Abstract
Attachment of cells to the extracellular matrix induces clustering of membrane receptor integrins which in turn triggers the formation of focal adhesions (FAs). The adaptor/scaffold proteins in FAs provide linkage to actin cytoskeleton, whereas focal adhesion kinase (FAK) and other FA-associated kinases and phosphatases transduce integrin-mediated signaling cascades, promoting actin polymerization and progression of cell spreading. In this study, we explored the role of OLA1, a newly identified member of Obg-like ATPases, in regulating cell adhesion processes. We showed that in multiple human cell lines RNAi-mediated downregulation of OLA1 significantly accelerated cell adhesion and spreading, and conversely overexpression of OLA1 by gene transfection resulted in delayed cell adhesion and spreading. We further found that OLA1-deficient cells had elevated levels of FAK protein and decreased Ser3 phosphorylation of cofilin, an actin-binding protein and key regulator of actin filament dynamics, while OLA1-overexpressing cells exhibited the opposite molecular alterations in FAK and cofilin. These findings suggest that OLA1 plays an important negative role in cell adhesion and spreading, in part through the regulation of FAK expression and cofilin phosphorylation, and manipulation of OLA1 may lead to significant changes in cell adhesion and the associated phenotypes.
Keywords: OLA1, cell-matrix adhesion, cell spreading, FAK, cofilin
1. Introduction
Cell adhesion plays key roles in embryonic development, immune response, tissue repair, and cancer metastasis [1,2,3]. Upon the initial attachment of cells to ECM components, the cell membrane receptor integrins are induced to form clusters at the attachment sites. In the inner surface of the cell membrane, the clustered integrins serve as a platform for the recruitment of various adaptor and signaling proteins, resulting in the formation of focal adhesions (FAs). The adaptor/scaffold proteins in FAs, including talin, paxillin, and α-actinin [4], provide strong linkages to the actin cytoskeleton, thus connecting the cells firmly to the ECM, whereas the signaling proteins, predominantly the focal adhesion kinase (FAK) and Src, play central roles in transduction of the integrin-mediated signaling. FAK is an ubiquitously expressed non-receptor tyrosine protein kinase [5,6] and has multiple functions within the cell, including contributing to FA maturation and turnover, phosphorylation of other focal adhesion proteins, and regulation of motility and survival [6].
Actin is the primary component of the cytoskeleton, and in the cell adhesion processes the dynamics of actin polymerization and depolymerization are required for the generation of cellular forces that lead to FA maturation, turnover, and cell spreading [7]. Cofilin is an actin-binding protein and plays an essential role in regulating actin filament dynamics by stimulating the severing and depolymerization of actin filaments, and as such providing abundant actin monomers for further actin polymerization and filament assembly. Cofilin’s severing activity is inhibited upon phosphorylation at the Ser3 residue by Lim family kinases (LIMKs) [8,9] and testicular protein kinases (TESKs), and restored when cofilin is dephosphorylated by various phosphatases [10,11].
To date nearly 200 different molecules have been described as components of a complex network, collectively referred to as the integrin adhesome, contributing to the structural and signaling functions of FAs [12,13]. However, current knowledge about the adhesome is just the tip of the iceberg; recent proteomics studies suggest that we still have much to learn about its complexity [14]. In the present study we describe the addition of a novel regulatory factor to this network: Obg-like ATPase 1 (OLA1). OLA1 belongs to the translation factor-related (TRAFAC) class, Obg family, and YchF subfamily of P-loop GTPases [15,16]. P-loop NTPases are the most abundant nucleotide-binding proteins [17], and involved in the regulation of diverse cellular processes including protein translation, intracellular transport, signal transduction, and cell proliferation [18,19]. The OLA1/YchF proteins are highly conserved from yeast to human [17,20], and unlike other Obg family members they bind and hydrolyze ATP more efficiently than GTP [21]. However, the physiological functions of these unconventional Obg-like ATPases are poorly understood. Recently, our group demonstrated that human OLA1 functions as an intrinsic regulator in cellular stress responses such as oxidative stress [22] and heat shock [23]. In another study, we reported that down-regulation of OLA1 causes changes in cell migration and invasiveness in cultured human cancer cells [15].
In this report we demonstrate that down-regulation of OLA1 results in significantly accelerated cell adhesion and spreading associated with increased levels of FAK and decreased cofilin Ser3 phosphorylation while overexpression of OLA1 causes delayed cell adhesion associated with the opposite molecular changes in FAK and cofilin. Our findings suggest that OLA regulates the essential cell-matrix adhesion processes by affecting multiple components of the adhesome.
