Abstract
A specific covalently cross-linked complex between redox partners yeast cytochrome c peroxidase (CCP) and cytochrome c (cyt. c) has been made by engineering cysteines into CCP and cyt. c that form an intermolecular disulfide bond in high yield. The crystal structure of the cross-linked complex has been solved to 1.88-Å resolution and closely resembles the structure of the noncovalent complex [Pellitier, H. & Kraut, J. (1992) Science 258, 1748–1755]. The higher resolution of the covalent complex has enabled the location of ordered water molecules at the peroxidase–cytochrome c interface that serve to bridge between the two proteins by hydrogen bonding. As in the noncovalent complex, direct electrostatic interactions between protein groups appear not to be critical in complex formation. UV–visible spectroscopic and stopped-flow studies indicate that CCP in the covalent complex reacts normally with H2O2 to give compound I. Stopped-flow kinetic studies also show that intramolecular electron transfer between the cross-linked ferrocytochrome c and the Trp-191 cation radical site in CCP compound I occurs fast and is nearly complete within the dead time (≈2 ms) of the instrument. These results indicate that the structure of the covalent complex closely mimics the physiological electron transfer complex. In addition, single-turnover and steady-state experiments reveal that CCP compound I in the covalent complex oxidizes exogenously added ferrocytochrome c at a slow rate (t1/2 ≈ 2 min), indicating that CCP does not have a second independent site for physiologically relevant electron transfer.
Understanding the mechanism by which electrons transfer between proteins has proven to be a challenging and complex problem. Of fundamental importance is specific recognition between redox partners, which involves complementary protein surfaces and, possibly, conformational changes. The process by which electrons flow between proteins also is complex and remains somewhat controversial. In one view electron transfer is mediated by the polypeptide backbone and amino acid side chains (1, 2). In effect the proteins are wired with preferred paths of electron transfer. An alternative view holds that it is sufficient for proper complex formation to enable redox cofactors to approach each other at a minimal distance for rapid electron transfer without the requirement for selective paths (3).
The complexity of interprotein electron transfer can be simplified by focusing on individual steps. One way of separating protein–protein binding from electron transfer is to site-selectively cross-link the redox pairs such that the covalent complex mimics the functionally important electron-transfer complex. Cross-linking provides a unimolecular system for studying electron transfer without the complication of second-order binding kinetics. To achieve this goal requires a redox pair that is structurally well characterized and readily amenable to protein engineering. The cytochrome c peroxidase (CCP)–cytochrome c (cyt. c) electron transfer system meets these criteria. Since the initial characterization of CCP by Yonetani and colleagues (4), the CCP–cyt.c system has been a paradigm for studying protein intermolecular electron transfer reactions. CCP first reacts with H2O2 to give compound I. In compound I the iron has been oxidized from Fe(III) to Fe(IV) and an active-site Trp has been oxidized to a cation radical (5). Then in two successive one-electron transfer reactions, ferrocytochrome c (ferrocyt. c) reduces compound I back to resting-state CCP.
The CCP mechanism just described is based on the assumption that CCP has only one binding site for cyt. c, which agrees well with titration calorimetry data (6, 7), although calorimetric methods would not detect a second site that exhibits a low heat of binding. Indeed, several investigations using a variety of methods indicate two cyt. c binding sites (8–12), although less clear is whether both sites are productive in electron transfer of physiological relevance.
As a step toward addressing these questions we engineered Cys residues into both CCP and yeast cyt. c that were designed to make a covalent complex that mimics the noncovalent complex crystal structure (13). In addition, another CCP–cyt.c crosslink (14) was produced that was designed to block the second site proposed by Northrup et al. (15). This work indicated that the complex designed to mimic the noncovalent crystal structure of Pelletier and Kraut (16) is productive for electron transfer, whereas the second site proposed by Northrup et al. (15) is not likely to be relevant for physiological electron transfer. However, this earlier cross-linking work was hindered owing to low yields of the cross-link and our inability to get suitable diffraction-quality crystals for structural characterization. We have now redesigned the location of the Cys residues in both CCP and cyt. c, which has enabled the isolation of a stable cross-linked complex that yields diffraction-quality crystals. Here we present the 1.88-Å crystal structure of the complex and its activities for compound I formation and oxidation of ferrocyt. c.
Experimental Procedures
Materials. Oligonucleotides were purchased from Qiagen Operon (Alameda, CA). Enzymes and reagents for site-directed mutagenesis were purchased from Stratagene and New England Biolabs. Wild-type yeast (Saccharomyces cerevisiae) cyt. c and horse heart cyt. c were purchased from Sigma. All other chemicals used were of molecular biology grade or better and were purchased from Fisher, Sigma, and EM Science.
