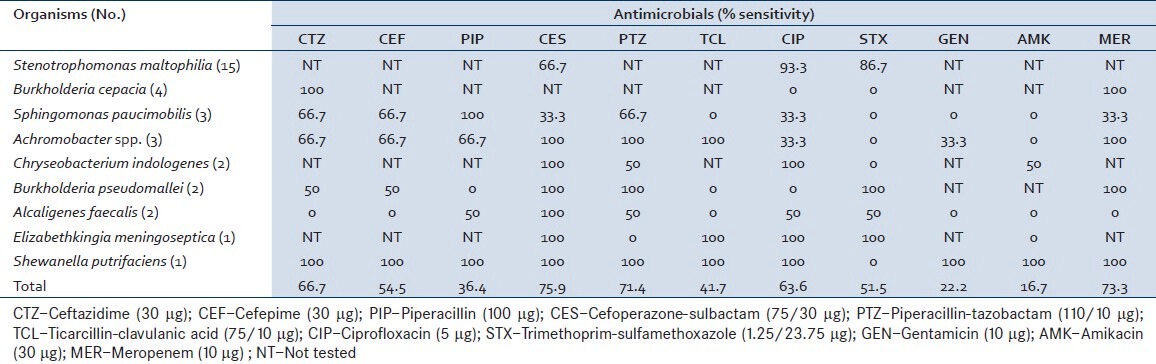
A total of 9363 respiratory specimens were received during the study period out of which 5056 (54%) cultures yielded a significant growth and 4307 (46%) cultures had growth of normal oropharyngeal flora. NFGNB were isolated in 830 (16.4%) out of 5056 sputum cultures. Out of 830 isolated NFGNB, 471 (56.7%) were P. aeruginosa, 326 (39.3%) were Acinetobacter spp., and 33 (4%) constituted others. The distribution of these nonfermenters is shown in Table 1. Stenotrophomonas maltophilia (15, 45.5%) was the most common among other NFGNB followed by Burkholderia cepacia (4, 12.1%) and others. Demographic parameters and various clinical presentations of the patients are shown in Table 2. On the basis of clinicomicrobiological correlation, among the 33 isolates, 69.7% (n = 23) were pathogens and 30.3% (n = 10) were colonizers. The pathogens were responsible for hospital-acquired pneumonia in 24.24% (n = 8) patients including S. maltophilia (n = 6), B. cepacia (n = 1) and Elizabethkingia meningoseptica (n = 1). Susceptibility pattern of the isolates to various antimicrobial agents is shown in Table 3.
Table 1.
Nonfermenting gram-negative bacilli other than Pseudomonas aeruginosa and Acinetobacter spp. isolated from clinical specimens

Table 2.
Demographic and clinical details of the patients

Table 3.
Susceptibility profile of non-fermenters to various antimicrobials
DISCUSSION
The current study has shown the isolation of NFGNB in 16.4% of respiratory samples. The importance of isolation of non-fermenters has increased in last decade, after more and more reports are correlating them with the either infection outbreaks in hospitals, or healthcare-associated infections.[1] Earlier the identification of non-fermenters, based on biochemical tests, was cumbersome and many non-fermenters were misidentified. But, now with the availability of commercial systems like Vitek-2 or API, the identification has become easier.[8] Malini et al.,[9] from Kolar in India have documented the isolation of 6.8% (25 of 365) of NFGNB in respiratory samples.