2. Materials and methods
2.1 Cells
MDA-MB-231, WI-38, and HeLa cells were obtained from ATCC and cultured in Dulbecco’s Modified Eagle’s medium (DMEM, Thermo Scientific) containing 10% fetal bovine serum (FBS, Thermo Scientific), 100 units/ml penicillin, and 100 mg/ml streptomycin at 37°C with 5% CO2.
2.2 RNAi of OLA1
Human OLA1 cDNA (NM_013341.3)-specific siRNA (SASI_Hs01_00244684) and the control siRNA (MISSION siRNA Universal Negative Control, #1, SIC001) were acquired from Sigma-Aldrich. Cells seeded in 6-well plates were transiently transfected with 5 μM siRNA with the DharmaFECT1 siRNA Transfection Reagent (Thermo Scientific) according to the manufacturer’s instructions. To establish stable OLA1-knockdown cell lines (HeLa) SMARTvector lentiviral shRNA particles (Thermo Scientific) containing a shRNA sequence specific for OLA1 (TGTTCGCTTCCAGATACTT) and the control shRNA sequence were used at the range of 5~20 TU/cell. Cell clones expressing the respective shRNAs were selected with puromycin (5μg/ml) for 1 month. The knockdown efficiency of the target gene was verified by Western blot analysis.
2.3 Cell adhesion assay
Cells were serum starved overnight before detaching with 0.25% trypsin for 1 min. The cells were re-suspended in fresh medium, and then seeded into 6-well dishes pre-coated with 10 μg/ml human fibronectin or 20 μg/ml laminin at the density of 1×105 cells/well. Attached cells were fixed in 1 ml of 4% paraformaldehyde at the indicated times and photographs were taken under a microscope.
2.4 Quantification of Cell adhesion
The 96 well plates were pre-coated with fibronectin (10 μg/ml), laminin (20 μg/ml), or BSA (1 mg/ml, negative control) and seeded with 2 × 104 cells/well for 30 min or 60 min. At the end of incubation, cells were fixed with 4% paraformaldehyde and non-adherent cells were washed off with phosphate buffered saline (PBS). The remaining attached cells were stained with crystal violet solution (Sigma-Aldrich, 5 mg/ml in 2% ethanol), and the final color was measured with a plate reader (Fluostar Optima) at 570 nM.
2.5 Overexpression of OLA1
cDNA fragments encoding full-length human OLA1 cDNA (NM_013341.3) were cloned into the pdEYFP-N1gen plasmid with a C-terminal YFP tag as previously described [22,23]. Cells in 6-well plates were transfected with the OLA1-YFP plasmid (4 μg DNA/well) or the pdEYFP-N1gen (YFP control) plasmid using Lipofectamine 2000 (10 μl/well, Invitrogen). The parental plasmids were a kind gift from Stefan Wiemann of European Molecular Biology Laboratory, Heidelberg, Germany [24].
2.6 Immunoblot analysis
Western immunoblot analysis was performed according to our standard procedures as previously described [22,23]. All antibodies used in these studies were purchased from Cell Signaling Technology except anti-OLA1 and anti-β-actin antibodies, which were from Sigma-Aldrich, and anti-rabbit IgG peroxidase linked whole antibody and anti-mouse Ig peroxidase linked whole antibody, from GE Healthcare.
2.7 Statistical Analysis
Statistical analysis was performed by a two-tailed Student’s t-test. Values of P < 0.05 were considered to be statistically significant.