Site-Directed Mutagenesis. V197C CCP and A81C cyt. c were constructed for the specific disulfide cross-link. Site-directed mutagenesis was carried out by using as the template C128A mutant DNA, which was designated as the wild-type CCP (13). The A197C/C128A CCP mutant was constructed by using a QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) and following the manufacturer's instructions. The pT-7 vector (expressing C128A CCP) and the oligonucleotides 5′-GGAGCCGCTAACAACTGCTTTACCAATGAG-3′ and its complementary strand were used for this construction.
The A81C cyt. c mutant was constructed by using as template the construct pBTR1 (a bacterial overexpression system for wild-type yeast iso-1-cytochrome c, which was a gift from Grant A. Mauk, University of British Columbia, Vancouver), which had its naturally occurring Cys at position 102 converted to Thr (17, 18). Construction of the A81C cyt. c mutant was carried out by PCR using the method of overlap extension (19). In brief, two overlapping complementary oligonucleotides, [5′-GGTACCAAGATGTGCTTTGGTGGGTTGAAG-3′] and its complementary strand (mutant primers), each containing the new mutation, were designed and synthesized, along with two external primers. The full-length product was recloned into the expression plasmid pBTR1 by using the two BamHI restriction sites. All mutations were confirmed by DNA sequencing at the automated DNA sequencing facility DBS Sequencing at the University of California, Davis.
Protein Expression and Purification. Mutant CCP was expressed in Escherichia coli BL-21(DE3) cells in terrific broth (Teknova, Hollister, CA) containing 100 μg/ml ampicillin. At OD600 ≈ 1.5, 0.75 mM isopropyl β-d-thiogalactoside (IPTG) was used for induction. The mutant protein V197C/C128A CCP was purified as previously described (20, 21) with modifications. High-purity protein (judged by SDS/PAGE) was obtained after an anion-exchange (DEAE-Sephacel) column and a gel-filtration column (Sephadex G-75). The protein was then crystallized twice by dialyzing against cold water and then stored as crystals at –80°C. The CCP concentrations were estimated spectrophotometrically by using an extinction coefficient at 408 nm (ε408) of 96 mM–1·cm–1.
The A81C cyt. c mutant was prepared from 7 liters of E. coli BL-21(DE3) culture, and purified as described previously (13, 14, 17, 18) with minor modifications. Phosphate buffer was used in all steps instead of cacodylate buffer. Briefly, cell pellet was thawed at 4°C and resuspended in 50 mM Tris·HCl buffer, pH 8.0, containing 1 mM EDTA, lysozyme (3 g/liter), 10 mg each of DNase and RNase, and then lysed by sonication on ice. Cell debris was removed by centrifugation. The supernatant was subjected to precipitation with ammonium sulfate (326 g/liter). After centrifugation, the pink-red supernatant was dialyzed overnight against 50 mM potassium phosphate, pH 6.8, then loaded onto a CM-Sepharose CL-6B column that had been preequilibrated with 50 mM potassium phosphate, pH 6.8. The column was washed with the same buffer containing 75 mM NaCl, and then eluted with the same buffer containing 250 mM NaCl. Fractions were checked by SDS/PAGE, and pure fractions were pooled. The cyt. c concentration was estimated spectrophotometrically by using an extinction coefficient of 106 mM–1·cm–1 at 410 nm.
Cross-Linking. CCP contains a naturally occurring Cys residue at position 128. This Cys first was changed to Ala to ensure that only Cys-197 was available for formation of an S–S bond. The conditions for cross-linking A81C cyt. c to V197C/C128A CCP are similar to those described previously (13, 14). Briefly, cyt. c was treated with dithiothreitol (DTT) (10 mM, 30 min on ice) before cross-linking. The excess of DTT was removed by passage through a PD10 column. The cross-linking reaction was performed by mixing of 0.5 mol eq of V197C/C128A CCP and A81C cyt. c in 50 mM potassium phosphate buffer, pH 6.0, containing 4 mM CuSO4 at 0–4°C for ≈0.5–1 h. The covalent complex was purified from CCP and cyt. c monomers and homodimers by cation-exchange chromatography (CM-Sepharose) as described previously (13, 14), except phosphate buffer was used in all of the steps instead of cacodylate buffer. The purity of the complex was checked by reducing and nonreducing SDS/PAGE. The concentration of the complex was estimated spectrophotometrically by using an extinction coefficient of 200 mM–1·cm–1 at the Soret absorption maximum.