S. maltophilia is considered now as a common non-fermenter to cause infection in hospital settings.[10,11] Correct identification of this NFGNB assumes importance as it shows inherent resistance to commonly used broad spectrum beta-lactam group antibiotics and even to imipenem.[12] Our study has shown the isolation of this bacterium in 45.5% cases. Earlier A’Court and Garrard[13] have reported S. maltophilia to account for 5% of nosocomial pneumonias. These nosocomial pneumonia are frequently associated with mechanical ventilation, tracheostomy, previous exposure to broad-spectrum antibiotics, the use of respiratory tract equipment such as nebulizers[14,15,16,17,18,19,20,21] and therapy with aerosolized polymyxin.[22] Majority of our isolates were sensitive to ciprofloxacin (93.3%) followed by trimethoprim-sulfamethoxazole (86.7%). Malini et al.,[9] have documented 100% sensitivity to ciprofloxacin and trimethoprim-sulfamethoxazole. This bacterium produces an unusual chromosomally encoded zinc-depended β-lactamase that confers broad resistance to carbapenems and other β-lactames.[23]
B. cepacia complex (BCC) is another NFGNB colonizing and infecting patients with chronic respiratory illness. It is known to cause disease in cystic fibrosis (CF) patients and once infected, it is very difficult to eradicate.[24] In our study BCC was isolated in 12.1% of cases, though its association with CF was not studied. Rahbar et al.,[25] have shown the isolation of BCC as 4.66% of all the nonfermenters isolated from different type of specimens (respiratory, blood, urine, wound, etc.). BCC is known for its intrinsic resistance to many beta-lactam drugs, aminoglycosides, colistin and polymixin B, the first-line therapeutics of choice against serious pseudomonal infections.[24] Our strains have demonstrated 100% sensitivity to ceftazidime and meropenem.
Achromobacter xylosoxidans is an aerobic, gram-negative bacillus found in a variety of aquatic environments and has proved to survive on inanimate surfaces in hospital settings, connected to its role as a nosocomial colonizer. It is generally considered an opportunistic pathogen and has attracted attention as an emerging pathogen in CF.[26] Reported prevalence rates of A. xylosoxidans have increased in recent years, although this may in part result from growing attention or improved microbiologic techniques. Our study has observed the isolation of A. xylosoxidans in 9.1% cases. These isolates have shown 100% sensitivity to beta-lactam-beta-lactamase inhibitor combination antimicrobials.
B. pseudomallei the causative agent of melioidosis, is an emerging NFGNB that causes lung infections. It is responsible for forming abscesses in multiple organs besides lungs like kidney, skin, musculoskeletal tissue, heart, etc. One of the nasty things about this bacterium is that the incubation period can be anywhere from a couple of days to couple of years. The other interesting fact about this pathogen is that it demands prolonged treatment first in intensive phase, followed by prolonged continuation phase.[27] The current study observed its isolation in 6.1% cases. Both the patients presented with cough and expectoration of 1-2 weeks duration. History of alcohol intake was present in both cases for last 15 years. One of them was diabetic for last 5 years and his blood culture also grew same pathogen. The patient was on piperacillin-tazobactam for 7 days. No improvement was observed in this patient and he expired. The other patient was empirically started on amoxicillin-clavulanic acid and amikacin but after culture report was changed to ceftazidime and trimethoprim-sulphamethoxazole and he showed improvement.
Differentiation between colonization and infection by these pathogens is of utmost importance. Otherwise, unnecessary institution of antibiotics will contribute to further increase in resistance. The present study observed that 69.7% of these NFGNB were pathogens, whereas 30.3% were colonizers. Most of the patients were having hospital stay for more than 9-10 days. It supports the fact that these patients may have acquired these pathogens in hospital settings from one source of other. But tracing of source giving rise to these infections was not observed in the present study.
Most of these NFGNB were sensitive to cefoperazone-sulbactam (75.9%), meropenem (73.3%), and piperacillin-tazobactam (71.4%) [Table 2]. The aminoglycosides that are considered as good option for life-threatening lower respiratory infections have shown high resistance in the present study for these non-fermenters wherever tested. It may be due to poor penetration of aminoglycosides from blood into infected respiratory tissues so as to reach the local drug concentration above the minimum inhibitory concentration necessary for the infecting organisms. This observation has also been discussed by earlier studies.[28] All these non-fermenters are known for their inherent resistance to multiple groups of antibiotics. Hence, correct identification of these non-fermenters is very important for choosing correct antibiotic so as to reduce the morbidity and mortality. In the present study, 29 (87.9%) cases showed improvement after institution of correct treatment, whereas 3 (9.1%) expired. The mortality may be due to underlying illness contributing to their immuno-compromised state or late institution of treatment.
