Abstract
Many secreted eukaryotic glycoproteins that play fundamental roles in development, hearing, immunity, and cancer polymerize into filaments and extracellular matrices through zona pellucida (ZP) domains. ZP domain proteins are synthesized as precursors containing C-terminal propeptides that are cleaved at conserved sites. However, the consequences of this processing and the mechanism by which nascent proteins assemble are unclear. By microinjection of mutated DNA constructs into growing oocytes and mammalian cell transfection, we have identified a conserved duplicated motif [EHP (external hydrophobic patch)/IHP (internal hydrophobic patch)] regulating the assembly of mouse ZP proteins. Whereas the transmembrane domain (TMD) of ZP3 can be functionally replaced by an unrelated TMD, mutations in either EHP or IHP do not hinder secretion of full-length ZP3 but completely abolish its assembly. Because mutants truncated before the TMD are not processed, we conclude that the conserved TMD of mammalian ZP proteins does not engage them in specific interactions but is essential for C-terminal processing. Cleavage of ZP precursors results in loss of the EHP, thereby activating secreted polypeptides to assemble by using the IHP within the ZP domain. Taken together, these findings suggest a general mechanism for assembly of ZP domain proteins.
The zona pellucida (ZP) domain is a protein polymerization module of ≈260 aa (1). Since its identification in mouse ZP glycoproteins (2), this domain has been found in many extracellular eukaryotic proteins of diverse molecular architecture and biological function (2, 3). These include egg coat proteins, inner ear proteins, urinary and pancreatic proteins, transforming growth factor-β receptors, immune defense proteins, nematode cuticle components, and fly proteins involved in transmission of mechanical stimuli and in wing and tracheal morphogenesis (4).
We study the ZP (3), an extracellular coat secreted by growing mouse oocytes, as a model for ZP domain protein maturation and assembly. The ZP consists of long filaments composed of ZP2 and ZP3, crosslinked by a third ZP domain protein, ZP1. Nascent ZP polypeptides have features in common with other ZP domain proteins (2–4) that include an N-terminal signal sequence (SP), a ZP domain, and a consensus furin cleavage site (CFCS). The latter is followed by a C-terminal propeptide containing a transmembrane domain (TMD) and short cytoplasmic tail (Fig. 1A) that are replaced by a glycosyl phosphatidylinositol anchor in some ZP domain proteins. During secretion, but before incorporation into the ZP, a furin-like enzyme excises the propeptide from ZP precursors by cleavage at the CFCS (5–8). C-terminal processing of precursors also occurs for ZP homologues from fish to birds (9–11), as well as for other ZP domain proteins (12–14). Recombinant ZP proteins synthesized from cDNAs mutated at the CFCS are not secreted and accumulate in the endoplasmic reticulum of transfected cells (15–17). Recently, assembly of ZP proteins was studied by microinjection of epitope-tagged cDNAs into growing oocytes followed by confocal microscopy (CM) (6). Analysis of deletion mutants revealed that, whereas the TMD is not required for secretion of ZP precursors, it is required, in conjunction with the ZP domain, for ZP assembly (1). Although these findings suggest that the C-terminal region of ZP domain protein precursors is essential for secretion and assembly, it is not known how the propeptide regulates the two processes.
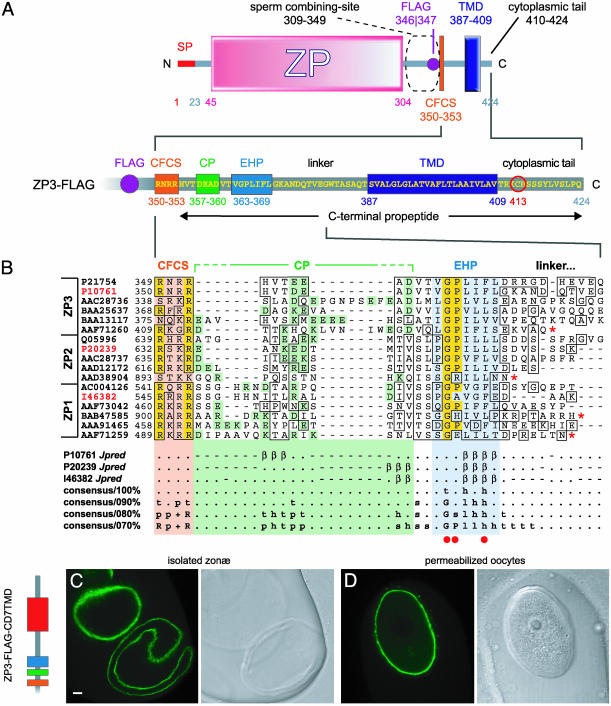
Fig. 1.