Crystallization and X-Ray Data Collection. Initial crystallization trials with PEG-Ion Kit (Hampton Research) yielded crystals with components from 200 mM KI and 20% (wt/vol) polyethylene glycol (PEG) 3350 in the well solution. After fine-tuning crystallization conditions, single crystals of the cross-linked complex were prepared by mixing 2 μl of protein (11 mg/ml) with 2 μl of the well solution, 200 mM KI, NaI, KSCN, NaSCN, or NH4SCN, and 20–25% PEG 3350. Single diffraction-quality crystals were obtained in a few days at 15 or 22°C. Single crystals can be produced by either hanging-drop or sitting-drop vapor-diffusion methods, but the latter yielded larger crystals as red thin plates. Crystals belong to space group P21 with unit cell dimensions a = 45.24 Å, b = 107.92 Å, c = 88.38 Å and β = 104.42° with two molecules of the CCP–cyt.c complex per asymmetric unit. For cryogenic data collection 25% (vol/vol) glycerol was used as the cryoprotectant. In-house x-ray diffraction data were collected from a single flash-frozen crystal by using an R-AXIS IV imaging plate detector equipped with a Rigaku rotating anode x-ray generator with Osmic optics. The crystal was maintained at –160°C in a cryogenic liquid N2 steam (Crystal Logic, Los Angeles). High-resolution data for the same crystal were collected at Stanford Synchrotron Radiation Laboratory (SSRL) beamline 9-1. Optimization of data collection was guided by the STRATEGY function of mosf lm (22). All data were indexed, integrated, and scaled by using the hkl2000 suite (23) and rejections were performed with endhkl (Louis Sanchez, California Institute of Technology, Pasadena) in conjunction with scalepack. For all data collection, a 220° scan using 1° frames was collected. A summary of data collection is provided in Table 1.
Table 1. Crystallographic data and refinement statistics.
| Resolution, Å | 50.0-1.88 |
| Total no. of observations | 245,413 |
| Completeness, % | 91.9 |
| Rsym* on I all data/1.91 to 1.88 Å | 0.054/0.57 |
| I/σ(I) all data/1.91 to 1.88 Å | 13.3/3.19 |
| Unique reflections | 60,538 |
| R/Rfree† | 0.19/0.24 |
| rms deviation for bond distances,‡ Å/angles, ° | 0.013/1.63 |
| No. of protein/heme atoms | 6,439/172 |
| No. of solvent molecules | 738 |
Rsym = Σ|Ih - 〈Ih〉|/Ih.
R = Σ|Fobs - Fcalc|/ΣFobs
The rms deviation for bond lengths and bond angles represent the rms deviations between the observed and ideal values.
Model Building and Refinement. Molecular replacement with amore (24) was used to obtain initial phases. Searches were carried out at 5 Å in the P21 space group by using the crystal structures of CCP and cyt. c (PDB accession number 2PCC) in the noncovalent complex (16), with all of the residues changed to alanine and water molecules removed. The best rotation and translation function solutions were fine tuned by rigid-body refinement. Searches using CCP as a probe gave two solutions, corresponding to the expected two CCP molecules per asymmetric unit. Searches using cyt. c while keeping CCP molecules fixed gave only one solution. The solutions of two CCP and one cyt. c were then displayed in setor (25) and a reasonable CCP–cyt.c cross-linked pair was identified. This pair was then used as the search model to look for the remaining CCP–cyt.c pair in the asymmetric unit. Two solutions were found, corresponding to the expected two CCP–cyt.c pairs per asymmetric unit. The R factor at the end of amore was 39.3% with a correlation coefficient of 0.532. The calculated electron density maps from the molecular replacement model phases were sufficiently clear to enable the fit of side chains of CCP and cyt. c manually with the graphics program o (version 8.0) (26). The model was refined further with cns (version 1.1) (27). Simulated annealing starting at 3000 K was implemented, followed by 100 cycles of conjugate gradient minimization. Water molecules were added by the automatic water picking routine in cns. Subsequent refinements involved iterations between model adjustment of the poorly fit protein atoms, ligands, and water molecules in o and 50 cycles of positional refinement followed by B-factor refinement implemented in cns. Noncrystallographic symmetry restraints were not applied. Throughout the refinement 5% of the data was set aside to calculate Rfree for cross-validation (27). The backbone geometry was checked in procheck (28) and none of the residues were in the disallowed region. Diffraction and refinement statistics are summarized in Table 1.
