Abstract
The Cu- and Zn-containing superoxide dismutase 1 (SOD1) largely obtains Cu in vivo by means of the action of the Cu chaperone CCS. Yet, in the case of mammalian SOD1, a secondary pathway of activation is apparent. Specifically, when human SOD1 is expressed in either yeast or mammalian cells that are null for CCS, the SOD1 enzyme retains a certain degree of activity. This CCS-independent activity is evident with both wild-type and mutant variants of SOD1 that have been associated with familial amyotrophic lateral sclerosis. We demonstrate here that the CCS-independent activation of mammalian SOD1 involves glutathione, particularly the reduced form, or GSH. A role for glutathione in CCS-independent activation was seen with human SOD1 molecules that were expressed in either yeast cells or immortalized fibroblasts. Compared with mammalian SOD1, the Saccharomyces cerevisiae enzyme cannot obtain Cu without CCS in vivo, and this total dependence on CCS involves the presence of dual prolines near the C terminus of the SOD1 polypeptide. Indeed, the insertion of such prolines into human SOD1 rendered this molecule refractory to CCS-independent activation. The possible implications of multiple pathways for SOD1 activation are discussed in the context of SOD1 evolutionary biology and familial amyotrophic lateral sclerosis.
Eukaryotic Cu- and Zn-containing superoxide dismutase 1 (SOD1) is a key superoxide scavenging enzyme that is largely localized in the cytosol but is also found in the intermembrane space of mitochondria and in other organelles as well (1–5). In humans, dominant mutations scattered throughout the SOD1 enzyme have been linked to the fatal motor neuron disease familial amyotrophic lateral sclerosis (FALS) (6, 7). However, the mechanism underlying SOD1 toxicity in FALS is still unresolved. Both protein aggregation and oxidative models have been invoked, and these models are not necessarily mutually exclusive (8). Perhaps most controversial has been the possible role of Cu ions bound to SOD1 in promoting toxicity (reviewed in refs. 9–12).
The catalytic Cu cofactor for SOD1 can be inserted into the enzyme in vivo by means of the action of a Cu chaperone known as CCS. CCS was originally discovered in yeast as the product of the Saccharomyces cerevisiae LYS7 gene (13), and this Cu chaperone is widely conserved throughout eukaryotes. S. cerevisiae SOD1 depends absolutely on CCS in vivo, as evidenced by the fact that yeast cells lacking CCS express a SOD1 polypeptide that is apo for Cu and is inactive (13–15).
Recently, a mouse strain bearing a homozygous deletion in CCS was created (16), and these CCS–/– mice were crossed to transgenic mouse models for SOD1-linked FALS. In each case, the absence of CCS did not preclude motor neuron disease effected by mutant SOD1, demonstrating that Cu inserted by means of CCS is not required for SOD1-linked FALS (17). These studies revealed also that a limited degree of SOD1 activity persists in mice lacking CCS (16, 17); hence, mammalian SOD1 can obtain some Cu independent of CCS. This secondary pathway for activating SOD1 may not be unique to the mammalian cell host. Corson et al. (18) have provided evidence that human wild-type and FALS mutant SOD1 molecules also retain activity when expressed in lys7Δ yeast cells lacking CCS. Although the mechanism for CCS-independent activation is not understood, it has been suggested as a factor worth considering in SOD1-linked FALS (19).
The studies presented here probe the mechanism responsible for CCS-independent activation of mammalian SOD1. We find that reduced glutathione (GSH) is needed for activating mammalian SOD1 with Cu in both yeast and murine cells null for CCS. Compared with mammalian SOD1, the SOD1 enzyme from S. cerevisiae cannot obtain Cu independent of CCS, due in part to sequences near the C terminus of the SOD1 polypeptide. In particular, the presence of dual prolines at positions 142 and 144 prohibit activation of yeast SOD1 in the absence of CCS.
Materials and Methods
Yeast Strains and Growth Conditions. All strains of S. cerevisiae were maintained by growth in GasPak anaerobic culture jars (BBL). Plasmid transformed cells were grown on SD synthetic selecting medium that was supplemented with 1 mM glutathione for cultivation of gsh1Δ mutants. Yeast strains KS107 (sod1Δ:TRP1) (20), LS101 (sod1Δ::TRP1 lys7Δ::URA3) (4), PS131 (lys7Δ::TRP1), PS132 (lys7Δ::TRP1 gsh1Δ::LEU2), and MC107 (sod1Δ::TRP1 gsh1Δ::LEU2) were all derived from the parental strain EG103 (MATα leu 2–3, 112 his3Δ1 trp1–289 ura3–52). Strains 2737 (glr1Δ::kanMX4) and 614 (lys7Δ::kanMX4) are derivatives of BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0; Research Genetics, Huntsville, AL). The glr1Δ::kanMX4 lys7Δ::LEU2 mutant MC112 was generated by deleting LYS7 in strain 2737.
Plasmids. Plasmid pLC1 (2μ URA3) expresses human wild-type SOD1 under the control of S. cerevisiae PGK1, as described (18). The human SOD1 mutant S142P,L144P was obtained by site-directed mutagenesis of pLC1 (according to the manufacturer's guidelines; QuikChange kit, Stratagene). The FALS SOD1 mutants E100G, E100K, L144F, and D101G were a gift from Jorge Rodriguez and Peter Doucette (University of California, Los Angeles) and were obtained by mutagenesis of YEp351-hSOD1 (21). Plasmids pLS101 (CEN LEU2) (4) and pLJ175 (identical to pLS101 except URA3) express S. cerevisiae SOD1 and were used to derive by mutagenesis ySOD1 mutants G122K (from pLS101), D130G,T131N (from pLS101), and P142S,P144L (from pLS101 and pLJ175). The DNA sequence integrity of all plasmids was verified by double-stranded DNA sequencing (The Johns Hopkins University Core facility).
