Abstract
Pattern formation depends on the acquisition of precise cellular identities due to the differential expression of transcription factors. Enhancers within regulatory regions integrate the positive and negative regulatory signals directing gene transcription. Here, we analyze the enhancer that drives expression of the Drosophila gene spalt in the wing blade. This enhancer integrates positive signals, mediated by the Decapentaplegic signaling effector protein Medea, with the repressor activity of Brinker. The enhancer functions in the absence of binding sites for the wing-specific factor Scalloped. The molecular analysis of this enhancer indicates that there are additional factors yet unknown involved in the activation of spalt in the wing blade and that the mechanism of repression by Brinker does not rely on competition with Mad–Medea overlapping sites. The comparisons with other enhancers that respond to Decapentaplegic suggest that there are different possibilities to integrate the positive and negative inputs triggered by this signaling pathway.
In the Drosophila wing blade, Decapentaplegic (Dpp), a member of the transforming growth factor-β superfamily, controls the patterning of the veins, through the regulation of different downstream genes (for comprehensive reviews on the Dpp pathway and vein patterning, see refs. 1 and 2). Briefly, secreted Dpp forms a gradient and binds to membrane receptors that propagate the signal via phosphorylation of Mothers against dpp (Mad). This protein binds to Medea (Med), and the Mad–Med complexes translocate to the nucleus, where they regulate transcription. Brinker (Brk), a repressor of Dpp target genes, is expressed in the most lateral parts of the wing blade, while it is repressed in the central part of the wing by Schnurri (Shn), a zinc finger transcription factor that forms complexes with Mad and Med (3). Thus, Brk repression depends on Dpp, and, at the same time, it represses Dpp target genes. It has been proposed that Brk displaces the activator Mad from target sequences (4–6).
The role of Dpp on wing and vein patterning is mainly mediated by the Spalt (Sal) zinc finger transcription factors (7, 8). The expression of sal in the wing blade is strictly dependent on Dpp signaling and occurs in a broad central domain that covers from the L2 provein until the anterior limit of the L5 provein (Fig. 1C). Thus, dpp mutant discs, or clones of cells mutant for the Dpp receptor Thick veins (Tkv), completely lose sal expression (7, 9, 10). Two main inputs of Dpp signaling have been identified affecting sal expression. First, Brk represses sal expression, and Dpp is necessary to repress brk expression in the central domain of the wing by means of Shn–Mad–Med complexes. A second input of Dpp is necessary to reach normal levels of sal expression and occurs independently of Brk. Thus, tkv/brk and mad/brk double mutant clones, which lack at the same time the repressor (Brk) and the activators of the pathway (Tkv or Mad), still express sal, although at levels lower than normal (11–13). These observations indicate that some activators of sal expression are operative in the absence of Dpp signaling. It has been proposed that this activation is provided by Vestigial (Vg), a factor essential for wing and haltere disc development (14–16). Vg forms complexes with Scalloped (Sd) that bind DNA in a sequence-specific manner to regulate the expression of downstream genes (17–19).
Fig. 1.
