Abstract
In this issue, Aarke et al. (2013) identify a toxin-antitoxin system in Caulobactercrescentus that acts by a unique mechanism. The toxin, which blocks DNA replication, is constitutively degraded by ClpXP and this degradation requires the antitoxin, a ClpXP adaptor.
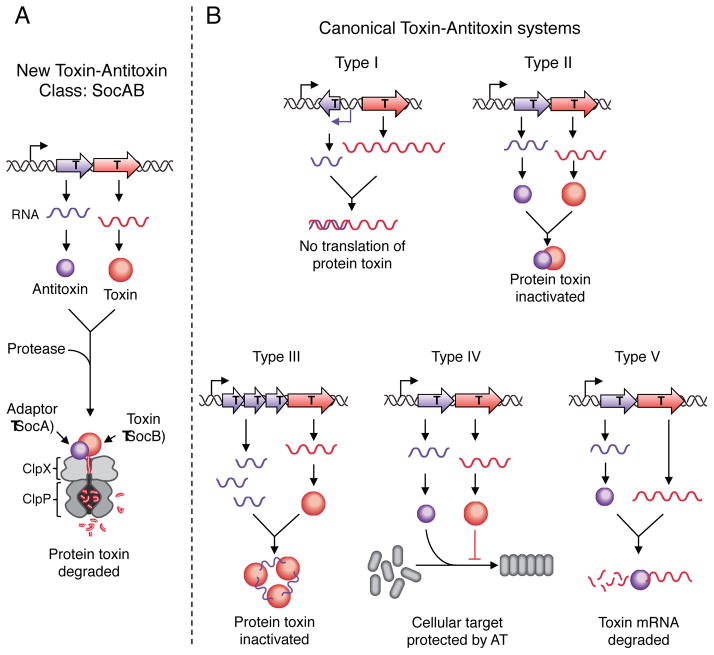
Toxin-antitoxin (TA) modules are highly abundant on the chromosomes of many bacteria. Traditional TA systems encode both a stable toxin and a labile small noncoding RNA or protein antitoxin. Under normal growth conditions, this antitoxin either prevents the translation of the toxic protein or counteracts its activity. Stress conditions can allow the toxic protein to be freed from the inhibitory effects of the antitoxin. In its active form, the toxin prevents cell growth by affecting essential cellular processes (Gerdes et al., 2005; Schuster and Bertram, 2013). Under environmental stress conditions, these TA systems may potentially function to modulate growth. In this issue of Molecular Cell, Aarke et al. (2013) identify a new TA locus, socAB, in Caulobactercrescentus and describe a novel, atypical mechanism of TA action. They find that the toxin, SocB, interacts with its antitoxin, SocA, and is delivered by SocA to the ClpXP protease where it is degraded (Figure 1a). In the absence of SocA or ClpXP, SocB forms a complex with the β sliding clamp, an essential component of the DNA replication elongation machinery, thereby inhibiting DNA replication.
Figure 1. The SocAB system is a new class of toxin-antitoxin modules.
(A) Unlike canonical toxin-antitoxin systems, the SocA antitoxin (purple) acts as a proteolytic adaptor for the degradation of SocB toxin (red) by the protease ClpXP (grey).
(B) The five types of toxin-antitoxin systems are classified based on the mechanism of action the antitoxin (purple) employs to inactivate toxin (red) activity.
AT, antitoxin; T, toxin.
All TA systems contain a protein toxin. However, they vary based on the nature of the antitoxin and the mechanism of action the antitoxin exerts on its toxin partner. To date, five TA classes have been identified (Figure 1b) (Mruk and Kobayashi, 2013; Schuster and Bertram, 2013). Type I TA systems transcribe a small noncoding RNA toxin that base pairs with the toxin mRNA and prevents its translation. In Type II TA systems, the protein antitoxin directly binds to the protein toxin to prevent toxin activity. The Type III class encodes numerous small noncoding RNAs that interact directly with the protein toxin to inactivate it. Type IV systems have a protein toxin that destabilizes cytoskeletal protein polymerization and a protein antitoxin that counteracts this effect. Finally, in Type V TA systems, the mRNA of the toxin-encoding gene is cleaved by the protein antitoxin.