Biochemical Analyses. For analysis of SOD1 activity from yeast cells, strains were grown in selecting SD medium to an OD600 of ≈1.5. With experiments involving gsh1Δ mutants, cells proceeded through five to six doublings in SD medium lacking glutathione before harvesting. As such, glutathione was depleted in these cells to levels beneath detection (see Fig. 3). Cell lysates were prepared by glass-bead homogenization in a buffer devoid of reducing agents as described (22), and SOD was activity assayed by nondenaturing gel electrophoresis at 4°C and staining with nitro blue tetrazolium (23, 24). With the exception of the nondenaturing gels of Fig. 2C, all experiments used 12% precast gels (NOVEX, San Diego) lacking EDTA. As shown in Fig. 2C, an 8% acrylamide gel solution was prepared and, where indicated, 0.1 mM EDTA was included in both the stacking and resolution compartments as well as the electrophoresis buffer. For immunoblot analyses, cell lysates were subjected to 14% denaturing PAGE (precast gels, NOVEX) and probed with antibodies specific to human (24) or yeast (4) SOD1 or an antibody that recognizes both murine and human SOD1 (25). The activity gels and immunoblots were subjected to densitometric tracings by using nih image software. Integrated density of pixel intensity was recorded, and the results are presented in Fig. 6, which is published as supporting information on the PNAS web site.
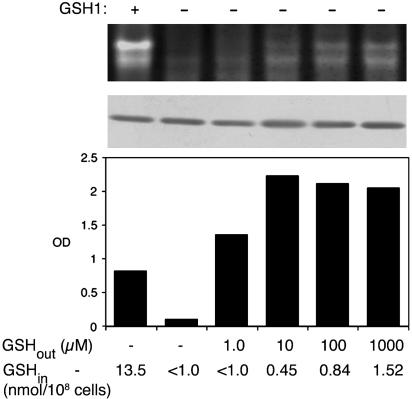
Fig. 3.
Distinct concentrations of glutathione are required to support cell growth and to stimulate hSOD1 activity in vivo. The indicated strains expressing hSOD1 from pLC1 were grown in SD medium supplemented with the designated concentrations of glutathione (GSHout). After five to six doublings, an aliquot of cells were harvested and assayed for hSOD1 activity by the native gel assay (Top), for SOD1 polypeptide levels by immunoblotting (Middle), and for intracellular glutathione levels (GSHin). Glutathione measurements represent the average of duplicate samples, in which range was <10%. To help correct for the increased levels of SOD1 protein in gsh1Δ cells¶, only approximately one-third as much total cellular protein was assayed in the native gel and immunoblot in the case of untreated GSH– cells. Cells that were not harvested were diluted to OD600 = 0.075 and cultured for an additional 14 h under the same glutathione-treatment conditions, and total cell growth was determined by absorbance at 600 nm (Bottom). The following strains were used: GSH1+, PS131 (lys7Δ); and GSH1–, PS132 (lys7Δ gsh1Δ).
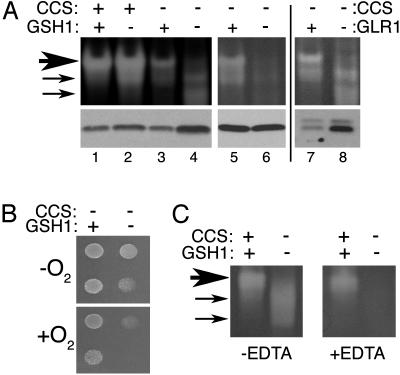
Fig. 2.
Glutathione depletion inhibits CCS-independent activation of human SOD1. The indicated yeast strains expressing wild-type hSOD1 from pLC1 were tested either for SOD activity and hSOD1 polypeptide levels, as shown in Fig. 1 (A); lysine independent growth in air (B); or effect of metal chelators during gel electrophoresis (C). The following strains were used: CCS+ GSH1+, KS107 (sod1Δ); CCS+ GSH1–, MC107 (sod1Δ gsh1Δ); CCS– GSH1+, PS131 (lys7Δ); CCS– GSH1–, PS132 (lys7Δ gsh1Δ); CCS– GLR1+, 614 (lys7Δ); and CCS– GLR1–, MC112 (lys7Δ glr1Δ). (A) Cells were grown for five to six doublings in SD medium before preparation of cell lysates. We assayed 40 μg of cellular protein, except for in lane 6, which contained 15 μg to help normalize hSOD1 polypeptide levels. (B) We spotted 2 × 104 and 2 × 103 cells of the indicated strains onto SD medium lacking lysine and allowed them to grow for 2 days either in air (+O2) or in anaerobic culture jars (–O2). (C) Native gel electrophoresis and nitro blue tetrazolium staining for SOD1 activity was carried out as described for A except that 8% polyacrylamide gels were poured that contained 0.1 mM EDTA where indicated. (All other native gels of this study used 12% precast gels that were not treated with EDTA.) Large arrow indicates position of major hSOD1 activity species, whereas smaller arrows indicate hSOD1 that was apparently acquired Cu during electrophoresis (see Discussion).