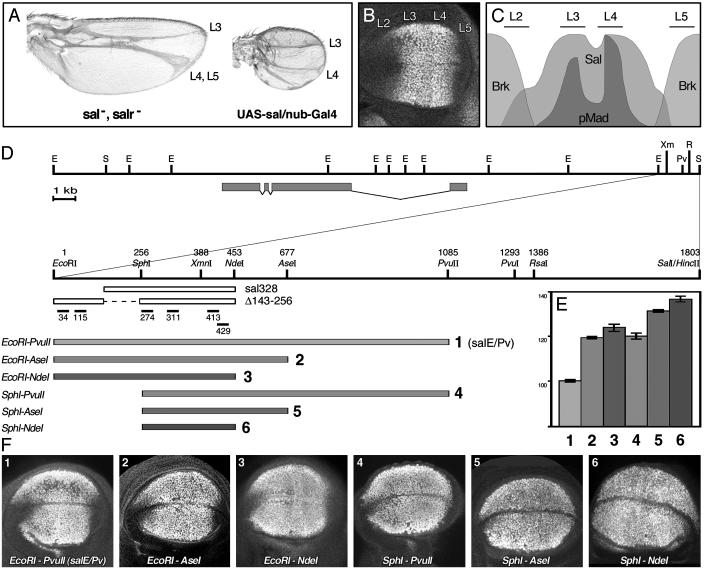
Characterization of the sal wing blade-specific enhancer. (A) Phenotypic consequences of changes in sal and salr expression in adult wings. (Left) When both genes are absent (sal-, salr-), vein L2 does not form and L4 and L5 are fused. The intervein regions L2/3 and L4/5 disappear. (Right) When sal is expressed in the whole wing blade (UAS-sal/nub-Gal4), the veins L2 and L5 do not form, and the size of the wing is reduced. (B) Immunostaining showing Sal expression in a wild-type third-instar wing imaginal disc. Vein L2 forms in the anterior compartment in the region, where sal is expressed at low levels. The high levels of Sal coincide with the presumptive veins L3 and L4. In the posterior compartment, sal expression is adjacent to the presumptive vein L5. (C) Schematic representation of the antero/posterior axis of the wing blade. Phosphorylated Mad (pMad) colocalizes with high levels of Sal protein in the presumptive veins L3 and L4. pMad and sal are less abundant at the antero/posterior border. Brk overlaps with the regions of low levels of expression of sal. (D) Genomic region surrounding the sal transcript (gray boxes) according to Kühnlein et al. (21). Below, close-up of the 1.8-kb DNA fragment that contains the sal wing blade enhancer, indicating the relevant restriction enzymes used in our analysis. The oligonucleotides used in EMSA experiments are shown below, as well as the constructs sal328 and salE/Pv-Δ143–256. In the lower part, gray bars indicate the DNA fragments cloned in front of the reporter gene lacZ. EcoRI–PvuII corresponds to the salE/Pv enhancer. (E) Measurements of the areas of β-gal expression relative to the endogenous sal expression domain. Gray tones and numbers are according to D. Bars represent mean values and standard errors. Eliminating the EcoRI–SphI fragment increases the β-gal area by 21%. Eliminating the AseI–PvuII region also increases the β-gal area by 20%. (F) Third-instar wild-type wing imaginal discs stained with anti-Sal and anti-β-gal antibodies. The pictures show only the anti-β-gal single channels. Abbreviations for restriction enzymes: E, EcoRI; Pv, PvuII; R, RsaI; S, SalI; Xm, XmnI.
sal genes show a complex pattern of expression regulated by independent enhancer regions (20, 21). Here, we characterize a sal wing blade-specific enhancer that includes all the information needed to generate the sal expression domain in the wing. This enhancer contains Brk binding sequences, responsible for the repression of sal in the lateral regions of the wing, and activator sequences that drive reporter gene expression in the sal domain. Because activation and repression regions do not overlap, a mechanism based on Mad displacement by Brk is, in this case, unlikely. Med and Sd bind to the activation region, although mutations in their binding sites do not abolish reporter expression. Furthermore, endogenous sal is still expressed in wing discs mutant for vg. Taken together, these results identify a region mediating Brk repression and corroborate that other wing-specific inputs beyond Mad–Med and Vg–Sd contribute to the activation of sal expression.
Materials and Methods
Drosophila melanogaster Strains and Clonal Analysis. Flies were raised on standard Drosophila medium at 25°C. Wing discs lacking vg were generated in larvae of genotype P[Gal4–638]; FRT42D vgnull/FRT42D M (2)l2P[arm-lacZ]; UAS-FLP. vgnull mutants are described in ref. 22. Wing discs lacking sal and salr expression in the wing blade were generated in larvae of genotype P[Gal4–638]; FRT40 Df(2L)32FP-5/FRT40 M (2)z P[arm-lacZ]; UAS-FLP (20). The Gal4 line 638 is expressed only in the wing blade (data not shown). We also used the UAS-sal (7) and nubbin (nub)-Gal4 (23). Information about strains not described in the text and balancer chromosomes can be found in Flybase (http://flybase.bio.indiana.edu).