Steady-State Activity. The C128A CCP mutant exhibited normal wild-type activity, and throughout this paper this mutant was used as the “wild-type” CCP control for activity measurements. The steady-state enzyme kinetics of wild-type CCP, mutant, and the cross-linked covalent complex, with horse heart and yeast ferrocyt. c as substrates, were determined as described previously (13, 14) in potassium phosphate buffer, pH 6.0. Activities were measured at two different ionic strengths of potassium phosphate buffer, pH 6.0, namely 200 mM and 20 mM. The oxidation of fully reduced ferrocyt. c (25 μM) was monitored by following the change of absorbance at 550 nm with a Cary3E UV-visible spectrophotometer at room temperature by using a Δε550 of 19,600 M–1·cm–1. Reactions were initiated by the addition of hydrogen peroxide (200 μM). The protein concentrations were typically 250–500 pM for the wide-type CCP and CCP mutants and 10 nM for the cross-link complex. Steady-state turnover values were derived by division of the initial velocity by enzyme concentration.
Single Turnover. The ability of compound I in the covalent complex to oxidize exogenous ferrocyt. c was carried out as follows. The cyt. c in the covalent complex was first fully oxidized to ferricytochrome c by potassium ferricyanide, which was subsequently removed by desalting on a PD10 column and then treated with 1 eq of H2O2 to generate compound I. This reaction was followed by the addition of 1 eq of yeast ferrocyt. c. The oxidation of ferrocyt. c was followed at 550 nm in 200 mM potassium phosphate buffer, pH 6.0, by using the Cary 3E UV-visible spectrophotometer. Control experiments consisted of free CCP compound I mixed with 1 eq of oxidized and reduced yeast cyt. c.
Stopped-Flow Kinetics. Transient pre-steady-state kinetic studies were performed with an Applied Photophysics SX 18MV-R stopped-flow spectrophotometer equipped with SpectraKinetic workstation (version 4.46) using a 10-mm pathlength cell. For rates of compound I formation, the CCP–cyt.c complex (4 μM) was mixed with various concentrations (4–20 μM) of hydrogen peroxide and the rate of compound I formation (kobs) was followed at 425 nm. The second-order rate constant was calculated from the slope of kobs vs. hydrogen peroxide concentration. All rates were determined in 200 mM potassium phosphate, pH 6.0. The intramolecular electron transfer rate in the CCP–cyt.c complex was determined by stopped flow using the procedure described previously (14) in degassed 200 mM potassium phosphate, pH 6.0. Briefly, one syringe was filled with the covalent complex in which the cyt. c had been reduced with ascorbate and the other syringe was filled with peroxide. Mixing results in the formation of CCP compound I followed by intramolecular electron transfer from ferrocyt. c to CCP compound I. Oxidation of ferrocyt. c was followed at 412 nm, an isosbestic point for CCP and CCP compound I in the complex. The CCP–cyt.c complex concentration was 3.4–5 μM, and the peroxide was added at 3- to 9-fold excess. To determine whether compound I Fe(IV)–O center or the Trp-191 cation radical accepts the electron from ferrocyt. c during intramolecular electron transfer, the reaction was followed at 430 nm, an isosbestic point for Fe(II)/Fe(III)cyt. c in the covalently cross-linked complex. A decrease at 430 nm indicates reduction of the Fe(IV)–O center.
Results and Discussion
Cross-Link Design. The main goal of engineering an interprotein disulfide bridge was to generate a CCP–cyt.c complex that mimics the crystal structure of the noncovalent complex. Our original cross-linking efforts required introducing a Cys residue at position 290 in CCP and 73 in cyt. c, which should form a covalent complex that mimics the noncovalent complex crystal structure (13). The S–S bridge formed is located at the edge of the noncovalent complex well removed from the hemes and, hence, was judged less likely to interfere with interactions at the site of electron transfer near the exposed edge of the cyt. c heme. However, this covalent complex could be produced only in low yields and consistently failed to give diffraction-quality crystals. Therefore, we sought other potential cross-linking sites that would give higher yields and, hopefully, crystallize. In the structure of the noncovalent complex (16), the Cα carbon atoms of Val-197 in CCP is 6.5 Å from Ala-81 in cyt. c. Computer modeling indicated that converting these two residues to Cys would generate a disulfide bond.
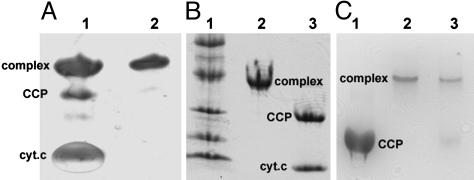
The mutants were prepared, purified, and cross-linked under mild oxidizing conditions in the presence of copper sulfate. The yield of cross-linked product was estimated from SDS/PAGE to be >60% (Fig. 1A). The disulfide bond was unexpectedly stable, because treatment with DTT followed by native PAGE showed that a majority of the complex remained cross-linked (Fig. 1C). Only under strongly denaturing conditions was the disulfide bond broken (Fig. 1B).