Total glutathione was measured by the 5,5′-dithiobis(2-nitrobenzoic acid)-glutathione reductase (DTNB-GR) recycling assay (26–28). For yeast, cells were grown for five to six doublings in SD medium, and 1 × 108 cells were harvested and subjected to glass-bead homogenization in the presence of 250 μl of 8 M HCl/1.3% sulfosalicylic acid. Proteins were precipitated on ice for 15 min, followed by centrifugation at 13,000 × g for 15 min. We subjected 40 μl of the clarified supernatants to glutathione analysis in a 1-ml reaction for DTNB-GR recycling, as described (26, 27). Analysis of total glutathione levels in fibroblasts involved a procedure that was similar except that sulfosalicylic acid was added to a final concentration of 1% after the isolation of cell lysates, and glutathione measurements were carried with 20 μl of sample.
Cell Lines and Tissue Culture Conditions. CCS+/+, CCS–/–, G37R;CCS+/+, and G37R;CCS–/– fibroblasts, derived from abdominal skin of adult mice, were cultured in DMEM, supplemented with 10% FBS/fungizone (1 μg/ml)/1× pencillin/streptomycin at 37°C/5% CO2. For immortalization, primary fibroblasts were trypsinized and plated at a density of 15,000 cells in 35-mm culture dishes and transfected with a plasmid containing SV40 large T-antigen (a kind gift from M. Tevethia, Pennsylvania State University Medical Center, Hershey; ref. 29) by using the Lipofectamine (Invitrogen) method. Transformed cells were collected and cloned by limiting dilution, and they were subsequently expanded in 24-well plates.
To culture cells for biochemical analyses, 5 × 105 cells were seeded in 75-cm2 flasks, and after a 2-h incubation, they were supplemented with buthionine sulfoximine (BSO) to a final concentration of 50 μM or the same volume of water vehicle control. After growth for 48 h (two to four generations), cells were split and allowed to grow for an additional 48 h before harvesting. Cells were scraped in PBS and subjected to two rounds of freeze–thaw lysis, followed by centrifugation at 13,000 × g for 5 min (30). A portion of each supernatant was acidified for glutathione determination, and the remaining portion was used for activity gel assay and Western blotting, as described above.
Results
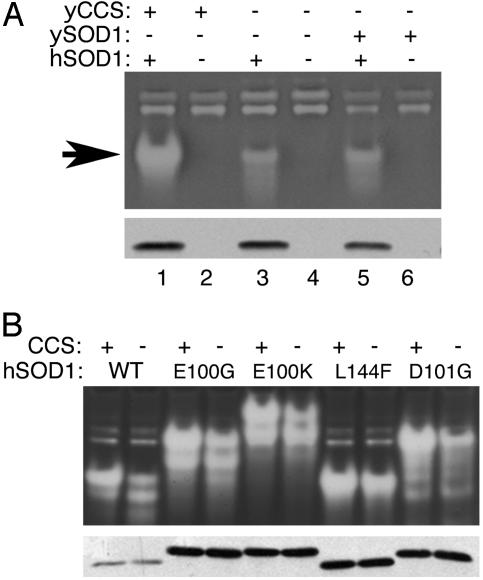
CCS-Independent Activation of Human Wild-Type and FALS Mutant SOD1 Expressed in Yeast. To study the requirement for CCS in activating human SOD1 (hSOD1), we first used a yeast expression system. Lysates from yeast cells expressing hSOD1 were subjected to the native gel assay for SOD activity. As shown in Fig. 1A, hSOD1 exhibited significant activity in both sod1Δ (Fig. 1 A, lane 1) and lys7Δ sod1Δ cells that lack CCS (Fig. 1 A, lane 3), although activity was reduced in the latter case. The same result was observed in a lys7Δ strain that lacks CCS but expresses endogenous yeast SOD1 (ySOD1) (Fig. 1 A, lane 5). The activation of wild-type hSOD1 in yeast cells lacking CCS is highly reproducible and ranges from ≈25% to 50% that of the CCS-dependent activity (see Fig. 6 for densitometric tracings for Figs. 1, 2, and 5). Compared with results obtained with hSOD1, there was no detectable activity from ySOD1 expressed in the strain lacking CCS (Fig. 1 A, lane 6). The complete dependence of ySOD1 on CCS for activity (also shown in Fig. 5B) is consistent with several studies (4, 13, 31, 32).
Fig. 1.
Activity of human wild-type and ALS mutant SOD1 in yeast cells lacking CCS. The indicated yeast strains were grown in SD medium to confluency over night. Forty micrograms of lysate protein was subject to either native gel electrophoresis and nitro blue tetrazolium staining for SOD activity (A Upper and B Upper) or to denaturing SDS gel electrophoresis and immunoblotting using an antibody directed against human SOD1 (A Lower and B Lower). (A) Strains were transformed with either pLC1 expressing human wild-type SOD1 (hSOD1, +) or pSM703 empty vector control (hSOD1, –). The following strains were used: yCCS+ ySOD1–, KS107 (sod1Δ); yCCS– ySOD1–, LS101 (sod1Δ lys7Δ); and yCCS– ySOD1+, PS131 (lys7Δ). Arrow indicates position of hSOD1. The activity band of slower migration represents S. cerevisiae SOD2. (B) Strains were transformed with either pLC1 (WT) or with the YEp351-hSOD1-based plasmids (21) for expression of the indicated FALS mutant alleles of SOD1. The following strains were used: CCS+, KS107; and CCS–, LS101. The YEp351-hSOD1-based plasmids generally yield higher overall expression levels than those of pLC1. The effects of point mutations on hSOD1 mobility on both native and denaturing gels are not unusual (30).