Immunohistochemistry. Immunohistochemistry was performed in third-instar larval imaginal wing discs following standard procedures using rat anti-Sal (dilution 1:200) (24), rabbit anti-β-galactosidase (β-gal) (dilution 1:200; Cappel), and anti-Vg antibodies (dilution 1:200) (16). Fluorescent secondary antibodies were from Jackson ImmunoResearch and were used at a final 1:200 dilution. Samples were analyzed in a Bio-Rad confocal microscope. The areas of β-gal and Sal expressions were quantified by using the histogram tool of photoshop software (Adobe Systems, Mountain View, CA). The areas of β-gal expression were normalized to the endogenous Sal expression for each disc. Measurements were taken at least three times on independent immunostainings, and graphics were generated by using Microsoft excel.
DNA Binding Assays. Recombinant proteins expressed in bacteria were used in the electrophoretic mobility-shift assays (EMSAs). M. Frasch provided MED DNA binding fragments, and band-shifts were performed as described (25). M. Affolter provided Brk-expressing construct, and bandshifts were according to ref. 6. Sd full-length expressing clone was provided by J. B. Bell (19), and bandshifts were done as described (17). Oligonucleotides were labeled radioactively with T4 polynucleotide kinase, hybridized with the corresponding complementary oligonucleotide, and purified in acrylamide gels. The sequences for oligonucleotides 34, 34c2, 115, and 115c1 are indicated in Figs. 2 and 5. The sequences for oligonucleotides 274, 274aaa, 413, 413t5, 429, and 429t6 are indicated in Figs. 3 and 5. Oligonucleotides 311, 311aa, 311tt, and 311.8n appear in Figs. 4 and 5.
Fig. 2.
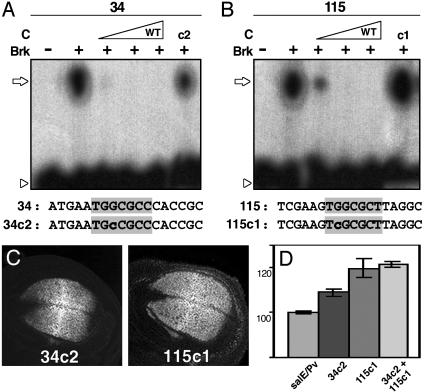
Brk is responsible for the repression of salE/Pv in the lateral regions of the wing blade. (A and B) EMSA experiments using radioactive oligonucleotides and bacterially expressed Brk protein at increasing concentrations. The sequences of the oligonucleotides used as probes and competitors are indicated under each panel. The Brk consensus binding sites are shaded in gray. Nucleotides mutated are indicated in lowercase. An open arrow indicates changes in mobility, whereas an arrowhead indicates the free probes. (A) Oligonucleotide 34 labeled radioactively shifts in the presence of recombinant Brk (+). The binding is competed for by increasing quantities of unlabeled wild-type oligonucleotide 34 (C). The mutant oligonucleotide 34c2 competes with the binding less efficiently (c2). (B) Oligonucleotide 115 binds recombinant Brk (+). Wild-type cold oligonucleotide 115 competes for the binding (C), whereas mutant 115c1 does not (c1). (C) β-gal expression driven by mutant salE/Pv enhancers in third-instar imaginal discs. Sal expression in the same discs is not shown. (D) Measurements of the areas of β-gal expression in relation to the endogenous sal expression. Note that mutation 115c1 increases β-gal expression area by 21%, whereas mutation 34c2 has a milder effect. When both sites are mutated, the reporter expression area increases similarly to single 155c1 mutant.
Fig. 5.