In this report, Aarke et al. (2013) identify a toxin-antitoxin system in Caulobactercrescentus that acts by a unique mechanism where the protein antitoxin functions as a proteolytic adaptor to target the toxin for degradation. They began the, study by asking why the conserved AAA+ protease, ClpXP (Sauer and Baker, 2011), is absolutely essential in Caulobacter. This requirement for ClpXP in C. crescentus has been puzzling for many years since homologous proteasome-like proteases are nonessential in other well-studied bacteria. They devised a transposon mutagenesis selection to identify suppressors that allowed cells to grow in the absence of ClpP and identified the socB locus. It was noted that the gene encoding SocB was in an operon with a gene that was predicted to be essential, which they named socA. They hypothesized that SocAB might encode a TA system. Indeed, although repression of SocB in ΔsocA cells allowed colonies to grow well, artificial induction of SocB in ΔsocA cells prevented colony formation, thus indicating that socAB behaves genetically like other TA systems.
Together these results suggested that SocB might be a ClpXP substrate. In an unorthodox twist to the TA story, Aarke et al. (2013) found that ClpXP degrades the SocB toxin. This ClpXP requirement for SocB degradation provides a compelling reason why ClpXP is essential for viability in Caulobacter: SocB is toxic to cells without ClpXP. Also, anovel component of the system is the SocA antitoxin. Instead of functioning as a specific inhibitor of SocB, SocA is an essential ClpXP adaptor necessary for the degradation of SocB. Adaptor proteins, such as SspB, RssB and ClpS, bind to a substrate and deliver the substrate to the Clp protease (Battesti and Gottesman, 2013). The SocAB TA module acts by a mechanism not previously described and thus is the founding member of a new class of TA systems (Figure 1a).
To pinpoint the cellular target of SocB, Aarke et al. (2013) screened for mutants that could tolerate high levels of the toxin. Aside from the expected mutations in clpX, they identified dnaN mutants that bypassed the toxicity of increased SocB. DnaN encodes the β sliding clamp, a central component of the DNA elongation machinery that binds DNA polymerase III (Pol III) and increases the processivity of Pol III during replication (Johnson and O’Donnell, 2005; Kurth and O’Donnell, 2013). Additionally DnaN plays roles in trans-lesion DNA synthesis and mismatch repair and is known to interact with several other proteins, including DNA Pol I, II, IV and V, a replication regulator, Hda, and mismatch repair proteins, MutS and MutL (Dalrymple et al., 2001; Fernandez-Fernandez et al., 2013). The dnaN mutations identified by Aarke et al. (2013) led to amino acid substitutions located in a hydrophobic groove shown to interact with Pol III and the other DnaN interacting proteins (Dalrymple et al., 2001) (Johnson and O’Donnell, 2005). One interpretation of the results is that SocB inhibits DNA replication elongation by directly interacting with DnaN and preventing its interaction of Pol III. This inhibition leads to replication fork collapse and induction of the DNA damage response.
TA systems are prevalent throughout bacteria, and a single organism generally encodes multiple TA systems. It has been hypothesized that these modules may play regulatory roles that allow the cell to withstand stress in the natural habitat of the organism (Gerdes et al., 2005). It is not known yet why this unique TA system is needed for Caulobacter. Could the SocAB TA system promote cellular adaptation to the ever-changing aquatic environment where Caulobacter is normally found and where nutrients may periodically be limited? By preventing DNA replication, SocB may initiate a global regulatory response that allows cells to adapt to environmental changes and stresses quickly.
This work raises some broader questions as well. What is the environmental signal or stressor that would activate a TA system of this type? Why are multiple TA systems so prevalent in bacteria? Perhaps elucidating the mechanism by which SocB becomes active in Caulobacter can provide us with insight into why TA systems are needed.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aarke CD, Phung TN, Huang D, Laub MT. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.10.014. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Gottesman S. Curr Opin Microbiol. 2013;16:140–147. doi: 10.1016/j.mib.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA. Proc Natl Acad Sci USA. 2001;98:11627–11632. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Fernandez C, Grosse K, Sourjik V, Collier J. Microbiology. 2013;159:2237–2248. doi: 10.1099/mic.0.068577-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Lobner-Olesen A. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Johnson A, O’Donnell M. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- Kurth I, O’Donnell M. Trends Biochem Sci. 2013;38:195–203. doi: 10.1016/j.tibs.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mruk I, Kobayashi I. Nuc Acids Res. 2013 doi: 10.1093/nar/gkt711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- Schuster CF, Bertram R. FEMS Microbiol Lett. 2013;340:73–85. doi: 10.1111/1574-6968.12074. [DOI] [PubMed] [Google Scholar]