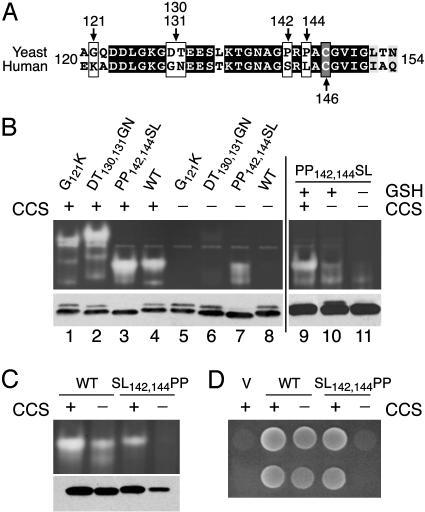
Fig. 5.
The role of C-terminal prolines in determining the dependence of SOD1 on CCS. (A) An alignment of the C-terminal region of S. cerevisiae and human SOD1. Black and gray residues represent amino acid identity and similarity, respectively. White boxes mark positions where amino acid substitutions were introduced. Gray box indicates conserved disulfide cysteine. (B) The indicated strains expressing wild-type (WT) or the mutant variants of yeast SOD1 were subject to the native gel assay for SOD1 activity (Upper) and immunoblot analysis for ySOD1 using an antibody directed against yeast SOD1 (Lower). To help correct for the higher SOD1 polypeptide levels seen in gsh1Δ mutants¶,1/3 as much total cellular protein was assayed in lane 11 compared with lanes 9 and 10. (C) Activity and polypeptide levels of human wild-type (WT) and mutant hSOD1 expressed in yeast was assayed as in Fig. 1. (D) The indicated strains transformed with empty pSM703 vector (V) or plasmids expressing wild-type (WT) or the indicated mutant hSOD1 were assayed for lysine-independent aerobic growth, as described for Fig. 2B. The following strains were used in B, lanes 1–8, C, and D: CCS+, KS107 (sod1Δ); and CCS–, LS101 (sod1Δ lys7Δ). The following strains were used in B, lanes 9–11: CCS+ GSH1+, KS107 (sod1Δ); CCS– GSH1+, LS101 (sod1Δ lys7Δ); and CCS– GSH1–, PS132 (lys7Δ gsh1Δ).
The CCS-independent activation of hSOD1 was also examined in a wide array of mutant molecules associated with FALS. Fig. 1B shows the results with mutants E100G, E100K, L144F, and D101G, and the same pattern was observed with mutants G37R, G93A, G41D, G41S, A4V, and I112T (data not shown). All mutants retained substantial activity in lys7Δ mutants lacking CCS. The FALS mutations as a whole do not appear to interfere with the CCS-independent pathway, at least in the case of mutants that retain enzymatic activity (33).
A Role for Glutathione in the CCS-Independent Pathway for Activating SOD1. We used a genetic approach to identify the transacting factor(s) responsible for CCS-independent activation of hSOD1. Small Cu-binding proteins were examined, including the Cu carriers encoded by S. cerevisiae ATX1 and COX17 (34), as well as the CUP1 and CRS5 Cu metallothioniens (35, 36). However, deletion of these various genes had no effect on CCS-independent activation of hSOD1 (data not shown).
Glutathione has been shown to donate Cu to SOD1 in vitro (37–39) and to play an important role in Cu homeostasis in vivo (40–43). In S. cerevisiae, glutathione depletion can be achieved by a gsh1Δ mutation encoding γ-glutamyl cysteine synthetase 1 (GSH1), catalyzing the rate-limiting step in glutathione biosynthesis (44). When hSOD1 was expressed in a lys7Δ gsh1Δ mutant (referred to throughout as CCS– GSH1–), there was a substantial effect on activity, as shown in the native gel. Specifically, the major hSOD1 activity species (Fig. 2, large arrow) was virtually abolished, although some minor activity species of faster migration (Fig. 2, small arrows) became evident (Fig. 2A, lane 4). We have noted that in gsh1Δ mutants, the SOD1 polypeptide generally accumulates to ≈3-fold higher levels compared with isogenic GSH1+ yeast∥ (Fig. 2A Lower, compare lanes 3 and 4). Therefore, to assess more accurately the effects of gsh1Δ on hSOD1 enzyme activity, we adjusted for differences in hSOD1 protein levels. When roughly similar hSOD1 protein levels were applied to the gel (Fig. 2A, lanes 5 and 6), hSOD1 activity was clearly inhibited in CCS– GSH1– cells, indicating that glutathione is somehow important for CCS-independent activation of hSOD1.
It is curious that some aberrant activity appears on gels with lysates from CCS– GSH1– cells expressing hSOD1 (Fig. 2, small arrows). However several lines of evidence indicate that these aberrant activity species do not represent hSOD1 activity in vivo. First, hSOD1 expressed in CCS– GSH1– cells does not readily support aerobic growth of yeast on medium lacking lysine (Fig. 2B). This is a very sensitive in vivo test for SOD1 that requires ≤2% of the normal endogenous SOD1 activity (18). Moreover, the aberrant in vitro activity associated with CCS– GSH1– lysates was not affected by metal chelators applied either to live yeast cultures or to cell-free lysates (data not shown), but this same activity was very sensitive to metal chelators applied during electrophoresis (Fig. 2C). Specifically, electrophoresis on gels containing 0.1 mM EDTA prevents apo SOD1 from acquiring Cu during electrophoresis (17), and this treatment eliminated SOD1 activity from lysates of CCS– GSH1– cells (Fig. 2C). We propose that these aberrant activity species represent a form of SOD1, which is capable of being activated by Cu during electrophoresis (see Discussion).