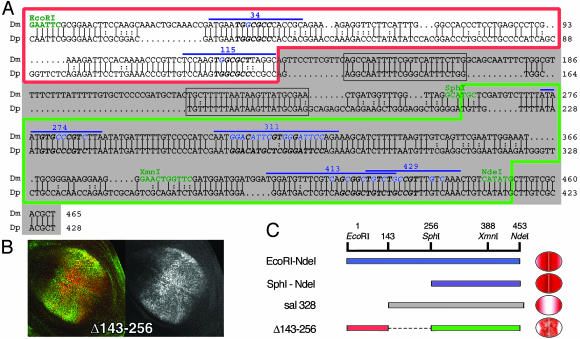
salE/Pv enhancer is conserved in D. pseudoobscura. (A) Alignment of D. melanogaster (Dm) and D. pseudoobscura (Dp) salE/Pv enhancers. Only the EcoRI–NdeI region of the enhancer is shown. Relevant restriction enzymes are shown in green. Oligonucleotides used in this analysis are indicated with a blue line. The nucleotides mutated in the different experiments are shown in blue. The red line boxes the repressor region, and the green line boxes the region important for activation. sal328 fragment is shaded in gray. The conserved regions eliminated in the mutant salE/Pv-Δ143–256 are boxed in gray. Italics and bold characters indicate consensus sequences for Brk, Med, or Sd. (B) Expression of Sal (green) and β-gal (red) in third-instar wing imaginal discs driven by the salE/Pv-Δ143–256 fragment. The corresponding red channel is in black and white. (C) Schematic representation of the EcoRI–NdeI fragment, which drives the reporter expression in the central part of the wing blade. When the EcoRI–SphI fragment is removed, the expression expands laterally (SphI–NdeI). The normal domain of expression is recovered when the repressor sites are added, albeit the levels of expression are heterogeneous (Δ143–256). However, the elimination of the repressor sites produces a change in the specificity of the enhancer that now drives reporter expression only in the lateral regions of the wing blade (sal328).
Fig. 3.
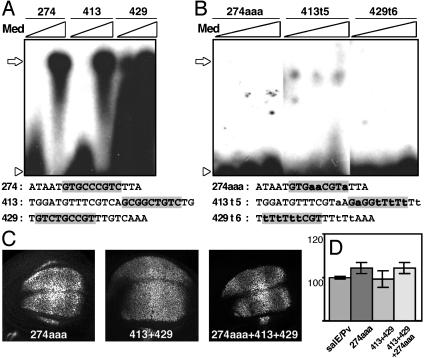
Med contributes to salE/Pv enhancer expression in certain regions of the wing blade. (A and B) EMSA experiments using radioactive oligonucleotides and bacterially expressed Med protein at increasing concentrations. The sequences of the oligonucleotides used as probes are indicated under each panel. The sequence consensus for Mad–Med binding is shaded in gray. The mutated nucleotides are indicated in lowercase. Open arrows indicate Med binding, whereas open arrowheads indicate the free probe. (A) Med binds to oligonucleotides 274, 413, and 429. (B) Mutated oligonucleotides 274aaa, 413t5, and 429t6 do not bind Med. (C) Expression of β-gal in third-instar discs driven by mutant salE/Pv enhancers. (D) Measurements of the areas of β-gal expression in relation to endogenous sal expression. The antero/posterior extension of the β-gal domain in the different mutants does not change in respect to the salE/Pv control.
Fig. 4.
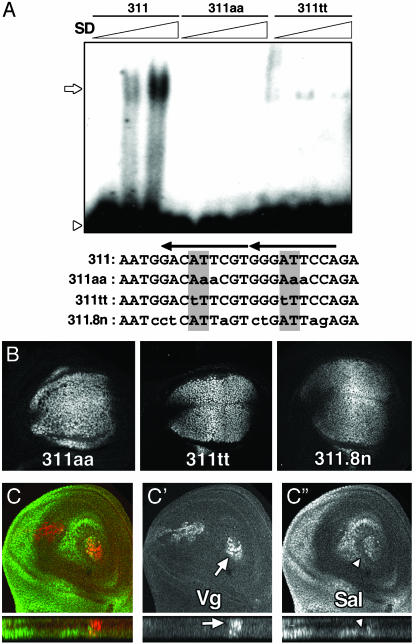
Sd–Vg complex is not essential for sal expression in the wing blade. (A) EMSA radiograms of bacterially expressed Sd protein. Wild-type oligonucleotide 311 shifts in the presence of increasing quantities of Sd. Binding is abolished by mutation 311aa or greatly diminished by mutation 311tt. Below the panel, the above oligonucleotide sequences are shown, as well as the sequence of the 311.8n mutant. The mutations are indicated in lowercase. The AT core from the TEA domain (TEF1, TEC1, ABAA) consensus sequences is shaded, and arrows indicate the Sd consensus binding sites. Open arrows indicate Sd binding, whereas open arrowheads indicate the free probe. (B) β-gal expression driven by mutant salE/Pv in third-instar imaginal discs. Sal expression in the same discs is not shown. (C) Expression of Sal (green) and Vg (red) in early third-instar wing imaginal discs with large territories mutant for vg. Red (C′) and green (C″) channels are shown in black and white. Despite the absence of Vg, Sal is expressed in the blade. Orthogonal sections below C′ show the remaining Vg-expressing cells (arrow). Around them, sal-expressing cells appear in a different plane (C″, arrowhead).