3. Results
3.1 Downregulation of OLA1 accelerates cell adhesion and spreading
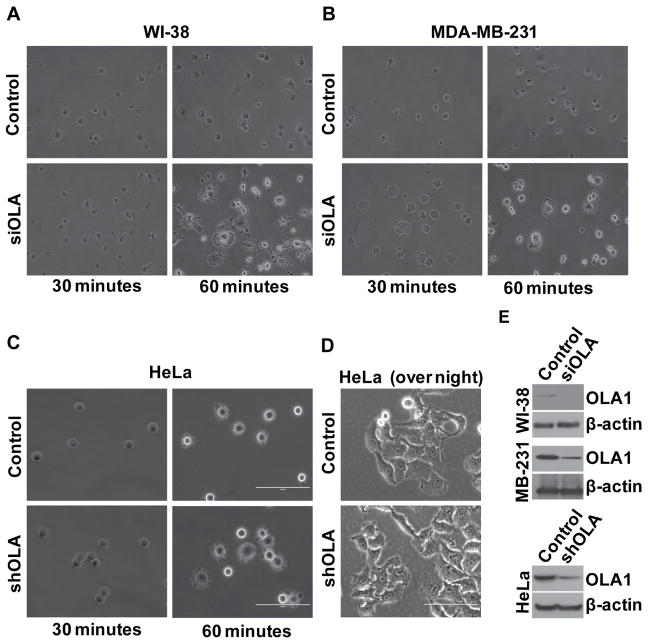
To investigate the role of OLA1 in cell adhesion and spreading, we manipulated the expression of OLA1 in multiple human cell lines, first by siRNA-mediated gene silencing. Human lung fibroblasts (WI-38) and breast cancer cells (MDA-MB-231) were transiently transfected with OLA1-specific siRNA or non-targeting siRNA, and at time 0 identical numbers of cells were plated on fibronectin-coated plates. Compared with the control transfected cells, OLA1-siRNA cells exhibited a much faster progression of attachment and cell body spreading (Fig. 1A & B). As early as 30 min, notably more OLA1-siRNA cells had attached to the plate and initiated spreading. Here spread cells were defined as cells with flat and extended cell margins lacking phase-brightness, whereas non-spread cells were rounded and phase-bright under microscope. More strikingly, at 60 min, while only a small portion of the control cells had entered the “intermediate stage” of spreading with the rest of the cells remaining small and rounded, almost all OLA1-siRNA cells had achieved maximum cell areas with well-developed membrane protrusions indicating “late-stage” spreading (Fig. 1A & B). After prolonged incubation the difference evened out as the control cells also completed the whole course of adhesion and spreading. After overnight incubation, the two groups of cells showed no apparent variation in either total adherent cells or the morphology of single cells (Fig. 1D). Consistent with our previous report [22], general cell growth was not affected by the RNAi treatment.
Figure 1. Down-regulation of OLA1 results in accelerated cell adhesion and spreading.
A–B, Human lung WI-38 cells (A) and breast cancer MDA-MB-231 cells (B) were transiently transfected with the control and OLA siRNA (siOLA), and 48 hours later the cells were plated on fibronectin-coated 6-well plates. Cell images were taken under an inverted light microscope at 30 and 60 minutes after plating. C, HeLa cells stably transfected with non-target shRNA (control), or OLA1-specific shRNA (shOLA) were plated on fibronectin-coated plates and imaged at 30 and 60 minutes after the plating. D, HeLa cells stably transfected with control or shOLA vectors were plated and imaged after an overnight incubation. All the above results represent at least three independent experiments. Scale bars: 100 μm. E, Cell lysates were immunoblotted to verify the effectiveness of OLA1-downregulation in the above knock-down experiments. The blot was re-probed with anti-β-actin antibody to check the equal loading of protein samples.
In order to rule out the possibility that the observed phenotype was dependent on the transient RNAi strategy, stable cell sub-lines were raised by transduction of lentiviruses expressing shRNA targeting to human OLA1 mRNA at a sequence region different from that of the siRNA, or expressing a non-targeting shRNA. When the HeLa-derived sub-lines were tested for their performance in cell adhesion, the OLA1-knockdown cells demonstrated significantly accelerated cell adhesion and spreading compared to the control cells (Fig. 1C).