(A) Predicted domain structure and major sequence features of FLAG-tagged mouse ZP3 precursor. The complete sequence of the CFCS/C-terminal propeptide region is shown below the overall domain architecture of ZP3. Polypeptide boundaries are marked by gray bars, with the signal peptide (SP) in red; the ZP domain, CFCS, CP, EHP, and TMD are depicted as pink, orange, green, cyan, and blue rectangles, respectively; FLAG-tag is a violet circle, and conserved C413 is circled in red. (B) Conserved motifs in the C-terminal propeptides of ZP/VE proteins. Multiple sequence alignments of part of the C-terminal propeptides of ZP1–3 homologues from human, mouse, marsupial, avian, amphibian, and fish (shown in that order from top to bottom). Sequence accession numbers are reported on the left and highlighted in red for mouse ZP proteins. Regions of homology are boxed, with residues identical in >65% of the sequences shaded in yellow. Conserved features are indicated and shaded by using the same colors as in A. Brackets indicate the boundaries of the alignment regions corresponding to the CP; however, for each sequence, only actual charged residues are shaded. Red asterisks mark the C-termini of ZP domain protein precursors lacking a TMD; red dots indicate residues within the EHP of ZP3 that were individually mutated. Below the alignment are jpred secondary structure predictions for mouse ZP proteins [(β), β-sheet; (.), coil], as well as consensus sequences calculated at different thresholds by using a nonredundant (90% sequence identity threshold; 77 sequences) database of aligned ZP1–3 homologues (see Materials and Methods). Note that ZP2 homologues have not been identified in bird and fish, and sequence AAD38904 belongs to a separate fish egg ZP domain protein class (“zpax”) whose significant homology to ZP2 homologues is limited to the ZP domain (46). (C) Detection of recombinant ZP3 incorporated into the ZP of oocytes microinjected with ZP3-FLAG-CD7TMD cDNA. A schematic diagram of the C-terminal propeptide, as well as representative confocal sections of both isolated ZP (C) and permeabilized oocytes (D), is shown. Matching light images are shown on the right side of each panel. (In C, bar = 10 μm.)
Mutations in genes encoding ZP domain proteins can result in severe human pathologies, including nonsyndromic deafness (18), vascular (19) and renal (20) diseases, and cancer (21, 22). Characterization of some of these mutations suggest that, by reducing or abolishing secretion, protein polymerization is affected only indirectly (1). Here, we describe authentic assembly mutants for ZP proteins; analysis of these mutants suggests a conserved function for the C-terminal propeptide. This finding leads, in turn, to a general assembly mechanism based on coupling between processing and polymerization.
Materials and Methods
DNA Constructs. Constructs were derived from previously described mammalian expression vectors carrying cDNAs for ZP3-FLAG and ZP2-Myc (1, 6). All ZP3 constructs contain a FLAG-tag sequence between the sperm combining site and the CFCS, except for protein ZP3–373-FLAG where the epitope was moved to the C terminus of the truncated propeptide. Point mutations were introduced with a QuikChange site-directed mutagenesis kit (Stratagene); deletion mutants were obtained by overlap extension PCR.
DNA Microinjection and CM Analysis of Microinjected Growing Oocytes. Mouse oocytes were collected, microinjected with DNA constructs, cultured, and analyzed by CM (1, 6).
Mammalian Cell Culture and Transient Transfections. Processing and secretion of recombinant ZP proteins were analyzed by using mammalian cell cultures because expression levels in microinjected mouse oocytes are too low for detection by immunoblotting (1). Chinese hamster ovary and human embryonic kidney 293 cells were cultured and transiently transfected as described (1, 6, 15). Immunoblotting was carried out by using monoclonal anti-FLAG or anti-Myc (1, 6, 15), or polyclonal anti-ZP3 (8811, 1:500; Pocono Rabbit Farm, Canadensis, PA). Densitometric analysis of immunoblots was performed with imagej (http://rsb.info.nih.gov/ij/), taking care to use exposures that did not saturate x-ray films.
Protein Sequence Analysis. Nonredundant protein sequence databases were generated by using scripts derived from nrdb90.pl (www.ebi.ac.uk/∼holm/nrdb90); sequences were aligned by using emma (www.hgmp.mrc.ac.uk/Software/EMBOSS). Sequence conservation was evaluated with scorecons (www.ebi.ac.uk/thornton-srv/databases/valdarprograms), consensus sequences were calculated by using consensus.pl (www.bork.emblheidelberg.de/Alignment/consensus.html). Lowercase characters within the consensus sequences of Figs. 1B and 3A correspond to the following amino acid sets: l, [I,V,L]; h, [F,Y,W,H,I,V,L]; +, [H,K,R]; –, [D,E]; p, [Q,N,S,T,C,H,K,R,D,E]; u, [G,A,S]; s, [G,A,S,V,T,D,N,P,C]; t, [G,A,S,Q,N,S,T,C,H,K,R,D,E]; (.), any amino acid (no consensus); uppercase letters within consensus sequences indicate the specific amino acids with the same one-letter code. Secondary structure was predicted with jpred (www.compbio.dundee.ac.uk/∼www-jpred), hmmstr (www.bioinfo.rpi.edu/∼bystrc/hmmstr/server.php), and psipred (http://bioinf.cs.ucl.ac.uk/psipred). Domains were identified with smart (http://smart.embl-heidelberg.de); degenerate pattern searches were carried out with nps@pattinprot (http://npsa-pbil.ibcp.fr) and fastscanner (L.J., unpublished observations).
Fig. 3.