Analysis of the Regulatory Regions and Generation of Mutant DNA. Genomic DNA was cloned in the enhancer tester vector C4PLZ (26). The constructs were introduced in the germ line by P-element transformation as described (27). The reporter expression was detected by immunostaining of third-instar larvae. For each construct, at least three independent lines were analyzed. For mutagenesis, we used the QuikChange site-directed mutagenesis kit (Stratagene). Mutations introduced in the salE/Pv enhancer are indicated in the figures. Altered nucleotides are indicated in lowercase. Mutation 413 + 429 is 5′-TGGATGTTTCGTaAGaGGtTtTtTttCGTTTtTtAAACTGTCATA-3′. To generate the deletion Δ143–256, a new SphI restriction site was created by PCR using the oligonucleotides 1(+)EcoRI (5′-CGAATTCGCGGAACTTCCAAG-3′) and 124(-)SphI (5′-TCGAGCATGCACGAAGGAACTGCCTAAGCG-3′). The resulting fragment was cloned into the EcoRI–SphI sites of salE/Pv.
DNA Comparison and Sequence Analysis. For DNA analysis, we used the GCG package (Wisconsin University) and blastn (National Center for Biotechnology Information). salE/Pv enhancer European Molecular Biology Laboratory accession number is AJ578459. For transcription factor binding site prediction, we used matinspector (Genomatix).
Results
Characterization of the sal Wing Blade-Specific Enhancer. The expression of sal in the wing blade is regulated at the level of transcription, and changes in the expression domain bring about major morphological consequences (7). Thus, when sal and salr are absent from the entire wing blade, the vein L2 does not form and the intervein regions L2/3 and L4/5 disappear, causing an anterior shift of the L3 and L5 veins (Fig. 1 A). When sal is ectopically expressed in the whole wing blade, the veins L2 and L5 do not form and the size of the wing is reduced (Fig. 1 A) (8). The normal domain of sal expression coincides with the region where Dpp signaling is active, as revealed by the presence of phosphorylated Mad (pMad; Fig. 1 B and C) (28). The lateral regions of low expression of sal are partially coincident with the expression of the brk–lacZ reporter (Fig. 1C) (11).
A fragment of genomic DNA 1.8 kb long, sal1.8S/E, located 5′ of sal, drives the expression of lacZ in the sal wing blade domain (data not shown) (21). By cutting down the original sal1.8S/E fragment, we generated the construct salEcoRI–PvuII that reproduces the sal endogenous expression and is not detected in the lateral parts of the blade (Fig. 1 D, row 1, E, lane 1, and F, image 1). The expression of β-gal is prominent in the lateral regions of the Sal domain, where sal is normally expressed at low intensity; this is likely due to the lifespan of the reporter protein. The constructs salEcoRI–AseI and salEcoRI–NdeI, which lack the AseI–PvuII and NdeI–PvuII regions, respectively, expand the reporter expression laterally (Fig. 1 D, rows 2 and 3, E, lanes 2 and 3, and F, images 2 and 3). When we eliminated the EcoRI–SphI region from these constructs, generating the sal-SphI–PvuII, salSphI–AseI, and salSphI–NdeI constructs, the expression of lacZ expanded further laterally (Fig. 1 D, rows 4–6, E, lanes 4–6, and F, images 4–6).