Loss of glutathione clearly inhibits hSOD1 activity in cells lacking CCS, but what is the effect of glutathione depletion in cells that express CCS? As shown in Fig. 2 A, lane 2, hSOD1 that was expressed in a gsh1Δ (CCS+ GSH1–) cell exhibited no loss of activity. If anything, activity was increased, and this increase correlated with an increase in hSOD1 polypeptide levels.¶ Therefore, under conditions in which CCS is active, glutathione does not appear to be essential for hSOD1 activity.
To test whether GSH or the oxidized form of glutathione (GSSG) is needed for CCS-independent activation, we used a glr1Δ mutant encoding glutathione reductase. Total levels of glutathione are not affected in glr1Δ mutants; however, the fraction of glutathione that is GSH is reduced drastically (45, 46). As shown in Fig. 2A, lanes 7 and 8, a glr1Δ mutant lacking CCS mimicked the effect of a gsh1Δ mutant and resulted in higher SOD1 polypeptide levels compared with wild type and in a loss of the major hSOD1 activity band. Hence, it is reduced GSH that appears to be necessary for CCS-independent activation of hSOD1.
The presence of GSH is not only important for CCS-independent activation of hSOD1 but is also needed for cell growth. In the experiment shown in Fig. 3, we supplemented cultures of CCS– GSH1– yeast with various concentrations of glutathione to test whether activation of hSOD1 followed GSH-stimulation of growth. As shown in Fig. 3 Top, significant hSOD1 activity required treatment with 100 μM to 1.0 mM glutathione, and even then, activity levels were less than that of the control GSH1+ cells (see densitometric tracings in Fig. 6). By comparison, cell growth was substantially restored by supplementing cultures with 1 μM glutathione (Fig. 3 Bottom). Measurements of total cellular glutathione revealed that CCS-independent activation of SOD1 requires at least 10 times more intracellular glutathione than is needed to support growth (Fig. 3 Bottom). Hence, the GSH effect on hSOD1 activity is not simply due to GSH– promotion of cell growth.
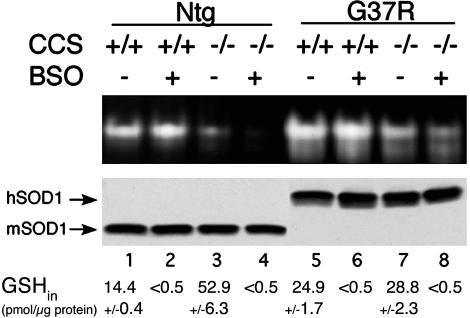
We also tested whether glutathione is important for CCS-independent activation of SOD1 in a mammalian cell host. Immortalized fibroblasts from mice that were either homozygous wild type (CCS+/+) or null (CCS–/–) for CCS (16), and expressing where indicated, FALS SOD1 mutant G37R (line 42, ref. 17) were cultured in the presence of 50 μM BSO to deplete intracellular glutathione. As shown in the top of Fig. 4, both the endogenous mouse SOD1 (lane 3) and the transgenic human G37R mutant (lane 7) retained some activity in cell lines null for CCS. This activity in CCS–/– cells was decreased in BSO treated cells, with no change in SOD1 polypeptide levels (Fig. 4, lanes 4 and 8). Although the effect of BSO treatment was not as dramatic as seen with a yeast gsh1Δ mutation, the trends were the same; i.e., glutathione depletion in mammalian cells resulted in inhibition of hSOD1 activity in CCS–/– cells (Fig. 4, lanes 4 and 8) but not in CCS+/+ cells (Fig. 4, lanes 2 and 6).
Fig. 4.
Glutathione depletion inhibits CCS-independent activation of mammalian SOD1 in fibroblasts. Immortalized fibroblasts from CCS+/+ and CCS–/– mice that were either nontransgenic for hSOD1 (Ntg) or transgenic for G37R hSOD1 (line 42, ref. 17) were cultured in the presence or absence of 50 μM BSO to deplete intracellular glutathione. We subjected 20 μg of lysate protein to SOD1 activity analysis by the native gel, as described for Fig. 1, and to immunoblot analysis to monitor SOD1 polypeptide levels by using an antibody that recognizes both human and mouse SOD1 (lanes 1–4) or is specific to hSOD1 (lanes 5–8). GSHin, total intracellular glutathione levels, where numbers represent the averages of at least two independent experimental trials; range is indicated below the blots.
SOD1 Sequences That Facilitate CCS-Independent Activation of SOD1. Even though mammalian SOD1 can acquire Cu independent of CCS, the yeast SOD1 polypeptide solely depends on CCS in vivo (14). A sequence comparison between human and fungal SOD1 proteins revealed that hSOD1 contains two cysteines (Cys-6 and Cys-111) that are absent in S. cerevisiae SOD1. However, these potential Cu binding residues are not critical because mutation of these to serines had no effect on CCS-independent activation of hSOD1 (data not shown).
Cu(I)–GSH complexes can donate Cu to SOD1 in vitro (38, 39), and by using modeling studies, a potential Cu–GSH docking site has been noted on SOD1 that approaches the electrostatic channel and C terminus (37). In this region of the polypeptide, a number of residues are not conserved between yeast and human SOD1 (Fig. 5A). By means of site-directed mutagenesis, we introduced a subset of the corresponding hSOD1 sequences into the yeast protein. As shown in Fig. 5B, all of the resultant yeast SOD1 mutants showed roughly equivalent activities when expressed in yeast cells that contain CCS (Fig. 5A, lanes 1–4). However, one mutation had a dramatic effect on SOD1 activity in cells lacking CCS. In particular, PP142,144SL imparted a gain-of-function to yeast SOD1, and this molecule exhibited significant activity without CCS (lane 7). Furthermore, PP142,144SL ySOD1 showed a dependence on cellular GSH similar to human SOD1 (Fig. 5B, lane 11). These results indicated that the two prolines near the C terminus of ySOD1 prevent CCS-independent activation. To confirm this notion, we created the reciprocal mutation in human SOD1. As shown in Fig. 5C, SL142,144PP hSOD1 was active in cells expressing CCS, but no activity could be detected in cells lacking CCS. In fact, SL142,144PP hSOD1 failed to support lysine-independent aerobic growth in CCS– cells (Fig. 5D), indicating that SOD1 activity is inhibited by >98% (18). Together, these studies demonstrate an important role for sequences near the C terminus of SOD1 in promoting CCS-independent activation of SOD1.