A second hydrophobic patch within the ZP domain is also involved in secretion and assembly of ZP3. (A) Comparison of EHP and IHP sequences of ZP1–3 and (B) schematic representation of their position within a minimal ZP domain protein. Amino acid numbers and sequences refer to human and mouse ZP proteins; secondary structure predictions and consensus sequences for individual ZP protein subfamilies (ZP1–3) were obtained as in Fig. 1B. The IHP is depicted as a yellow rectangle, with all other elements as in Fig. 1, except for the CP, which was omitted. Amino acid F171 within the IHP of ZP3 is indicated by a red dot. (C) IHP mutation F171S does not affect secretion of full-length ZP3 (ZP3-FLAG-F171S; lanes 1 and 2) but abolishes secretion of protein constructs lacking a TMD (ZP3-FLAG-370-F171S; lanes 3 and 4). (D and E) The IHP mutant ZP3-FLAG-F171S is packaged into secretory vesicles by microinjected oocytes (E) but is not assembled into the ZP (D). Red crosses mark mutated IHPs.
Results
The TMD of ZP Proteins Ensures Correct Localization for Assembly. Truncation of their polypeptides before the TMD does not prevent packaging of ZP2 and ZP3 into secretory vesicles whereas it completely prevents incorporation into the ZP (1). Because ZP proteins truncated just before the TMD are secreted by mammalian cells as efficiently as wild-type counterparts (Fig. 2A, lanes 1, 3, and 5, and refs. 1 and 23), inhibition is not due to reduced secretion. These findings suggest that the TMD and short cytoplasmic tail are not required for secretion but do contain elements that are essential for assembly.
Fig. 2.
(A and B) The EHP is sufficient for secretion, but not for C-terminal processing of truncated ZP3 precursors lacking a TMD. (A) Immunoblot analysis with an anti-FLAG monoclonal antibody reveals that ZP3 constructs truncated in the C terminus up to the EHP are secreted into the medium (M) of transfected cells as efficiently as wild-type ZP3 (compare lanes 3 and 5 with lane 1) although they are not cleaved (lane 13). Further truncations of the ZP3 C terminus cause the protein to be retained in cell lysate (L) (lanes 7–10). Unexpectedly, the EHP is not required for secretion of a ZP3 construct retaining a TMD (lane 17), and mutation of the conserved CP does not affect protein secretion either in the presence (lane 11) or absence (lane 15) of a TMD. (B) Introduction of single amino acid mutations within the EHP of a secreted construct truncated before the TMD (ZP3-FLAG-370; lanes 1 and 2) abolishes (G364A mutant; lanes 3 and 4) or severely impairs (P365A and F368S mutants; lanes 5–8) ZP3 secretion. (C and D) The EHP is required for ZP3 incorporation into the ZP. Confocal sections of isolated, immunostained ZP reveal that deletion of the EHP abolishes protein incorporation into the ZP (C) although mutant ZP3 is packaged into secretory vesicles (D). Numbers within the names of truncated constructs in A and B indicate their C-terminal amino acid; outline and conventions of C and D are as in Fig. 1 C and D.
Aside from 2–4 basic juxtamembrane amino acids commonly found in type I transmembrane proteins (24), a single C residue is the only feature somewhat conserved in the short cytoplasmic tail of ZP proteins (Fig. 1A). Because the other 12 C residues of ZP3 are present as intramolecular disulfides in the extracellular portion of the protein (7, 8), and because cytoplasmatic C residues can be important for assembly of transmembrane protein complexes (25, 26), we asked whether mutation of the C residue to an S residue would affect secretion and assembly of mouse ZP3. The mutant C413S protein was secreted and assembled to the same extent as wild-type ZP3; also, its secretion was unaffected in transfected cell lines (data not shown). These findings suggest that C413 is not essential for secretion or assembly of ZP3.
Assembly of extracellular complexes can be mediated by specific interactions between their TMDs (27). To assess whether this was the case for ZP proteins, the TMD of ZP3 (amino acids 387–409) was replaced by the single-spanning C-terminal TMD of human CD7 (amino acids 178–201), which is very different in sequence and is not involved in specific interactions (ref. 28; A. Ting, personal communication). The resulting construct was efficiently secreted by transfected cells (data not shown) and was incorporated into the ZP of microinjected oocytes as well as wild-type ZP3 (Fig. 1 C and D). These results suggest that the conserved TMD is not involved in specific interactions but ensures proper localization and/or topological orientation of nascent proteins so that assembly can occur.
The TMD Is Required for Proteolytic Processing of ZP Proteins. Cleavage of the C-terminal portion of ZP domain protein precursors is required for secretion and assembly into extracellular structures (5–14). To determine whether truncation of ZP3 before its TMD affected proteolytic processing, a truncated construct with a C-terminal FLAG-tag was produced (ZP3–373-FLAG). As shown Fig. 2A (lane 13), immunoblotting of transfected cells revealed that the tag was retained in the secreted protein. Therefore, the absence of a TMD prevents cleavage of the C-terminal region of the ZP3 precursor.