Two main conclusions emerge from this analysis. First, the region responsible for the activation of the reporter is located in the 197-bp-long SphI–NdeI fragment. Second, the repression of the reporter in the lateral regions of the blade resides in the EcoRI–SphI and NdeI–PvuII fragments. All of the necessary activator and repressor sequences would be included in the salEcoRI–PvuII construct (henceforth referred to as salE/Pv).
Brk Represses the Expression of salE/Pv in the Lateral Regions of the Wing Blade. Brk is the main repressor of sal in the most lateral parts of the wing blade. To analyze whether Brk repression is direct, we examined the binding of Brk protein to probes along the salE/Pv enhancer (data not shown). The EcoRI–SphI fragment contains three consensus GGCG(c/t)(c/t) sequences (6). From these, only sequences 34 and 115 are able to bind Brk in vitro (Fig. 2 A and B and data not shown). The binding is competed for by wild-type oligonucleotides but not by the mutated forms 34c2 and 115c1 (Fig. 2 A and B). Furthermore, Brk does not bind to 34c2 and 115c1 mutant oligonucleotides (data not shown). The fragment AseI–PvuII contributes to the repression of the reporter in the lateral regions of the blade (compare Fig. 1E, lanes 1 to 2 or lanes 4 to 5). This fragment contains two GGCG(c/t)(c/t) sequences that show low affinity for Brk in vitro under our experimental conditions (data not shown). The mechanism of repression exerted through the AseI–PvuII fragment remains unexplored.
To analyze the functionality of the 34 and 115 Brk sites in vivo, we introduced the 34c2 or 115c1 mutations in the salE/Pv construct by directed mutagenesis and analyzed the salE/Pv-34c2 and salE/Pv-115c1 constructs for reporter expression in transgenic flies. The 115c1 mutation expands the domain of β-gal expression similarly to the elimination of the EcoRI–SphI fragment (Fig. 2 C and D, compare with Fig. 1 E, lane 4, and F, image 4). The mutation 34c2 produces a milder lateral expansion of the reporter gene (Fig. 2 C and D). The two mutations together produce an expansion similar to the 115c1 mutation alone (Fig. 2D). In summary, we conclude that Brk can repress reporter expression in the lateral regions of the blade through the EcoRI–SphI fragment.
Med Is Necessary for Normal Levels of Expression of salE/Pv in the Wing Blade. Dpp signaling is necessary for the activation of sal in the wing blade. Heteroligomers of Mad and Med transmit the signaling into the nucleus through their binding to target DNAs. The SphI–NdeI fragment, which is able by itself to activate reporter expression in the wing blade and thus should respond to Dpp signaling (Fig. 1 E and F), contains Mad and Med binding sequences [CGCCGC(g/c)G(c/a)C] (25). Oligonucleotides 274, 413, and 429, containing consensus sites, bind Med in vitro (Fig. 3A). When mutated, the resulting oligonucleotides 274aaa, 413t5, and 429t6, lose their capacity to bind Med (Fig. 3B).
To analyze the functional relevance of these Med binding sites, we introduced the above mutations in the salE/Pv fragment. salE/Pv-274aaa shows β-gal expression in a region similar to the control salE/Pv (Fig. 3 C and D). Interestingly, the expression of the reporter is reduced in the regions where Sal is detected at maximal levels in the wild-type discs, that is the presumptive veins L3 and L4, where pMad is highly expressed (Figs. 1C and 3C). This effect is not observed when the 413 and 429 sites are mutated (Fig. 3C). However, when the three sites are mutated at the same time, salE/Pv-274aaa + 413 + 429 construct, the effect of the 274aaa mutation increases (Fig. 3C). None of the three mutations shows expansion or reduction of expression of the reporter laterally (Fig. 3D). We conclude that the Med binding sites analyzed contribute to achieve the correct pattern of expression of the reporter gene despite the fact that the reporter is still expressed in the wing blade when these sites are mutated.