Discussion
The CCS Cu chaperone represents a major source of Cu for SOD1 in vivo. Yet CCS is not the only means by which SOD1 can be activated. Specifically, mammalian SOD1 can acquire Cu independent of CCS, and we show here that this auxiliary pathway depends on the presence of GSH. This GSH-dependent pathway is conserved in S. cerevisiae and mammalian cell hosts and can act on both wild-type and FALS mutant variants of human SOD1.
Although GSH is clearly important for SOD1 activation in CCS null cells, is this also true in cells that express CCS? Under the laboratory conditions studied here, there was no inhibitory effect on human SOD1 when cells with abundant CCS were depleted of glutathione (Figs. 2 and 4). Yet it is conceivable that under certain in vivo conditions CCS activity may be limiting, placing more emphasis on the GSH pathway. For an enzyme as critical as SOD1, having multiple pathways for activation should represent an evolutionary advantage.
It has long been known that glutathione can participate in intracellular Cu homeostasis. In mammalian cells, newly imported Cu appears to be complexed first by glutathione, before appearing in Cu-containing molecules such as metallothionein and SOD1 (40, 41). Glutathione also plays an important role in Cu sequestration and detoxification in vivo (40, 42, 43). In cell-free systems, Cu(I) complexes of GSH have been shown to transfer Cu to both metallothionein (47) and to SOD1 (37–39). As such, it is possible that Cu(I)–GSH may also serve as a Cu donor for human SOD1 in vivo. However, we cannot exclude alternative or more indirect roles for GSH in the CCS-independent activation of SOD1. For example, GSH may provide the reducing equivalents necessary for Cu activation by a separate molecule. Treatment of S. cerevisiae cells with other reducing agents such as DTT or ascorbate failed to mimic the effects of glutathione in activating SOD1 (data not shown). Yet these negative results should be interpreted with caution because high levels of intracellular GSH are required for SOD1 activation (Fig. 3), and these alternative reductants may not be taken up efficiently by the cell.
The maturation of SOD1 into a fully active enzyme requires several steps including the insertion of Cu, the insertion of Zn, and oxidation of a critical disulfide (48, 49). The exact order by which these events occur in vivo is not completely clear. It is likely that without Cu insertion, the SOD1 polypeptide can still exist in a partially maturated state. For example, hSOD1 from CCS– GSH1– cells can acquire activity during electrophoresis (Fig. 2). In cells lacking CCS and depleted for glutathione, hSOD1 might still harbor an oxidized disulfide, particularly under the oxidizing conditions of low intracellular glutathione. It is not clear whether Zn is present in the “gel-activated” form of SOD1, but the absence of Zn might explain the aberrant migration seen on native gels (Fig. 2).
Perhaps one of the most surprising findings of these studies is the role of C-terminal SOD1 sequences in dictating the pathway for Cu activation. Specifically, the presence of dual prolines at positions 142 and 144 in S. cerevisiae SOD1 preclude this molecule from acquiring Cu independent of CCS. These prolines are indeed critical because introduction of these residues into human SOD1 rendered this molecule refractory to Cu activation without CCS. In the crystallographic analysis of structure, these two trans configuration prolines may be limiting the flexibility of this portion of the protein. It is noteworthy that these prolines are closely juxtaposed to Cys-146 of the critical disulfide in SOD1 (Fig. 5A); these prolines might then impinge on the mechanism of formation of the disulfide bond.
Even more surprising is the infrequent occurrence of these prolines across eukaryotes. An inspection of SOD1 sequences from nearly 80 distinct organisms (including fungi, plants, insects, fish, avians, and mammals) showed that P142 and/or P144 are largely restricted to Ascomycota fungi (see Table 1, which is published as supporting information on the PNAS web site). SOD1 molecules from other eukaryotes largely show G/A/S and L/A/V/I at positions corresponding to 142 and 144, respectively (Table 1). A major exception is avian SOD1, which contains a proline at position 144 in both of the available bird SOD1 sequences (from the chicken Gallus gallus, GenBank accession no. U28407; and from the Australia parakeet Melopsittacus undulates, GenBank accession no. AAO72711); yet the effect of a single proline is uncertain. Based on our findings with dual prolines (Fig. 5), it seems reasonable to predict that only a small fraction of SOD1 molecules have evolved to depend completely on CCS. A total dependence on CCS, such as seen in specific fungi, might reflect the organism's need to tightly control the pool of Cu that populates SOD1.
Finally, is the CCS-independent pathway relevant to SOD1-linked FALS? It is too early to predict, but it appears that GSH might also damage or alter the SOD1 protein because SOD1 typically shows evidence of decreased accumulation or decreased stability in cells with abundant GSH.∥ As such, it is possible that the glutathione-dependent pathway might play some role in toxicity related to specific mutant alleles of SOD1.