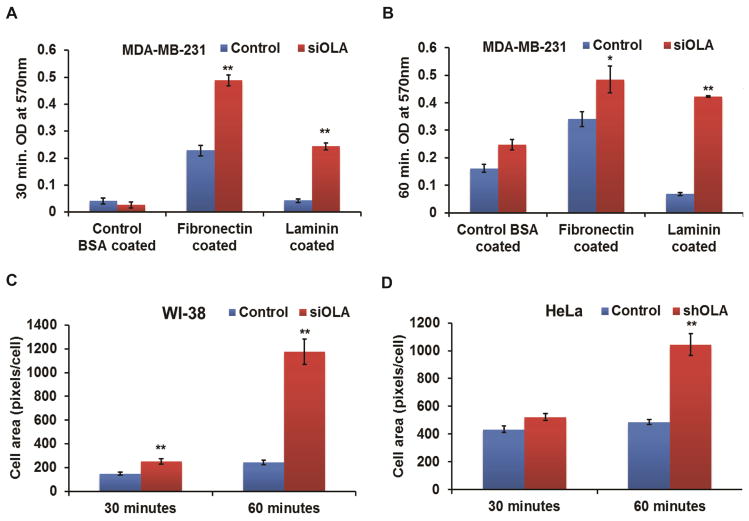
In addition to the above microscopic examination, two quantitative measurements were employed to evaluate OLA1-knockdown mediated cell adhesion and spreading. First, the number of total adherent cells (MDA-MB-231) on fibronectin- or laminin-coated surfaces at different time points were estimated by crystal violet staining after the non-adherent cells were washed away. At 30 min post-plating, 2.1-fold- and 5.7-fold- more OLA1-siRNA cells than the control transfected cells were attached on fibronectin- or laminin-coated plates, respectively (Fig. 2A). The increases were also discernibly significant at 60 min (1.4-fold and 6.1-fold, respectively) (Fig. 2B). Second, cell images taken at 30 and 60 min post-plating were analyzed with ImageJ software for average cell areas. As shown in Fig. 2 C–D, compared with the control transfected cells, OLA1 downregulated cells (HeLa and WI-38) showed a significant increase in mean cell area at 30 min: 68% in WI-38 cells and 20% in HeLa cells, respectively, and an even more dramatic increase at 60 min post plating (384% in WI-38 and 114% in HeLa, respectively).
Figure 2. Quantitative analysis of cell adhesion and spreading.
MDA-MB-231 cells transiently transfected with the control and OLA siRNA (siOLA) were analyzed for adhesion rate on BSA-, fibronectin-, or laminin-coated plates. Attached cells were fixed at 30 minutes (A) and 60 minutes (B) after plating, and quantified by crystal violet staining. Data represent mean ± SEM values from 3 independent experiments. *, p < 0.05; **, p < 0.001. Spreading of the control-transfected and OLA1-knockdown WI-38 (C) and HeLa (D) cells were quantified using ImageJ software based on images of cell adhesion taken at 30 and 60 minutes. Data represent the mean ± SEM of cell area, expressed as pixels per cell, of cells pooled from 5 different microscopic fields for each group. **, p < 0.001 (n = 25).
3.2 OLA1-deficient cells have increased basal FAK protein and decreased cofilin phosphorylation
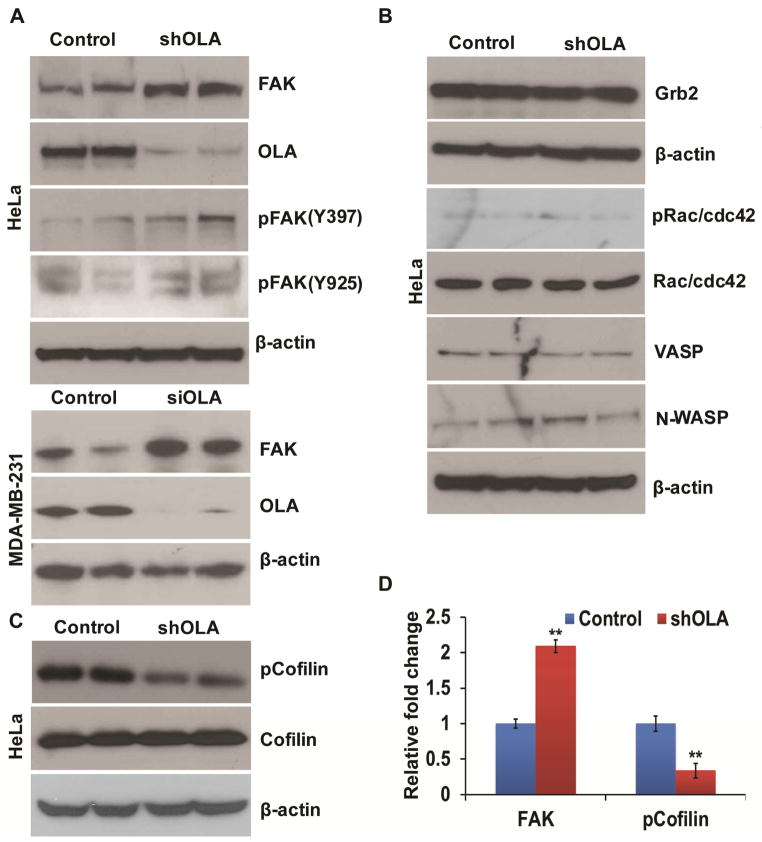
In order to identify possible molecular mechanisms that underlie the enhanced cell-matrix adhesion, we investigated FAK and other signaling proteins known to regulate cell adhesion and actin dynamics, first in stably transfected HeLa cells. Using immunoblot techniques, we screened FAK, Grb2, Rac/cdc42, VASP, and N-WASP for their total protein levels and phosphorylation status. The OLA1-knockdown cells were found to express a notably increased level of FAK protein compared to the control cells, although there was no consistent corresponding change in phosphorylation of the protein at Tyr397 or Tyr925 (Fig. 3A,D). We failed to detect any discernible changes for other proteins between the two types of cells (Fig. 3B). Furthermore, we examined some major actin cytoskeleton binding/regulatory proteins, including cofilin, ARP2, ARP3, and Ezrin, in stably transfected HeLa cells. This led to the identification of a markedly under-phosphorylated cofilin