Fig. 1.
Electrophoretic analysis of cross-linked product. (A) SDS/PAGE under nonreducing conditions. Lanes 1 and 2 are the cross-linked product before and after purification, respectively, under nonreducing conditions. (B) Lane 1 is molecular weight standards, lane 2 is SDS/PAGE under nonreducing conditions, and lane 3 is SDS/PAGE of complex after boiling for 10 min in the presence of 150 mM DTT. (C) Native PAGE. Lane 1 is CCP alone, lane 2 is the complex not treated with DTT, and lane 3 is the complex treated with DTT (10 mM),4hat4°C.
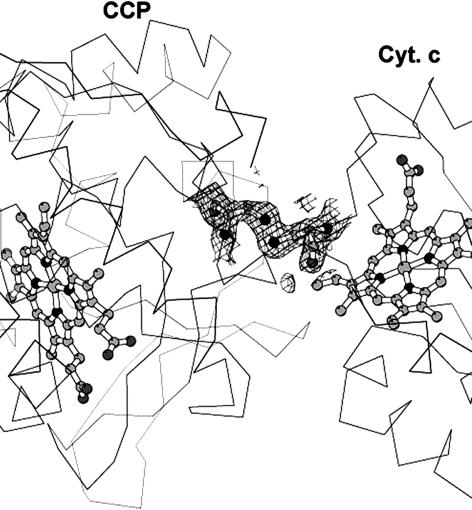
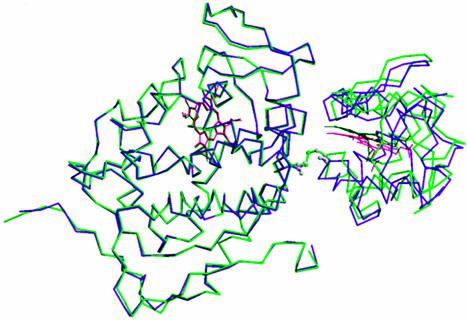
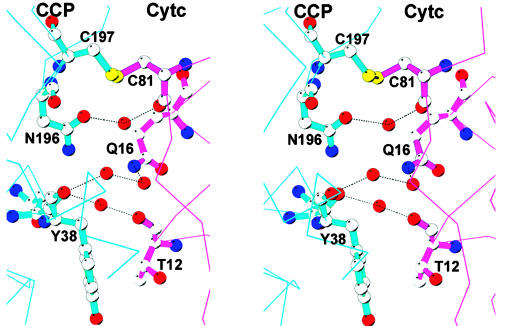
Crystal Structure. The covalent complex crystallized and the structure refined to Rcryst of 0.19 (Rfree of 0.24) at 1.88 Å (Table 1). The omit electron density map clearly shows that the S–S bond between the residues Cys-197 of CCP and Cys-81 of cyt. c is present (Fig. 2). Fig. 3 shows a superimposition of the covalent and noncovalent complexes. This figure was generated by superimposing the CCP molecules, thus providing a view of how the orientation of cyt. c relative to CCP differs in the two structures. It is clear that the cyt. c experiences a slight reorientation in the covalent complex. The difference in cyt. c Cα carbons ranges from 1 Å to 4 Å, with an overall rms deviation of 2.5 Å. A closer view of the CCP–cyt.c interface is shown in Fig. 4. The intermolecular contacts noted in the noncovalent complex (16) are maintained in the covalent complex. As in the noncovalent complex there are no strong intermolecular hydrogen bonds or ion pairs at the interface between CCP and cyt. c.
Fig. 2.
2Fobs – Fcalc composite omit electron density map contoured at 1σ showing the S–S cross-link between CCP and cyt. c. Figures were prepared withmolscript (34) and raster3d (35).
Fig. 3.
Superimposition of the covalent (green) and noncovalent (purple) CCP–cyt.c complexes. The CCP molecules were superimposed to show the slight difference in the orientation of cyt. c relative to CCP.
Fig. 4.
Stereo model showing the CCP–cyt.c interface. Note the ordered solvent molecules that bridge between polar groups in the two protein molecules. The thin lines represent the peptide backbone not in the interface, and dotted lines indicate hydrogen bonds.