A Sequence Between the CFCS and TMD Regulates Secretion of Truncated ZP Proteins. ZP protein constructs truncated just before the TMD are efficiently secreted by transfected cells (Fig. 2A, lanes 3, 5, and 13; refs. 1 and 23). On the other hand, polypeptides corresponding to mature protein species (i.e., truncated immediately after the CFCS) are retained in the endoplasmic reticulum and not secreted (Fig. 2A, lanes 9 and 10; ref. 15). Collectively, these findings suggest that elements crucial for secretion are located between the CFCS and TMD.
Sequence alignments of the C-terminal propeptides of ZP1–3 homologues (Fig. 1B) reveal that the CFCS is followed by a short stretch rich in charged amino acids (“charged patch,” CP). Because the CP sequence is relatively conserved in mammalian ZP3 homologues, we assessed whether this motif plays a role in protein secretion. The CP of ZP3 was mutated to AAAA in the context of the full-length protein (ZP3-FLAG-ΔCP) and a secreted construct truncated before the TMD (ZP3-FLAG-370-ΔCP). As seen in Fig. 2A (lanes 11 and 15), proteins encoded by both mutant constructs are secreted at levels comparable to wild-type counterparts, suggesting that the CP is not required for secretion of ZP3.
A second short conserved motif, consisting of an almost invariant GP sequence immediately followed by 4–5 hydrophobic amino acids, is found C-terminal to the CP (Fig. 1B). This element, designated as an “external hydrophobic patch” (EHP), is connected to the TMD by a linker that is not conserved in sequence or length (Fig. 1B; and Fig. 5, which is published as supporting information on the PNAS web site). Interestingly, fish homologues of ZP1 and ZP3 that are synthesized by the liver and travel in the blood to the oocyte vitelline envelope (VE) (29, 30) lack a TMD and end just after the EHP (Fig. 1B). The C-terminal propeptides of these proteins, which are missing from assembled VE proteins (9), are essentially equivalent to construct ZP3-FLAG-370 (Fig. 2A). Similarly, precursors of avian ZP1 homologues, also secreted by the liver (31, 32), terminate with the EHP (Fig. 1B). Finally, we identified EHPs in sequences of precursors of other ZP domain proteins, both with and without C-terminal TMDs (Fig. 5).
To determine whether the conserved EHP plays a role in secretion, a ZP3 construct truncated immediately before the EHP was produced (ZP3-FLAG-362). Mammalian cells transfected with this construct failed to secrete ZP3 (Fig. 2A, lanes 7 and 8). Identical results were obtained by using a corresponding deletion mutant of mouse ZP2 (data not shown). However, when the entire EHP was replaced by a single R residue in the context of full-length ZP3 (ZP3-FLAG-ΔEHP), secretion was comparable to that of wild-type protein (Fig. 2A, lane 17). Collectively, these results suggest that the linker between the EHP and TMD is not required for secretion; this finding is consistent with the lack of sequence and length conservation for the linker (Fig. 1B). On the other hand, the results demonstrate that the EHP sequence is required for secretion of constructs lacking a TMD whereas it is dispensable when the TMD is present. The latter finding contrasts with a recent report that substitution of the hydrophobic patch with a FLAG-tag abolishes secretion of a ZP3-EGFP fusion construct (33).
Conclusions just described were confirmed by analyzing the effect of single amino acid changes in the EHP of truncated construct ZP3-FLAG-370. A ZP3-FLAG-370-G364A mutant is equivalent to construct ZP3-FLAG-362 in that it was not secreted (Fig. 2B, lanes 3 and 4) whereas mutants P365A and F368S were secreted at 1–2% of the levels of wild-type ZP3 (Fig. 2B, lanes 5–8; see Materials and Methods). The severity of these mutations correlates with the degree of conservation of the corresponding amino acids (Fig. 1B). Collectively, these results suggest that ZP proteins must contain either an EHP or a TMD to be secreted.
The EHP Is Required for ZP Protein Assembly. Results of CM of growing oocytes microinjected with ZP3-FLAG-353 were consistent with cell transfection experiments described above; i.e., ZP3 was not detected in secretory vesicles or the ZP (data not shown). Similar analyses were carried out by using oocytes microinjected with ZP3-FLAG-ΔCP and ZP3-FLAG-ΔEHP mutant constructs. In the former case, mutant ZP3 was incorporated into the ZP to the same extent as wild-type ZP3, suggesting that the CP does not play a role in assembly. On the other hand, deletion of the EHP from ZP3 did not prevent packaging of mutant protein into vesicles (Fig. 2D) but completely abolished incorporation into the ZP (Fig. 2C). Therefore, both the EHP and TMD must be present for protein assembly into the ZP.
A Sequence Similar to the EHP Is Found Within the ZP Domain. Alignments of ZP2 and ZP1 protein homologues revealed that a sequence similar to the EHP is conserved within the ZP domain (Fig. 3 A and B) and is designated as an “internal hydrophobic patch” (IHP). Although alignments of ZP3 homologues showed no conservation of the GP subsequence within the IHP, a short stretch containing hydrophobic amino acids was found at an equivalent location (Fig. 3A). These hydrophobic residues are highly conserved in a comprehensive, nonredundant ZP domain protein sequence database (95% threshold consensus sequence for residues corresponding to ZP3 amino acids 171–175: h.hth; see Materials and Methods and ref. 4). Amino acids following the GP within the EHP and IHP are predicted to form β-strands (Figs. 1B and 3A and ref. 4), and F171, one of the few absolutely conserved residues of ZP3, is predicted to lie within the β-strand formed by the IHP. As seen in Fig. 3A, F171 corresponds to F368, the most conserved hydrophobic amino acid within the EHP of ZP3. Mutation of F368 severely impairs secretion of truncated construct ZP3-FLAG-370 (Fig. 2B).