at its Ser3 residue in the OLA1-knockdown cells though without changes in the total protein level (Fig. 3C, D). These data demonstrated that knockdown of OLA1 in HeLa cells was associated with at least two molecular alterations: the increased basal total protein of FAK and the decreased phosphorylation of cofilin. Additionally, we confirmed that the same molecular changes (increased FAK) also occurred to growing cultures of MDA-MB-231 cells transiently transfected with OLA1 siRNA (Fig. 3A).
Figure 3. Molecular alterations detected in OLA1-deficient cells.
A, OLA1-knockdown HeLa cells (stably transfected by shRNA, upper panel) and MDA-MB-231 cells (transiently transfected by siRNA, lower panel) were analyzed by immunoblotting for their expression of FAK and its phosphorylation. The effectiveness of OLA1-knockdown was evidenced by anti-OLA1 antibody probing and equivalent protein loading was verified with anti-β-actin antibody. B, Immunoblot analysis of OLA1-knockdown HeLa cells for detecting cytoskeletal signaling proteins including Crb2, Rac/cdc42, VASP, N-WASP, and their phosphorylation modification (pRac/cdc42, Ser71; pVASP, Ser157), with the corresponding antibodies. C, Additional immunoblot analysis with anti-cofilin and anti-phospho-cofilin (Ser3) antibodies. β-actin was used as a loading control. Note that in these experiments (A–C) duplicate samples prepared from independently transfected cells were loaded and analyzed on the same blot (to emphasize technical consistency). D, Densitomeric analysis of immunoblots shows fold changes of the FAK and pCofilin levels in OLA1 deficient cells compared to control cells. Data represent mean ± SD values from 3 independent experiments. **, p < 0.001.
3.3 Overexpression of OLA1 delays cell adhesion and correlates with decreased FAK expression and increased cofilin phosphorylation
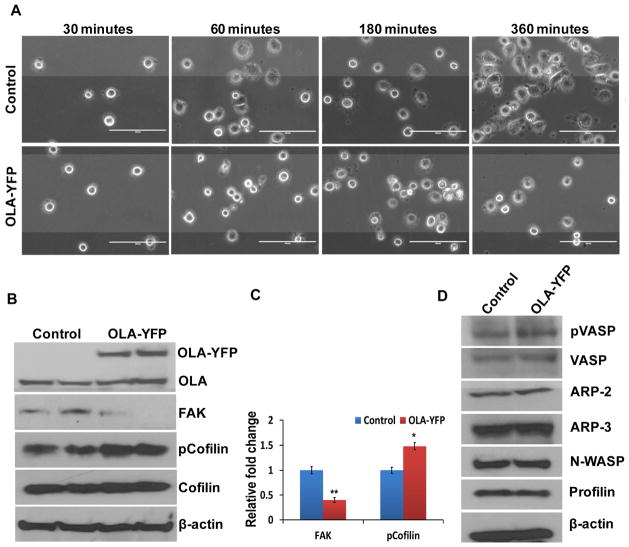
In addition to the above investigations based on downward-manipulation of OLA1 expression, we explored the effects of up-regulation of OLA1 on cell adhesion. As a result of ectopic expression of OLA1, cell adhesion on fibronectin-coated plates was significantly delayed (Fig. 4). By 360 min post-plating, mature adhesion morphology was observed in most of the control cells, but less than 50% of OLA1-overexpressing cells showed mature adhesion (Fig. 4A). We further examined the status of FAK and cofilin using immunoblot analysis. In OLA1-transfected cells, FAK was found to be markedly reduced (Fig. 4B–C). On the other hand, while phosphorylation of cofilin at Ser3 site was significantly increased, the amount of total cofilin protein was unchanged (Fig. 4B–C). Additional cytoskeletal signaling proteins were screened without producing positive findings (Fig. 4D). Based on the data from these knockdown and overexpression experiments, we conclude that OLA1 plays a negative role in the regulation of cell adhesion and spreading, likely through modulation of the levels of FAK expression and the phosphorylation status of cofilin.