Any NFGNB culture isolate from respiratory tract infection should not be ignored as just contaminant but correlated clinically for its pathogenic potential and identified using standard methods, so as to institute appropriate and timely antibiotic coverage.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.McGowan JE., Jr Resistance in nonfermenting gram-negative bacteria: Multidrug resistance to the maximum. Am J Med. 2006;119(6 Suppl 1):S29–36. doi: 10.1016/j.amjmed.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 2.Mellmann A, Bimet F, Bizet C, Borovskaya AD, Drake RR, Eigner U, et al. High interlaboratory reproducibility of matrix-assisted laser desorption ionization-time of flight mass spectrometry-based species identification of nonfermenting bacteria. J Clin Microbiol. 2009;47:3732–4. doi: 10.1128/JCM.00921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fass RJ, Barnishan J, Solomon MC, Ayers LW. In vitro activities of quinolones, beta-lactams, tobramycin, and trimethoprim-sulfamethoxazole against nonfermentative gram-negative bacilli. Antimicrob Agents Chemother. 1996;40:1412–8. doi: 10.1128/aac.40.6.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartlett RG. Medical microbiology: Quality, costs and clinical relevance. New York: John Wiley and Sons, Inc; 1974. pp. 24–31. [Google Scholar]
- 5.Chastre J, Combes A, Luyt CE. The invasive (quantitative) diagnosis of ventilator-associated Pneumonia. Respir Care. 2005;50:797–807. [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. Twentieth informational supplement M100-S20. Wayne: Clinical and Laboratory Standards Institute; 2010. Performance standards for antimicrobial susceptibility testings. [Google Scholar]
- 7.Horan T, Gaynes R. Surveillance of noscomial infections. In: Mayhall C, Gaynes R, editors. Hospital Epidemiology and Infection Control. 3rd ed. Philadelphia: Lippincott Williams and Wilkins; 2004. pp. 1659–702. [Google Scholar]
- 8.Funke G, Funke-Kissling P. Evaluation of the new VITEK 2 card for identification of clinically relevant gram-negative rods. J Clin Microbiol. 2004;42:4067–71. doi: 10.1128/JCM.42.9.4067-4071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malini A, Deepa E, Gokul B, Prasad S. Nonfermenting gram-negative bacilli infections in a tertiary care hospital in Kolar, Karnataka. J Lab Physicians. 2009;1:62–6. doi: 10.4103/0974-2727.59701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang TN, Wang FD, Wang LS, Liu CY, Liu IM. Xanthomonas maltophilia bacteremia: An analysis of 32 cases. J Formos Med Assoc. 1992;91:1170–6. [PubMed] [Google Scholar]
- 11.Verweij PE, Meis JF, Christmann V, Van der Bor M, Melchers WJ, Hilderink BG, et al. Nosocomial outbreak of colonization and infection with Stenotrophomonas maltophilia in preterm infants associated with contaminated tap water. Epidemiol Infect. 1998;120:251–6. doi: 10.1017/s0950268898008735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbolla R, Catalano M, Orman BE, Famiglietti A, Vay C, Smayevsky J, et al. Class 1 integrons increase trimethoprim-sulfamethoxazole MICs against epidemiologically unrelated Stenotrophomonas maltophilia isolates. Antimicrob Agents Chemother. 2004;48:666–9. doi: 10.1128/AAC.48.2.666-669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A’Court C, Garrard CS. Nosocomial pneumonia in the intensive care unit: Mechanisms and significance. Thorax. 1992;47:465–73. doi: 10.1136/thx.47.6.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elting LS, Khardori N, Bodey GP, Fainstein V. Nosocomial infection caused by Xanthomonas maltophilia: A case-control study of predisposing factors. Infect Control Hosp Epidemiol. 1990;11:134–8. doi: 10.1086/646136. [DOI] [PubMed] [Google Scholar]