A remarkable feature of our current work is the ordered water molecules trapped at the interface of CCP and cyt. c. This was not observed in the noncovalent complex, possibly because of the lower resolution of the data, 1.88 Å for the covalent complex vs. 2.3 Å for the noncovalent complex. These water molecules bridge between polar groups in CCP and cyt. c (Fig. 4). For example, a single ordered water forms an H-bonding bridge between the peptide carbonyl O atoms of Thr-12 in cyt. c and Tyr-38 of CCP, whereas a single water bridges between the peptide O atom of the mutant Cys-81 in cyt. c and the CCP Asn-196 side chain. Trapped solvent effectively solvates polar groups at the interface with no direct electrostatic intermolecular interactions between protein groups. Such water-mediated interactions are similar to what was observed in the noncovalent crystal structure of cytochrome P450-BM3 complexed with its electron-donating FMN domain (29). The complex ionic strength behavior of the CCP–cyt.c reaction (30) thus may reflect the nature of solvent structure at the interface rather than the more traditional view of protein–protein electrostatic interactions controlling complex formation.
Steady-State and Single-Turnover Experiments. Ferrocyt. c peroxidation activity by the covalently cross-linked complex has been examined by using steady-state and stopped-flow kinetics. The complex was assayed for its ability to support peroxide-dependent oxidation of both horse heart and yeast ferrocyt. c at both low and high ionic strengths. It is well known that horse heart and yeast cyt. c exhibit quite different ionic strength dependence. For horse heart ferrocyt. c, activity decreases with increasing ionic strength, whereas with yeast ferrocyt. c, activity first increases with ionic strength and then decreases. Such complex behavior has been interpreted as an ionic strength-dependent change in the rate-limiting step (30) as well as a multisite model with CCP having both high-affinity and low-affinity sites (8, 10–12). As shown in Table 2, the complex is essentially inactive (exhibits <7.2% activity compared with wild type) toward both substrates at low and high ionic strengths.
Table 2. Steady-state activity of the covalent complex (PCXL) compared with C128A (wild-type CCP) and the V197C/C128A mutant at pH 6.
|
Kcat, sec-1
|
||||
|---|---|---|---|---|
| 20 mM potassium phosphate
|
200 mM potassium phosphate
|
|||
| Enzyme | Horse cyt. c | Yeast cyt. c | Horse cyt. c | Yeast cyt. c |
| WT CCP | 850.3 ± 51.0 (100) | 219.1 ± 11.8 (100) | 175.1 ± 9.5 (100) | 1361.8 ± 57.4 (100) |
| V197C/C128A | 803.4 ± 17.0 (95) | 240.9 ± 7.3 (109) | 166.8 ± 7.2 (95) | 1167.2 ± 17.7 (86) |
| PCXL | 20.7 ± 2.0 (2.4) | 15.7 ± 0.9 (7.2) | 4.2 ± 0.09 (2.4) | 76.2 ± 4.2 (5.6) |
The values in parentheses are percent of wild-type activity.
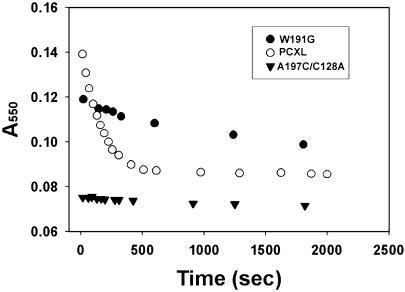
We also carried out single-turnover experiments using equal amounts of the covalent complex and exogenous ferrocyt. c. CCP in the complex was first converted to compound I by the addition of H2O2 and immediately followed by the addition of 1 eq of yeast ferrocyt. c. The oxidation of the exogenously added ferrocyt. c was followed at 550 nm on a conventional spectrophotometer. As a wild-type control, the V197C/C128A CCP mutant compound I and 1 eq of oxidized yeast cyt. c was mixed with 1 eq of yeast ferrocyt. c. As shown in Fig. 5, the ferrocyt. c is completely oxidized immediately after mixing the samples. Indeed, the reaction between CCP compound I and yeast ferrocyt. c is too fast for conventional stopped-flow spectroscopy (31). As an “inactive” control, a CCP mutant where the essential Trp-191 has been converted to Gly was used. As with the wild-type control, the mutant compound I with 1 eq of oxidized yeast cyt. c was mixed with 1 eq of yeast ferrocyt. c. In this case, ferrocyt. c oxidation was slow and not complete even after 30 min (Fig. 5). The oxidation of ferrocyt. c by compound I in the covalent complex also was very slow and required >10 min for complete oxidation of ferrocyt. c.
Fig. 5.
The rate of oxidation of exogenously added ferrocyt. c by compound I in the covalently cross-linked complex (PCXL; 3.6 μM) and by three controls: 3.4 μM wild-type CCP (data not shown), 3.4 μM V197C/C128A (C197), and 3.2 μM inactive CCP mutant W191G (G191). For wild-type CCP or V197C/C128A CCP, the ferrocyt. c is fully oxidized immediately after mixing.