Figure 4. Overexpression of OLA1 results in delayed cell adhesion and spreading.
A, HeLa cells were transfected with an OLA1-expressing plasmid (OLA-YFP) or the YFP-expressing (control) vector for 48 hours, and re-plated on fibronectin-coated plates for cell adhesion assay. Cell images were taken at 30, 60, 180, and 360 minutes post-plating. Scale bar indicates 100 μm. B, Immunoblot analysis of the HeLa cells transfected with OLA1-YFP and the control vectors. The ectopic expression of OLA1 was confirmed by the presence of a higher molecular weight OLA1-YFP band on top of the endogenous OLA1 band. The same blot was re-probed for total FAK, total cofilin, and Ser3 phosphorylation of cofilin levels. β-actin was used as a loading control.. C, Densitomeric analysis of immunoblots shows fold changes of the FAK and pCofilin levels in OLA1 deficient cells compared to control cells. Data represent mean ± SD values from 3 independent experiments. **, p < 0.001; *, p < 0.05. D, Additional immunoblot analysis of the transfected HeLa cells for cytoskeletal signaling proteins as indicated. These data represent at least three independent experiments.
4. Discussion
In this report, we demonstrated that downregulation of OLA1, a recently identified Obg-like ATPase, can dramatically accelerate cell-matrix adhesion and spreading, and that this phenotypic change is associated with at least two molecular alterations: increased total protein levels of FAK, a major signaling component of FAs, and decreased phosphorylation of cofilin, a key regulator of actin filament dynamics (Fig. 1–3). Our finding was further supported by overexpression studies that produced the opposite phenotypes and molecular alterations (Fig. 4).
FAK plays a central role in the formation and turnover of FAs, and increased FAK expression promotes FA formation, cell spreading, and migration [25,26]. As demonstrated by Serrels et al., FAK-null (FAK −/−) mouse embryonic fibroblasts were less well spread than wild-type FAK-expressing cells [27]. Moreover, when FAK expression was re-established through a conditional gene expression strategy, the mutant cells gained enhanced cell spreading along with accelerated formation and turnover of FAs [6,28,29]. We believe that the enhanced cell adhesion phenotype observed in OLA1-defficient cells is attributable at least in part to the increased basal FAK levels.
In addition, we showed that down-regulation of OLA1 is associated with markedly decreased phosphorylation of cofilin at Ser3 (Fig. 3C), indicating a hyperactive cofilin, whereas overexpression of OLA1 is linked with increased phosphorylation of cofilin, corresponding to its inactive form (Fig. 4C). Although cofilin was initially identified as an actin severance and depolymerization factor (ADF), recent studies have emphasized its central role in the generation of free barbed ends and recycling of actin monomers that are required for actin polymerization driven membrane protrusion [30,31]. We reason that in our current systems, reduced phosphorylation of cofilin elevates the basal level of actin polymerization dynamics, and vice versa.
Understanding OLA1-mediated cell adhesion phenotypes should also contribute to the understanding of the fundamental biological functions of OLA1. The OLA1/YchF protein is predicted to be a regulatory protein that interacts with downstream client protein(s) by switching between its ADP- and ATP-bound forms [21]. Our current study provides the first evidence showing that OLA1 regulates at least two key molecules in the FA and actin cytoskeleton signaling pathways. The involvement of YchF proteins in the protein synthesis or degradation machineries has been recently reported in, for example, the association of YchF with the S70 ribosome in E. coli [32,33], the co-fractionation of TcYchF with polysomes in Trypanosoma cruzi [34], the association of YBR025c/Ola1 with the 26S proteasome in yeast [25,26], and the interaction of OLA1 with HSP70 in mammalian cells [23]. Our laboratory is currently exploring mammalian OLA1’s role in general protein translation and degradation as well as the synthesis/turnover of specific proteins including FAK. On the other hand, regarding the way OLA1 affects the phosphorylation of cofilin, we consider the following two interpretations: (1) it is merely a downstream event reflecting the alteration of FAK signaling (2) OLA1 possesses a specific enzymatic activity that directly modulates cofilin phosphorylation. However, additional studies are needed to identify the “clients” of OLA1 from among the kinases or phosphatases that either directly act on cofilin or those at the upstream levels within the relevant pathways.