IHP Mutants of ZP Proteins Have the Same Phenotype as EHP Mutants. To determine whether the IHP is also involved in secretion and assembly of ZP3, F171 was mutated in the context of full-length (ZP3-FLAG-F171S) and truncated (ZP3-FLAG-370-F171S) ZP3. Transfection of cells with these constructs revealed that mutant ZP3-FLAG-F171S was secreted as efficiently as wild-type ZP3 (Fig. 3C, lanes 1 and 2) whereas no secretion of ZP3-FLAG-370-F171S was detected (Fig. 3C, lanes 3 and 4). Furthermore, CM analyses of oocytes microinjected with the ZP3-FLAG-F171S cDNA indicated that mutant protein was packaged into secretory vesicles (Fig. 3E) but was not incorporated into the ZP (Fig. 3D). Therefore, IHP mutants exhibit the same phenotypes as corresponding EHP mutants (Fig. 6, which is published as supporting information on the PNAS web site).
Discussion
Many extracellular eukaryotic proteins with mosaic architecture assemble into filaments and matrices through ZP domains (1, 4). Features that regulate their assembly are expected to lie within the polypeptide region that they share; i.e., between the start of the ZP domain and the C-terminal TMD of ZP precursors (Fig. 1A). Here, we identified two such elements, one external (EHP) and another internal (IHP) to the ZP domain, that are essential for incorporation of ZP3 into the ZP. In addition to being related in sequence (Fig. 3A) and giving rise to identical phenotypes when mutated (Fig. 6), the hydrophobic patches share a common topology within the structure of a minimal ZP domain protein precursor (≈300 aa; Fig. 3B). The only invariant residues within the ZP domain are eight conserved C residues (2, 4); apparently, the domain consists of two halves, one containing the first four C residues and the other containing the remaining C residues (7, 8). This finding is supported by the fact that the sequence between the two groups of C residues can be glycosylated (7, 8) and is susceptible to protease digestion (34), as expected for a solvent-exposed polypeptide fragment linking independently folded regions. Moreover, PLAC1 (35) and OOSP1 (36) are proteins whose N-terminal sequences share homology with the first half of the ZP3 ZP domain, including C residues 1–4 (Fig. 3B).
It is striking that the IHP and EHP identified here are located equivalently relative to the first and second half of the ZP domain (Fig. 3B). This finding is particularly relevant in that conserved exon-intron boundaries in mammalian ZP1–3 genes are found immediately after their IHP-encoding sequences (at T405, P494, and E177, respectively; Fig. 3A). Furthermore, protease-sensitive sites were mapped to the short linker between the first half of the ZP domain and IHP (L.J. and P.M.W., unpublished results; Fig. 3B), such that they are topologically equivalent to the CFCS between the second half of the ZP domain and EHP. Thus, the minimal ZP domain protein precursor depicted in Fig. 3B essentially consists of two subdomains of ≈120–130 aa each, connected by a ≈60- to 70-residue linker. Each subdomain contains four conserved C residues, separated from a C-terminal IHP or EHP by a short protease-sensitive hinge.
What are the functional implications of two subdomains within the ZP domain? What is the role of a duplicated hydrophobic motif in secretion and assembly of ZP precursors? Several clues come from our EHP/IHP mutation experiments in the presence and absence of a TMD (Fig. 6). Just like fish VE homologues synthesized by the liver, ZP precursors can be efficiently secreted in the absence of a TMD as long as they retain an EHP (Fig. 2A). Indeed, this result is likely to be a general property of ZP domain proteins. For example, a GP-2 construct truncated before the putative glycosyl phosphatidylinositol anchor attachment site (i.e., C-terminal to the EHP; Fig. 5) was also secreted by transfected cells as efficiently as wild-type GP-2 (37). Moreover, different ZP domain proteins with short propeptides ending with an EHP, such as DMBT1 and LZP (Fig. 5), are also secreted (21, 38). However, the EHP cannot simply be a secretion signal: even if the EHP ultimately dissociates from the mature protein, its presence at the plasma membrane is required for assembly of secreted, full-length ZP3 constructs (Fig. 2C). Similarly, although cleavage at the CFCS causes dissociation of membrane-bound EHP from secreted ZP proteins (6), there is no obvious reason why the short EHP-containing propeptides of fish VE proteins should be removed before incorporation (9) if their only function is to target precursors to assembly sites on the oocyte surface.
Here, we suggest that the EHP functions as a control switch for assembly by preventing the premature polymerization of ZP precursors (Fig. 4). This finding is consistent with the fact that ZP protein homologues from fish to mammals are all cleaved between the ZP domain and EHP before incorporation into the ZP/VE (5–11). Mammalian ZP protein constructs that lack a TMD are not C-terminally processed, such that the EHP is retained after secretion (Fig. 2A, lane 13) and are not incorporated into the ZP (1). Cleavage at the CFCS eliminates the EHP from secreted proteins, thereby activating them for assembly in the extracellular space.