Supplementary Material
Acknowledgments
We thank Soshanna Zittin Potter for computer analysis of SOD1 trans prolines; D. R. Borchelt for helpful discussions and for the mammalian SOD1 antibodies; E. B. Gralla and members of the O'Halloran laboratory for critical review of the manuscript; and L. Jensen for valuable discussions and for assistance in preparation of this article. This work was supported by The Johns Hopkins University National Institute of Environmental Health center; the Robert Packard Center for Amyotrophic Lateral Sclerosis Research at The Johns Hopkins University; and National Institutes of Health Grants GM 50016 (to V.C.C.), NS 40014 (to P.C.W.), and GM28222 (to J.S.V.). J.B.G. and J.L.U. are supported by National Institute of Environmental Health Sciences Training Grant ES 07141.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SOD1, superoxide dismutase 1; CCS, Cu chaperone for SOD1; FALS, familial amyotrophic lateral sclerosis; GSH, reduced glutathione; GSH1, γ-glutamyl cysteine synthetase 1; BSO, buthionine sulfoximine.
Footnotes
In yeast gsh1Δ mutant cells, the human and yeast SOD1 polypeptides generally accumulate to ≈3-fold higher levels, and the effect is even more dramatic with apo SOD1. High levels of intracellular glutathione result in lower levels of SOD1 polypeptide without changes in gene transcription, suggesting that GSH can affect the posttranslational stability of SOD1 in vivo (M.C.C. and V.C.C., unpublished data).
References
- 1.Keller, G. A., Warner, T. G., Steimer, K. S. & Hallewell, R. A. (1991) Proc. Natl. Acad. Sci. USA 88, 7381–7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, L., Slot, J. W., Geuza, H. J. & Crapo, J. D. (1988) J. Cell. Biol. 107, 2169–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weisiger, R. A. & Fridovich, I. (1973) J. Biol. Chem. 248, 4793–4796. [PubMed] [Google Scholar]
- 4.Sturtz, L. A., Diekert, K., Jensen, L. T., Lill, R. & Culotta, V. C. (2001) J. Biol. Chem. 276, 38084–38089. [DOI] [PubMed] [Google Scholar]
- 5.Okado-Matsumoto, A. & Fridovich, I. (2001) J. Biol. Chem. 276, 38388–38393. [DOI] [PubMed] [Google Scholar]
- 6.Deng, H. X., Hentati, A., Tainer, J. A., Iqbal, Z., Cayabyabi, A., Hung, W. Y., Getzoff, E. D., Hu, P., Herzfeld, B., Roos, R. P., et al. (1993) Science 261, 1047–1051. [DOI] [PubMed] [Google Scholar]
- 7.Gurney, M. E., Pu, H., Chiu, A. U., Canto, M. C. D., Polchow, C. Y., Alexander, D. D., Caliendo, J., Hentati, A., Kwon, Y., Deng, H. S., et al. (1994) Science 264, 1772–1775. [DOI] [PubMed] [Google Scholar]
- 8.Rakhit, R., Cunningham, P., Furtos-Matei, A., Dahan, S., Qi, X., Crow, J. P., Cashman, N. R., Kondejewski, L. H. & Chakrabartty, A. (2002) J. Biol. Chem. 277, 47551–47556. [DOI] [PubMed] [Google Scholar]
- 9.Cleveland, D. W. & Liu, J. (2000) Nat. Med. 6, 1320–1321. [DOI] [PubMed] [Google Scholar]
- 10.Cleveland, D. W. & Rothstein, J. D. (2001) Nat. Rev. Neurosci. 2, 806–819. [DOI] [PubMed] [Google Scholar]
- 11.Valentine, J. S. & Hart, P. J. (2003) Proc. Natl. Acad. Sci. USA 100, 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guegan, C. & Przedborski, S. (2003) J. Clin. Invest. 111, 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Culotta, V. C., Klomp, L., Strain, J., Casareno, R., Krems, B. & Gitlin, J. D. (1997) J. Biol. Chem. 272, 23469–23472. [DOI] [PubMed] [Google Scholar]
- 14.Rae, T. D., Schmidt, P. J., Pufhal, R. A., Culotta, V. C. & O'Halloran, T. V. (1999) Science 284, 805–808. [DOI] [PubMed] [Google Scholar]
- 15.Lyons, T. J., Nerissian, A., Goto, J. J., Zhu, H., Gralla, E. B. & Valentine, J. S. (1998) J. Biol. Inorg. Chem. 3, 650–662. [Google Scholar]
- 16.Wong, P. C., Waggoner, D., Subramaniam, J. R., Tessarollo, L., Bartnikas, T. B., Culotta, V. C., Price, D. L., Rothstein, J. & Gitlin, J. D. (2000) Proc. Natl. Acad. Sci. USA 97, 2886–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramaniam, J. R., Lyons, W. E., Liu, J., Bartnikas, T. B., Rothstein, J., Price, D. L., Cleveland, D. W., Gitlin, J. D. & Wong, P. C. (2002) Nat. Neurosci. 5, 301–307. [DOI] [PubMed] [Google Scholar]
- 18.Corson, L. B., Strain, J., Culotta, V. C. & Cleveland, D. W. (1998) Proc. Natl. Acad. Sci. USA 95, 6361–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beckman, J. S., Estvez, A. G., Barbeito, L. & Crow, J. P. (2002) Free Radical Biol. Med. 33, 1433–1435. [DOI] [PubMed] [Google Scholar]