Vg/Sd Function Is Not Required for sal Expression in the Wing Blade. Previous reports suggested that the Vg/Sd complex is necessary through DNA interactions for the activation of sal in the wing blade. It has been shown that a large 10.2-kb region upstream of sal (sal10.2S/C, which includes salE/Pv and drives the expression of a reporter gene in a manner similar to sal) depends on Sd for its expression in the wing blade (21, 29). Moreover, a smaller fragment of 328 bp (sal328), which is expressed in the lateral regions of the wing blade, binds Sd in vitro (see Figs. 1D and 5C). Mutations in this fragment that diminish Sd binding affinity eliminate the expression of the reporter in the wing blade (29), suggesting its dependence on Sd expression.
To analyze the relevance of Vg/Sd for the expression of the reporter, we analyzed the affinity of salE/Pv enhancer for this complex. Within the salE/Pv enhancer, the oligonucleotide 311 contains two Sd consensus sequences able to bind Sd (aCATTcc; Fig. 4A). We generated different mutant forms of 311 that abolish (311aa) or greatly diminish (311tt) Sd binding capacity (Fig. 4A). To analyze the significance in vivo of the described mutations, we introduced them in the salE/Pv enhancer and monitored the expression of β-gal in transgenic discs. None of these mutations eliminates reporter expression in the wing blade (Fig. 4B). Furthermore, the same mutations that diminish Sd binding and abolish reporter expression in the sal328 context (29) do not eliminate expression of salE/Pv-311.8n (Fig. 4B).
The results exposed above do not support a requirement for Sd in the regulation of the salE/Pv wing enhancer. To analyze the requirement for Vg in sal regulation, we generated mutant discs lacking Vg function in large territories of the wing blade (see Materials and Methods). Third-instar larval discs that lack Vg are still able to express sal (Fig. 4C). Similarly, smaller clones lacking Vg in proximal regions of the wing blade maintain sal expression (data not shown). These clones are hardly detected in distal regions of the wing blade because of low viability (L. A. Baena-López, personal communication). Therefore, we conclude that lack of Vg function does not eliminate sal expression in the wing blade.
salE/Pv Enhancer Is Conserved in Drosophila pseudoobscura. The D. pseudoobscura homologous sal gene, as well as the salE/Pv enhancer homologous region, was identified by blast search. The comparison between salE/Pv from D. melanogaster versus D. pseudoobscura shows that the oligonucleotides 34 and 115, involved in the repression of sal in the lateral regions of the blade, are highly conserved (Fig. 5A). In D. pseudoobscura, the Sd binding sequences in oligonucleotide 311 diverge from D. melanogaster, although the core binding sequences for TEA domains (TEF-1, TEC1, ABAA) are conserved. The other regions important for the activation of salE/Pv in the wing blade, corresponding to oligonucleotides 274, 413, and 429, are highly conserved between the two species, which supports their relevance for the activation of sal genes.
Our analysis demonstrates that the SphI–NdeI region (boxed in green in Fig. 5A) activates reporter expression in the wing blade, although expanded laterally (Fig. 1 D, row 6, E, lane 6, and F, image 6). However, the sal328 fragment (shaded in gray in Fig. 5A) drives the expression of the reporter gene only laterally, despite that it contains the SphI–NdeI region (Fig. 5C). salSphI–NdeI and sal328 fragments differ only in the nucleotides 143–246, which include two conserved regions (boxed in Fig. 5A). We analyzed the relevance of these sequences by removing them from salE/Pv enhancer. When analyzed in transgenic larvae, salE/Pv-Δ143–256 drives reporter expression in a manner comparable to salE/Pv (Fig. 5B). However, this expression does not seem homogenous. In conclusion, the conserved boxes in the region 143–256 are dispensable for the expression of the enhancer, although they confer consistency to its activity. Furthermore, the areas shown in our analysis to be relevant for the activation and repression of the salE/Pv enhancer are conserved in D. pseudoobscura.
Discussion
The enhancers are key elements in the regulation of gene expression, as they integrate activator and repressor signals from different transcription factors. Thus, the analysis of regulatory regions helps us to understand the molecular networks behind developmental decisions. The region that directs sal expression in the wing blade is especially interesting, as sal plays a fundamental role in wing growth and pattern formation and is transcribed there in response to the Dpp signaling pathway.