In conclusion, this study reveals an important role of OLA1 in the regulation of cell-matrix adhesion and spreading. Cell adhesion machineries, collectively known as the “adhesome”, not only provide a structural connection between the ECM and cytoskeleton but also act as signaling hubs enabling a cell to sense and respond to its environment, and thus control cell behavior for achieving cellular homeostasis [12]. The findings described in this report would promote our understanding of fundamental biological processes that involve or require cell adhesion, as well as pathological conditions that result from dysregulation of cell adhesion, including cancer, immunological disorders, osteoporosis, and fibrotic diseases.
Highlights.
Functions of OLA1 in mammalian cells are poorly understood.
Downregulation of OLA1 enhances cell adhesion and spreading while overexpression of OLA1 inhibits these processes.
OLA1 regulates protein levels of FAK and phosphorylation of cofilin.
OLA1 plays a negative role in cell-matrix adhesion and spreading.
Acknowledgments
We thank Drs. Renduo Song and Dong Xu for their technical assistance, and Luchang Wang for help with manuscript preparation. This work was financially supported by NIH grant R01CA155069 (ZS), the HMRI Cornerstone Award (ZS), and the National Natural Science Foundation of China #81101477 (JZ).
Abbreviations
- OLA1
Obg-like ATPase 1
- FAs
focal adhesions
- FAK
focal adhesion kinase
- ECM
extracellular matrix
- ROCK
Rho-associated kinase
- F-actin
filamentous actin
- LIMKs
Lim family kinases
- TESK
testicular protein kinases
- pFAK(Y397)
Tyr 397 phosphorylated FAK, pFAK(Y925), Tyr 925 phosphorylated FAK, pCofilin, Ser 3 phosphorylated cofilin, pRac/cdc42, Ser 71 phosphorylated Rac/cdc42
- pVASP
Ser 157 phosphorylated VASP
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 3.Daley WP, Yamada KM. ECM-modulated cellular dynamics as a driving force for tissue morphogenesis. Curr Opin Genet Dev. 2013;23:408–414. doi: 10.1016/j.gde.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 5.Parsons JT. Focal adhesion kinase: the first ten years. J Cell Sci. 2003;116:1409–1416. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- 6.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 7.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 8.Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- 9.Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- 10.Bamburg JR, Wiggan OP. ADF/cofilin and actin dynamics in disease. Trends Cell Biol. 2002;12:598–605. doi: 10.1016/s0962-8924(02)02404-2. [DOI] [PubMed] [Google Scholar]
- 11.Huang TY, DerMardirossian C, Bokoch GM. Cofilin phosphatases and regulation of actin dynamics. Curr Opin Cell Biol. 2006;18:26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Zaidel-Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byron A. Analyzing the anatomy of integrin adhesions. Sci Signal. 2011;4:jc3. doi: 10.1126/scisignal.2001896. [DOI] [PubMed] [Google Scholar]
- 14.Geiger T, Zaidel-Bar R. Opening the floodgates: proteomics and the integrin adhesome. Curr Opin Cell Biol. 2012;24:562–568. doi: 10.1016/j.ceb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Zhang JW, Rubio V, Zheng S, Shi ZZ. Knockdown of OLA1, a regulator of oxidative stress response, inhibits motility and invasion of breast cancer cells. J Zhejiang Univ Sci B. 2009;10:796–804. doi: 10.1631/jzus.B0910009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun H, Luo X, Montalbano J, Jin W, Shi J, Sheikh MS, Huang Y. DOC45, a novel DNA damage-regulated nucleocytoplasmic ATPase that is overexpressed in multiple human malignancies. Mol Cancer Res. 8:57–66. doi: 10.1158/1541-7786.MCR-09-0278. [DOI] [PubMed] [Google Scholar]
- 17.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 18.Paduch M, Jelen F, Otlewski J. Structure of small G proteins and their regulators. Acta Biochim Pol. 2001;48:829–850. [PubMed] [Google Scholar]