The steady-state and single-turnover experiments show that the oxidation of exogenously added ferrocyt. c by CCP in the covalent complex is far too slow to be of physiological significance. This slowness implies that the cross-linked cyt. c is occupying the physiologically relevant electron transfer site and raises doubts on the relevance of a second electron transfer site. The most favored second site was proposed after using computational methods (15). This site is available for interaction with cyt. c in the covalent complex, but clearly if cyt. c does bind at this second site, the rate of compound I reduction is very slow. Because the weight of the evidence favors two cyt. c binding sites (8, 10, 11), then the second site identified by various physical probes must not be important for electron transfer. Our results do not exclude the possibility that CCP does have a second electron transfer site but if so, then this second site must overlap with the primary site identified in the crystal structures.
Stopped-Flow Experiments. Stopped-flow methods were also used to determine the reaction of the complex with peroxide and the electron transfer from the covalently tethered ferrocyt. c to CCP compound I in the complex. CCP in the complex reacts normally with H2O2 with a second-order rate constant of 1.5 × 107 M–1·sec–1 compared with 1.2 × 107 M–1·sec–1 for wild-type CCP (Fig. 6, which is published as supporting information on the PNAS web site). For the electron-transfer studies, the method we previously used for CCP–cyt.c cross-links was used (13, 14). cyt. c in the complex was first reduced with ascorbate, followed by mixing the CCP-ferrocyt. c complex with excess H2O2. With excess H2O2 compound I will form quickly, followed by electron transfer from ferrocyt. c to CCP. If the formation of compound I is faster than the intramolecular electron transfer reaction, then the stopped-flow trace will be a measure of the unimolecular electron-transfer rate. Following the reaction at 412 nm monitors the oxidation of ferrocyt. c, whereas following the reaction at 430 nm is a measure of the CCP compound I Fe(IV)–O center (32). Approximately 80% of the ferrocyt. c is oxidized in the dead-time of the instrument, ≈2 msec (Fig. 7, which is published as supporting information on the PNAS web site). This behavior is very similar to what we previously observed (13, 14) and shows that the intramolecular electron transfer from ferrocyt. c to CCP compound I in the complex is too fast to measure by using stopped-flow methods. This result was expected on the basis of photo-induced methods used to follow electron transfer in the noncovalent complex (31, 33). In addition, the absorbance at 430 nm first increases owing to the formation of Fe(IV)–O and remains constant while ferrocyt. c is oxidized. This means that the Trp-191 cation radical and not Fe(IV)–O is reduced by ferrocyt. c (32). Overall, the covalent complex exhibits functional characteristics expected if the complex mimics the physiologically relevant electron-transfer complex.
Summary. A site-specifically cross-linked CCP–cyt.c complex has been prepared and its crystal structure has been solved to 1.88 Å. The structure of the covalent complex is very similar to that of the noncovalent complex (16). The higher resolution reveals that ordered solvent molecules bridge between the two protein molecules. The stopped-flow results indicate that electron transfer from the cross-linked ferrocyt. c to the Trp-191 cation radical in CCP compound I is very fast and that CCP in the complex reacts normally with H2O2 to give compound I. These results support the view that the CCP–cyt. c interaction observed in the crystal structure of the noncovalent complex (16) represents the physiologically important electron transfer complex. The very low rate of oxidation of exogenously added ferrocyt. c by CCP compound I in the covalent complex indicates that the previously characterized second cyt. c binding site is not efficient at reducing compound I.
Supplementary Material
Acknowledgments
Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Laboratory Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. This work was supported by National Institutes of Health Grant GM 42614.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CCP, cytochrome c peroxidase; cyt. c, oxidized cytochrome c; ferrocyt. c, reduced cytochrome c; CCP–cyt.c, CCP–cyt.c covalent complex; V197C/C128A, CCP with Val-197 replaced by Cys and Cys-128 replaced by Ala; W191G, CCP with Trp-191 replaced by Gly; A81C cyt. c, cyt. c with Ala-81 replaced by Cys.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1S6V).