Fig. 4.
A mechanism for ZP domain protein assembly. Nascent precursors anchored to the plasma membrane by their TMD are not competent for assembly due to interaction between the EHP and IHP (Upper Left). Proteolytic processing of precursors at the CFCS (Upper Right) leads to dissociation of the EHP and IHP, thereby releasing activated mature proteins into the medium (Lower Left). Interactions involving the IHP within the ZP domain drive polymerization of proteins into filaments or matrices (Lower Right). EHP-IHP interactions could involve more than one copy of the same precursor, and assembly of heteromeric systems may depend on interactions mediated by IHPs from different proteins. Although precursors of ZP domain proteins with short C-terminal propeptides lacking a TMD are not necessarily associated with plasma membrane, their assembly would still depend on proteolytic removal of the EHP. Relative domain orientation and protein arrangements are hypothetical. Elements are depicted as in previous figures, except for the linker between the two putative subdomains of the ZP domain, which is shown as a magenta line. Green scissors represent the protease(s) responsible for C-terminal cleavage of ZP domain protein precursors. CP is not shown, and elements are not drawn to scale.
How does the EHP inhibit assembly of ZP precursors? Because endogenous ZP1 and ZP2 are not present in our expression system, the phenotype of ZP3 EHP mutants implies that this motif acts in cis. The EHP could transiently mask a complementary hydrophobic sequence within the ZP domain that is required for interactions between ZP proteins. If there are two subdomains within ZP precursors, the structural and functional similarities between the EHP and the IHP, and the lack of assembly of secreted mutant ZP3-FLAG-F171S (Fig. 3D), raises the possibility that the IHP may be a binding partner for the EHP (Fig. 4). Alternatively, the EHP and IHP within the ZP3 precursor might compete for an as yet unidentified common element involved in assembly. If so, protein incorporation into the ZP may rely on CFCS cleavage-dependent swapping between the IHP and EHP.
Database searches with degenerate consensus sequences failed to identify a similar IHP-CFCS-EHP(-TMD) organization in proteins lacking a ZP domain. However, cleavage of inhibitory protein fragments is important for polymerization of fibrillin-1 (39), tau (40) and fibrin (41) and a short motif that prevents premature self-polymerization of complement component C9 has been described (42). Furthermore, thrombin has been shown to initiate assembly of fibrin by exposure of a polymerization site whose GPRVV sequence resembles EHP/IHP (41) and structures of p13suc1 and homologues suggest that these proteins can dimerize through swapping of hydrophobic β-strands preceded by P residue-containing hinge regions (HVPEPHILLFRR in yeast suc1; underlined residues form the swapped β-strand) (43).
Interaction between the EHP and IHP would explain why ZP3 constructs mutated in either motif and lacking a TMD are not secreted (Figs. 2 A and B and 3C). Presumably, these mutations lead to premature exposure of complementary binding sequences and/or other conformational changes that occur during assembly. This result would cause the mutants to be retained in the cell, most likely in the endoplasmic reticulum (15). However, constructs ZP3-FLAG-ΔEHP and ZP3-FLAG-F171S are secreted (Figs. 2A and 3C), suggesting that they are rescued by the presence of a TMD. The TMD could prevent intracellular aggregation of the mutants by constraining them to the bilayer. Regardless of the specific details of precursor cleavage, it is the combination of a CFCS upstream of the EHP and a TMD downstream that ensures that ZP precursors lose their EHP sequences and can interact at the assembly site. This finding is supported by TMD swapping experiment (Fig. 1 C and D), suggesting that the TMD of the ZP3 precursor is not directly involved in specific interactions between ZP proteins or between ZP3 and other factors required for assembly. It is also compatible with the observation that several other ZP domain proteins are linked to membrane through glycosyl phosphatidylinositol anchors rather than TMDs (2–4).
Events preceding protein assembly must differ in fish because VE precursors lacking a TMD incorporate into the egg coat (9, 29, 44) whereas secreted, truncated mouse constructs do not (1). However, our findings resolve this apparent contradiction by demonstrating that the TMD of mammalian ZP proteins is only indirectly required for incorporation into the ZP by ensuring cleavage of precursors at the CFCS (Figs. 1 C and D and 2A). Thus, regardless of the strategy by which ZP/VE precursors reach the oocyte and are activated, we suggest that the basic mechanism of egg coat assembly is conserved from fish to humans. Indeed, whereas fish precursors are not internalized by the egg (45), they incorporate into the innermost layer of the VE (44), as is the case in mammals (6). Furthermore, in certain fish, some of the VE precursors are synthesized by the liver and others by the oocyte (46). Finally, it should be noted that mouse ZP proteins are able to incorporate into the VE of Xenopus eggs (47).