- 15.Khardori N, Elting L, Wong E, Schable B, Bodey GP. Nosocomial infections due to Xanthomonas maltophilia (Pseudomonas maltophilia) in patients with cancer. Rev Infect Dis. 1990;12:997–1003. doi: 10.1093/clinids/12.6.997. [DOI] [PubMed] [Google Scholar]
- 16.Kollef MH, Silver P, Murphy DM, Trovillion E. The effect of late-onset ventilator-associated pneumonia in determining patient mortality. Chest. 1995;108:1655–62. doi: 10.1378/chest.108.6.1655. [DOI] [PubMed] [Google Scholar]
- 17.Laing FP, Ramotar K, Read RR, Alfieri N, Kureishi A, Henderson EA, et al. Molecular epidemiology of Xanthomonas maltophilia colonization and infection in the hospital environment. J Clin Microbiol. 1995;33:513–8. doi: 10.1128/jcm.33.3.513-518.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison AJ, Jr, Hoffmann KK, Wenzel RP. Associated mortality and clinical characteristics of nosocomial Pseudomonas maltophilia in a university hospital. J Clin Microbiol. 1986;24:52–5. doi: 10.1128/jcm.24.1.52-55.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orr K, Gould FK, Sisson PR, Lightfoot NF, Freeman R, Burdess D. Rapid inter-strain comparison by pyrolysis mass spectrometry in nosocomial infection with Xanthomonas maltophilia. J Hosp Infect. 1991;17:187–95. doi: 10.1016/0195-6701(91)90230-6. [DOI] [PubMed] [Google Scholar]
- 20.Villarino ME, Stevens LE, Schable B, Mayers G, Miller JM, Burke JP, et al. Risk factors for epidemic Xanthomonas maltophilia infection/colonization in intensive care unit patients. Infect Control Hosp Epidemiol. 1992;13:201–6. doi: 10.1086/646510. [DOI] [PubMed] [Google Scholar]
- 21.Zuravleff JJ, Yu VL. Infections caused by Pseudomonas maltophilia with emphasis on bacteremia: Case reports and a review of the literature. Rev Infect Dis. 1982;4:1236–46. doi: 10.1093/clinids/4.6.1236. [DOI] [PubMed] [Google Scholar]
- 22.Fass RJ, Prior RB. Comparative in vitro activities of piperacillin-tazobactam and ticarcillin-clavulanate. Antimicrob Agents Chemother. 1989;33:1268–74. doi: 10.1128/aac.33.8.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paton R, Miles RS, Amyes SG. Biochemical properties of inducible beta-lactamases produced from Xanthomonas maltophilia. Antimicrob Agents Chemother. 1994;38:2143–9. doi: 10.1128/aac.38.9.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Govan JR, Brown AR, Jones AM. Evolving epidemiology of Pseudomonas aeruginosa and the Burkholderia cepacia complex in cystic fibrosis lung infection. Future Microbiol. 2007;2:153–64. doi: 10.2217/17460913.2.2.153. [DOI] [PubMed] [Google Scholar]
- 25.Rahbar M, Mehragan H, Akbari NH. Prevalence of drug resistance in nonfermenter gram-negative bacilli. Iran J Pathol. 2010;5:90–6. [Google Scholar]
- 26.Amoureux L, Bador J, Siebor E, Taillefumier N, Fanton A, Neuwirth C. Epidemiology and resistance of Achromobacter xylosoxidans from cystic fibrosis patients in Dijon, Burgundy: First French data. J Cyst Fibros. 2012 doi: 10.1016/j.jcf.2012.08.005. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 27.Cheng AC, Currie BJ. Melioidosis: Epidemiology, pathophysiology, and management. Clin Microbiol Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navaneeth BV, Belwadi MR. Antibiotic resistance among gram-negative bacteria of lower respiratory tract secretions in hospitalized patients. Indian J Chest Dis Allied Sci. 2002;44:173–6. [PubMed] [Google Scholar]