- 19.Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 20.Czyz A, Wegrzyn G. The Obg subfamily of bacterial GTP-binding proteins: essential proteins of largely unknown functions that are evolutionarily conserved from bacteria to humans. Acta Biochim Pol. 2005;52:35–43. [PubMed] [Google Scholar]
- 21.Koller-Eichhorn R, Marquardt T, Gail R, Wittinghofer A, Kostrewa D, Kutay U, Kambach C. Human OLA1 defines an ATPase subfamily in the Obg family of GTP-binding proteins. J Biol Chem. 2007;282:19928–19937. doi: 10.1074/jbc.M700541200. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Rubio V, Lieberman MW, Shi ZZ. OLA1, an Obg-like ATPase, suppresses antioxidant response via nontranscriptional mechanisms. Proc Natl Acad Sci U S A. 2009;106:15356–15361. doi: 10.1073/pnas.0907213106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao RF, Rubio V, Chen H, Bai L, Mansour OC, Shi ZZ. OLA1 protects cells in heat shock by stabilizing HSP70. Cell Death Dis. 2013;4:e491. doi: 10.1038/cddis.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000;1:287–292. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim Y, Lim ST, Tomar A, Gardel M, Bernard-Trifilo JA, Chen XL, Uryu SA, Canete-Soler R, Zhai J, Lin H, Schlaepfer WW, Nalbant P, Bokoch G, Ilic D, Waterman-Storer C, Schlaepfer DD. PyK2 and FAK connections to p190Rho guanine nucleotide exchange factor regulate RhoA activity, focal adhesion formation, and cell motility. J Cell Biol. 2008;180:187–203. doi: 10.1083/jcb.200708194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilic D, Furuta Y, Kanazawa S, Takeda N, Sobue K, Nakatsuji N, Nomura S, Fujimoto J, Okada M, Yamamoto T. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature. 1995;377:539–544. doi: 10.1038/377539a0. [DOI] [PubMed] [Google Scholar]
- 27.Serrels B, Serrels A, Brunton VG, Holt M, McLean GW, Gray CH, Jones GE, Frame MC. Focal adhesion kinase controls actin assembly via a FERM-mediated interaction with the Arp2/3 complex. Nat Cell Biol. 2007;9:1046–1056. doi: 10.1038/ncb1626. [DOI] [PubMed] [Google Scholar]
- 28.Owen JD, Ruest PJ, Fry DW, Hanks SK. Induced focal adhesion kinase (FAK) expression in FAK-null cells enhances cell spreading and migration requiring both auto- and activation loop phosphorylation sites and inhibits adhesion-dependent tyrosine phosphorylation of Pyk2. Mol Cell Biol. 1999;19:4806–4818. doi: 10.1128/mcb.19.7.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagano M, Hoshino D, Koshikawa N, Akizawa T, Seiki M. Turnover of focal adhesions and cancer cell migration. Int J Cell Biol. 2012;2012:310616. doi: 10.1155/2012/310616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tania N, Condeelis J, Edelstein-Keshet L. Modeling the synergy of cofilin and arp2/3 in lamellipodial protrusive activity. Biophys J. 2013;105:1946–1955. doi: 10.1016/j.bpj.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bravo-Cordero JJ, Magalhaes MA, Eddy RJ, Hodgson L, Condeelis J. Functions of cofilin in cell locomotion and invasion. Nat Rev Mol Cell Biol. 2013;14:405–415. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomar SK, Kumar P, Prakash B. Deciphering the catalytic machinery in a universally conserved ribosome binding ATPase YchF. Biochem Biophys Res Commun. 2011;408:459–464. doi: 10.1016/j.bbrc.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 33.Becker M, Gzyl KE, Altamirano AM, Vuong A, Urban K, Wieden HJ. The 70S ribosome modulates the ATPase activity of Escherichia coli YchF. RNA Biol. 2012;9:1288–1301. doi: 10.4161/rna.22131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gradia DF, Rau K, Umaki AC, de Souza FS, Probst CM, Correa A, Holetz FB, Avila AR, Krieger MA, Goldenberg S, Fragoso SP. Characterization of a novel Obg-like ATPase in the protozoan Trypanosoma cruzi. Int J Parasitol. 2009;39:49–58. doi: 10.1016/j.ijpara.2008.05.019. [DOI] [PubMed] [Google Scholar]