Consistent with a unified view of ZP domain protein polymerization, we described a conserved duplicated motif (EHP/IHP) that plays an essential role in assembly of oocyte ZP proteins. Our findings support the idea that the ZP domain consists of two subdomains, provide a rationale for conservation of short C-terminal propeptides in ZP domain protein precursors lacking a TMD or a glycosyl phosphatidylinositol anchor, reveal a critical function for proteolytic processing of ZP domain proteins, and explain why TMDs are required for assembly of mammalian ZP proteins.
Supplementary Material
Acknowledgments
L.J. dedicates this paper to the memory of his father, Gian Jovine. We thank C. Darie and K. Quadrini for comments. Confocal microscopy was performed at the Mount Sinai School of Medicine Microscopy Shared Resource Facility. L.J. was supported by a Human Frontier Science Program long-term fellowship. This research was supported in part by National Institutes of Health Grant HD35105.
Abbreviations: CFCS, consensus furin cleavage site; CP, charged patch; EHP, external hydrophobic patch; IHP, internal hydrophobic patch; TMD, transmembrane domain; VE, vitelline envelope; ZP, zona pellucida; CM, confocal microscopy.
Note Added in Proof. After release of the first avian genome draft (http://genome.wustl.edu/projects/chicken), we identified conserved EHP and IHP motifs within the sequence of a putative chicken ZP2 homologue. The corresponding gene lies within contig 164.44.1.32133.291.12014 and encodes a protein that is 43% identical to human ZP2 in a 633-aa overlap (E value: 6.6e-141).
References
- 1.Jovine, L., Qi, H., Williams, Z., Litscher, E. & Wassarman, P. M. (2002) Nat. Cell Biol. 4, 457–461. [DOI] [PubMed] [Google Scholar]
- 2.Bork, P. & Sander, C. (1992) FEBS Lett. 300, 237–240. [DOI] [PubMed] [Google Scholar]
- 3.Wassarman, P. M., Jovine, L. & Litscher, E. S. (2001) Nat. Cell Biol. 3, E59–E64. [DOI] [PubMed] [Google Scholar]
- 4.Jovine, L., Litscher, E. & Wassarman, P. M. (2002) in Gene Expression at the Beginning of Animal Development, ed. DePamphilis, M. L. (Elsevier, Amsterdam), Vol. 12, pp. 31–54. [Google Scholar]
- 5.Litscher, E. S., Qi, H. & Wassarman, P. M. (1999) Biochemistry 38, 12280–12287. [DOI] [PubMed] [Google Scholar]
- 6.Qi, H., Williams, Z. & Wassarman, P. M. (2002) Mol. Biol. Cell 13, 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boja, E. S., Hoodbhoy, T., Fales, H. M. & Dean, J. (2003) J. Biol. Chem. 278, 34189–34202. [DOI] [PubMed] [Google Scholar]
- 8.Yonezawa, N. & Nakano, M. (2003) Biochem. Biophys. Res. Commun. 307, 877–882. [DOI] [PubMed] [Google Scholar]
- 9.Sugiyama, H., Murata, K., Iuchi, I., Nomura, K. & Yamagami, K. (1999) J. Biochem. (Tokyo) 125, 469–475. [DOI] [PubMed] [Google Scholar]
- 10.Kubo, H., Matsushita, M., Kotani, M., Kawasaki, H., Saido, T. C., Kawashima, S., Katagiri, C. & Suzuki, A. (1999) Dev. Genet. 25, 123–129. [DOI] [PubMed] [Google Scholar]
- 11.Sasanami, T., Pan, J., Doi, Y., Hisada, M., Kohsaka, T. & Toriyama, M. (2002) Eur. J. Biochem. 269, 2223–2231. [DOI] [PubMed] [Google Scholar]
- 12.Killick, R., Legan, P. K., Malenczak, C. & Richardson, G. P. (1995) J. Cell Biol. 129, 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuoka, S., Freedman, S. D., Yu, H., Sukhatme, V. P. & Scheele, G. A. (1992) Proc. Natl. Acad. Sci. USA 89, 1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki, K., Sato, K., Akiyama, Y., Yanagihara, K., Oka, M. & Yamaguchi, K. (2002) Cancer Res. 62, 4894–4898. [PubMed] [Google Scholar]
- 15.Williams, Z. & Wassarman, P. M. (2001) Biochemistry 40, 929–937. [DOI] [PubMed] [Google Scholar]
- 16.Kiefer, S. M. & Saling, P. (2002) Biol. Reprod. 66, 407–414. [DOI] [PubMed] [Google Scholar]
- 17.Sasanami, T., Toriyama, M. & Mori, M. (2003) Biol. Reprod. 68, 1613–1619. [DOI] [PubMed] [Google Scholar]
- 18.Steel, K. P. & Kros, C. J. (2001) Nat. Genet. 27, 143–149. [DOI] [PubMed] [Google Scholar]