References
- 1.Mayo, S. L., Ellis, W. R., Crutchley, R. J. & Gray, H. B. (1986) Science 233, 948–952. [DOI] [PubMed] [Google Scholar]
- 2.Winkler, J. R. & Gray, H. B. (1992) Chem. Rev. 92, 369–379. [Google Scholar]
- 3.Moser, C. C., Keske, J. M., Wanrcke, K., Farid, R. S. & Dutton, P. P. (1991) Nature 355, 796–802. [DOI] [PubMed] [Google Scholar]
- 4.Yonetani, T. (1976) in The Enzymes, ed. Boyer, P. (Academic, New York), Vol. 13, pp. 345–361. [Google Scholar]
- 5.Sivaraja, M., Goodin, D. B., Smith, M. & Hoffman, B. M. (1989) Science 245, 738–740. [DOI] [PubMed] [Google Scholar]
- 6.Wang, X. & Pielak, G. J. (1999) Biochemistry 38, 16878–16881. [DOI] [PubMed] [Google Scholar]
- 7.Pielak, G. J. & Wang, X. (2001) Biochemistry 40, 422–428. [DOI] [PubMed] [Google Scholar]
- 8.Kornblatt, J. A. & English, A. M. (1986) Eur. J. Biochem. 155, 505–511. [DOI] [PubMed] [Google Scholar]
- 9.Stemp, E. D. & Hoffman, B. M. (1993) Biochemistry 32, 10848–10865. [DOI] [PubMed] [Google Scholar]
- 10.Zhou, J. S. & Hoffman, B. M. (1994) Science 265, 1693–1696. [DOI] [PubMed] [Google Scholar]
- 11.Zhou, J. S., Nocek, J. M., DeVan, M. L. & Hoffman, B. M. (1995) Science 269, 204–207. [DOI] [PubMed] [Google Scholar]
- 12.Mauk, M. R., Ferrer, J. C. & Mauk, A. G. (1994) Biochemistry 33, 12609–12614. [DOI] [PubMed] [Google Scholar]
- 13.Pappa, H. S. & Poulos, T. L. (1995) Biochemistry 34, 6573–6580. [DOI] [PubMed] [Google Scholar]
- 14.Pappa, H. S., Tajbaksh, S., Saunders, A. J., Pielak, G. J. & Poulos, T. L. (1996) Biochemistry 35, 4837–4845. [DOI] [PubMed] [Google Scholar]
- 15.Northrup, S. C., Boles, J. O. & Reynolds, J. C. (1988) Science 241, 67–70. [DOI] [PubMed] [Google Scholar]
- 16.Pelletier, H. & Kraut, J. (1992) Science 258, 1748–1755. [DOI] [PubMed] [Google Scholar]
- 17.Cutler, R. L., Pielak, G. L., Mauk, A. K. & Smith, M. (1987) Protein Eng. 1, 95–99. [DOI] [PubMed] [Google Scholar]
- 18.Pollock, W. B., Rosell, F. I., Twitchett, M. B., Dumont, M. E. & Mauk, A. G. (1998) Biochemistry 37, 6124–6131. [DOI] [PubMed] [Google Scholar]
- 19.Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. (1989) Gene 77, 61–68. [DOI] [PubMed] [Google Scholar]
- 20.Fishel, L. A., Farnum, M. F., Mauro, M., Miller, M. A., Kraut, J., Liu, Y. L. & Scholes, C. P. (1991) Biochemistry 30, 1986–1996. [DOI] [PubMed] [Google Scholar]
- 21.Choudhury, K., Sundaramoorthy, M., Hickman, A., Yonetani, T., Woehl, E., Dunn, M. F. & Poulos, T. L. (1994) J. Biol. Chem. 269, 20239–20249. [PubMed] [Google Scholar]
- 22.Leslie, A. G. W. (1992) Jt. CCP4/ESF-EACMB Newslett. Protein Crystallogr. No. 26.
- 23.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 24.Navaza, J. (1994) Acta Crystallogr. A 50, 157–163. [Google Scholar]
- 25.Evans, S. V. (1993) J. Mol. Graphics 11, 134–138. [DOI] [PubMed] [Google Scholar]
- 26.Jones, T. A. & Kjeldgaard, M. (1994) o, The Manual (Uppsala Univ. Press, Uppsala).
- 27.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 28.MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1991) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 29.Sevrioukova, I. F., Li, H., Zhang, H., Peterson, J. A. & Poulos, T. L. (1999) Proc. Natl. Acad. Sci. USA 96, 1863–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, M. A. (1996) Biochemistry 35, 15791–15799. [DOI] [PubMed] [Google Scholar]
- 31.Geren, L., Hahm, S., Durham, B. & Millett, F. (1991) Biochemistry 30, 9450–9457. [DOI] [PubMed] [Google Scholar]
- 32.Hahm, S., Geren, L., Durham, B. & Millett, F. (1993) J. Am. Chem. Soc. 115, 3372–3373. [Google Scholar]
- 33.Hahm, S., Durham, B. & Millett, F. (1992) Biochemistry 31, 3472–3477. [DOI] [PubMed] [Google Scholar]
- 34.Kraulis, P. J. (1994) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 35.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.