- 20.Slekar, K. H., Kosman, D. & Culotta, V. C. (1996) J. Biol. Chem. 271, 28831–28836. [DOI] [PubMed] [Google Scholar]
- 21.Liu, H., Zhu, H., Eggers, D. K., Nersissian, A. M., Faull, K. F., Goto, J. J., Ai, J., Sanders-Loehr, J., Gralla, E. B. & Valentine, J. S. (2000) Biochemistry 39, 8125–8132. [DOI] [PubMed] [Google Scholar]
- 22.Luk, E. & Culotta, V. C. (2001) J. Biol. Chem. 276, 47556–47562. [DOI] [PubMed] [Google Scholar]
- 23.Flohe, L. & Otting, F. (1984) in Methods in Enzymology: Oxygen Radicals in Biological Systems, ed. Packer, L. (Academic, New York), Vol. 105, pp. 93–104. [Google Scholar]
- 24.Schmidt, P. J., Ramos-Gomez, M. & Culotta, V. C. (1999) J. Biol. Chem. 274, 36952–36956. [DOI] [PubMed] [Google Scholar]
- 25.Borchelt, D. R., Guarnieri, M., Wong, P. C., Lee, M. K., Slunt, H. S., Xu, Z., Sisodia, S. S., Price, D. L. & Cleveland, D. W. (1995) J. Biol. Chem. 270, 3234–3238. [DOI] [PubMed] [Google Scholar]
- 26.Anderson, M. E. (1985) Methods Enzymol. 113, 548–555. [DOI] [PubMed] [Google Scholar]
- 27.Cuozzo, J. W. & Kaiser, C. A. (1999) Nat. Cell Biol. 1, 130–135. [DOI] [PubMed] [Google Scholar]
- 28.Grant, C. M., Perrone, G. & Dawes, I. W. (1998) Biochem. Biophys. Res. Comm. 253, 893–898. [DOI] [PubMed] [Google Scholar]
- 29.Kierstead, T. D. & Tevethia, M. J. (1993) J. Virol. 67, 1817–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borchelt, D. R., Lee, M. K., Slunt, H. H., Guarnieri, M., Xu, Z., Wong, P. C., Brown, R. H., Price, D. L., Sisodia, S. S. & Cleveland, D. W. (1994) Proc. Natl. Acad. Sci. USA 91, 8292–8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Portnoy, M. E., Schmidt, P. J., Rogers, R. S. & Culotta, V. C. (2001) Mol. Gen. Genet. 265, 873–882. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt, P., Rae, T. D., Pufahl, R. A., Hamma, T., Strain, J., O'Halloran, T. V. & Culotta, V. C. (1999) J. Biol. Chem. 274, 23719–23725. [DOI] [PubMed] [Google Scholar]
- 33.Ratovitski, T., Corson, L. B., Strain, J., Wong, P., Cleveland, D. W., Culotta, V. C. & Borchelt, D. R. (1999) Hum. Mol. Gen. 8, 1451–1460. [DOI] [PubMed] [Google Scholar]
- 34.O'Halloran, T. V. & Culotta, V. C. (2000) J. Biol. Chem. 275, 25057–25060. [DOI] [PubMed] [Google Scholar]
- 35.Butt, T. R., Sternberg, E. J., Gorman, J. A., Clark, P., Hamer, D., Rosenberg, M. & Crooke, S. T. (1984) Proc. Natl. Acad. Sci. USA 81, 3332–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Culotta, V. C., Howard, W. R. & Liu, X. F. (1994) J. Biol. Chem. 269, 25295–25302. [PubMed] [Google Scholar]
- 37.Ciriolo, M. R., Battistoni, A., Falconi, M., Filomeni, G. & Rotilio, G. (2001) Eur. J. Biochem. 268, 737–742. [DOI] [PubMed] [Google Scholar]
- 38.Ciriolo, M. R., Desideri, A., Paci, M. & Rotilio, G. (1990) J. Biol. Chem. 265, 11030–11034. [PubMed] [Google Scholar]
- 39.Ascone, I., Longo, A., Dexpert, H., Ciriolo, M. R., Rotilio, G. & Desideri, A. (1993) FEBS Lett. 322, 165–167. [DOI] [PubMed] [Google Scholar]
- 40.Freedman, J. H., Ciriolo, M. R. & Peisach, J. (1989) J. Biol. Chem. 264, 5598–5605. [PubMed] [Google Scholar]
- 41.Ferruzza, S., Sambuy, Y., Ciriolo, M. R., Martino, A. D., Santaroni, P., Rotilio, G. & Scarino, M. L. (2000) Biometals 13, 179–185. [DOI] [PubMed] [Google Scholar]
- 42.White, A. R., Bush, A. I., Beyreuther, K., Masters, C. L. & Cappai, R. (1999) J. Neurochem. 72, 2092–2093. [DOI] [PubMed] [Google Scholar]
- 43.White, A. R. & Cappai, R. (2003) J. Neurosci. Res. 71, 889–897. [DOI] [PubMed] [Google Scholar]
- 44.Grant, C. M., MacIver, F. H. & Dawes, I. W. (1996) Curr. Genet. 29, 511–515. [DOI] [PubMed] [Google Scholar]
- 45.Outten, C. E. & Culotta, V. C. (2004) J. Biol. Chem., 279, 7785–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller, E. (1996) Mol. Biol. Cell. 7, 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferreira, A. M., Ciriolo, M. R., Marcocci, L. & Rotilio, G. (1993) Biochem. J. 292, 673–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Field, L. S., Furukawa, Y., O'Halloran, T. V. & Culotta, V. C. (2003) J. Biol. Chem. 278, 28052–28059. [DOI] [PubMed] [Google Scholar]
- 49.Bordo, D., Djinovic, K. & Bolognesi, M. (1994) J. Mol. Biol. 238, 366–386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.