- 19.Marchuk, D. A., Srinivasan, S., Squire, T. L. & Zawistowski, J. S. (2003) Hum. Mol. Genet. 12, R97–R112. [DOI] [PubMed] [Google Scholar]
- 20.Serafini-Cessi, F., Malagolini, N. & Cavallone, D. (2003) Am. J. Kidney Dis. 42, 658–676. [DOI] [PubMed] [Google Scholar]
- 21.Kang, W. & Reid, K. B. (2003) FEBS Lett. 540, 21–25. [DOI] [PubMed] [Google Scholar]
- 22.Copland, J. A., Luxon, B. A., Ajani, L., Maity, T., Campagnaro, E., Guo, H., LeGrand, S. N., Tamboli, P. & Wood, C. G. (2003) Oncogene 22, 8053–8062. [DOI] [PubMed] [Google Scholar]
- 23.Harris, J., Seid, C., Fontenot, G. & Liu, H. (1999) Protein Expression Purif. 16, 298–307. [DOI] [PubMed] [Google Scholar]
- 24.Boyd, D. & Beckwith, J. (1990) Cell 62, 1031–1033. [DOI] [PubMed] [Google Scholar]
- 25.Locker, J. K. & Griffiths, G. (1999) J. Cell Biol. 144, 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozanov, D. V., Deryugina, E. I., Ratnikov, B. I., Monosov, E. Z., Marchenko, G. N., Quigley, J. P. & Strongin, A. Y. (2001) J. Biol. Chem. 276, 25705–25714. [DOI] [PubMed] [Google Scholar]
- 27.Harrison, P. T. (1996) Mol. Membr. Biol. 13, 67–79. [DOI] [PubMed] [Google Scholar]
- 28.Schanberg, L. E., Fleenor, D. E., Kurtzberg, J., Haynes, B. F. & Kaufman, R. E. (1991) Proc. Natl. Acad. Sci. USA 88, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyllner, S. J., Westerlund, L., Olsson, P. E. & Schopen, A. (2001) Biol. Reprod. 64, 805–811. [DOI] [PubMed] [Google Scholar]
- 30.Arukwe, A. & Goksøyr, A. (2003) Comp. Hepatol. 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasanami, T., Pan, J. & Mori, M. (2003) J. Steroid Biochem. Mol. Biol. 84, 109–116. [DOI] [PubMed] [Google Scholar]
- 32.Bausek, N., Waclawek, M., Schneider, W. J. & Wohlrab, F. (2000) J. Biol. Chem. 275, 28866–28872. [DOI] [PubMed] [Google Scholar]
- 33.Zhao, M., Gold, L., Dorward, H., Liang, L. F., Hoodbhoy, T., Boja, E., Fales, H. M. & Dean, J. (2003) Mol. Cell. Biol. 23, 8982–8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosiere, T. K. & Wassarman, P. M. (1992) Dev. Biol. 154, 309–317. [DOI] [PubMed] [Google Scholar]
- 35.Cocchia, M., Huber, R., Pantano, S., Chen, E. Y., Ma, P., Forabosco, A., Ko, M. S. & Schlessinger, D. (2000) Genomics 68, 305–312. [DOI] [PubMed] [Google Scholar]
- 36.Yan, C., Pendola, F. L., Jacob, R., Lau, A. L., Eppig, J. J. & Matzuk, M. M. (2001) Genesis 31, 105–110. [DOI] [PubMed] [Google Scholar]
- 37.Colomer, V., Lal, K., Hoops, T. C. & Rindler, M. J. (1994) EMBO J. 13, 3711–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, Z. G., Du, J. J., Zhang, X., Cheng, Z. H., Ma, Z. Z., Xiao, H. S., Yu, L., Wang, Z. Q., Li, Y. Y., Huo, K. K., et al. (2003) Hepatology 38, 735–744. [DOI] [PubMed] [Google Scholar]
- 39.Handford, P. A., Downing, A. K., Reinhardt, D. P. & Sakai, L. Y. (2000) Matrix Biol. 19, 457–470. [DOI] [PubMed] [Google Scholar]
- 40.Gamblin, T. C., Chen, F., Zambrano, A., Abraha, A., Lagalwar, S., Guillozet, A. L., Lu, M., Fu, Y., Garcia-Sierra, F., LaPointe, N., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 10032–10037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mosesson, M. W., Siebenlist, K. R. & Meh, D. A. (2001) Ann. N.Y. Acad. Sci. 936, 11–30. [DOI] [PubMed] [Google Scholar]
- 42.Taylor, K. M., Trimby, A. R. & Campbell, A. K. (1997) Immunology 91, 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bourne, Y., Watson, M. H., Arvai, A. S., Bernstein, S. L., Reed, S. I. & Tainer, J. A. (2000) Struct. Fold. Des. 8, 841–850. [DOI] [PubMed] [Google Scholar]
- 44.Hamazaki, T. S., Nagahama, Y., Iuchi, I. & Yamagami, K. (1989) Dev. Biol. 133, 101–110. [DOI] [PubMed] [Google Scholar]
- 45.Hyllner, S. J. & Haux, C. (1992) J. Endocrinol. 135, 303–309. [DOI] [PubMed] [Google Scholar]
- 46.Kanamori, A., Naruse, K., Mitani, H., Shima, A. & Hori, H. (2003) Gene 305, 35–45. [DOI] [PubMed] [Google Scholar]
- 47.Doren, S., Landsberger, N., Dwyer, N., Gold, L., Blanchette-Mackie, J. & Dean, J. (1999) Dev. Genes Evol. 209, 330–339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.