General Structure of the Wing Blade sal Enhancer. The starting point of our analysis is the salE/Pv enhancer, which drives expression of a reporter gene in the domain of sal expression. This fragment can be subdivided into two regions. The EcoRI–SphI fragment appears to confer repression of sal in the most anterior and posterior regions of the wing blade. Consequently, elimination of this region results in the expansion of reporter expression. This fragment contains two Brk binding sites, and their elimination abolishes repressor activity, suggesting that Brk acts through these sites to repress sal transcription. The adjacent region is necessary for reporter activation in the wing blade, and we expect it to contain the binding sites for the transcriptional activators that regulate the endogenous sal gene. The existence of separate repressor and activator regions within the sal enhancer impose several restrictions to possible models of regulation. For example, competition of Mad–Med for Brk binding sites as a way to alleviate transcriptional repression is, in this case, unlikely. Moreover, it is not likely that Brk and Mad–Med complexes coexist in the same cells in the wing blade.
The Activator Region of the sal Enhancer. Our results show that the Med binding sites 274 and 413–429 are necessary for the normal expression of salE/Pv enhancer, even though when mutated they do not abolish the enhancer activity. The effects of mutant Med binding sites are observed only in some wing territories, implicating that sal is regulated distinctly in particular subdomains of its normal territory of expression.
Sd binding sites have been shown to be required for the expression of the sal328 fragment in the wing blade (29). However, in the context of the salE/Pv enhancer, modifications that prevent Sd binding do not abolish the activity of the enhancer. Moreover, the analysis of expression of sal in vg mutant territories confirms that the Sd/Vg complex is dispensable for the expression of sal in the wing blade. This observation does not exclude that these factors may be important to achieve normal levels of sal expression.
It is interesting to note that the DNA fragment between nucleotides 143 and 256 (deleted in the salE/Pv-Δ143–256 construct) is not necessary for reporter expression, but changes its specificity (Fig. 5 B and C). Thus, when this fragment is placed next to the activator segment SphI–NdeI (as is the case for the sal328 fragment), the reporter is no longer expressed in the central part of the wing blade but in the sides, where sal is not normally expressed and where the repressor Brk is expressed (Fig. 5C). Two DNA boxes within the 143–256 DNA fragment are conserved between D. melanogaster and D. pseudoobscura. The possible interaction or interactions mediated by these boxes confer consistency to the salE/Pv enhancer. The switch from the central to the lateral domains of the wing blade mediated by the 143–256 DNA fragment remains unexplored.
Other Enhancers Regulated by Dpp. The study of the regulatory regions of several Dpp target genes indicates that enhancers responding to this pathway can be organized in different ways. The UBX midgut-specific enhancer contains Mad–Med and Brk overlapping binding sequences (4, 30). In this enhancer, Cre binding sequences are adjacent to the Mad–Med site, contributing to the activation of the enhancer. Similarly, the zerknüllt (zen) Dpp responding enhancer contains multiple Mad binding sites overlapping with Brk binding sites (5). Here, occupancy of all Mad sites when pMad is highly concentrated is responsible for activation, whereas occupancy of Brk sites is responsible for repression. Among the wing blade-specific enhancers studied, optomotor blind (omb) activator and repressor sequences also overlap. omb is also repressed by Brk in the lateral parts of the wing, but it does not need Mad–Med for its activation (12). Interestingly, point mutations in the Brk binding site abolish reporter expression in the wing blade, indicating the presence of overlapping activator sequences (6). In the case of sal, activation and repression regions are adjacent. Here, activation involves Med binding but requires additional unknown factors. Thus, although the identity of the positive regulators driving the expression of omb or sal is unknown, positive regulatory regions can be adjacent to repressor regions (sal) or overlap with them (omb). One fundamental question that arises from this analysis is the nature of the factor or factors activating sal beyond the Dpp signaling pathway. At present, we cannot answer this question, as additional candidate genes involved in the regulation of sal have not yet been identified. The different mechanisms used to integrate positive and negative inputs in each Dpp-responding enhancer probably confer the flexibility necessary to regulate gene expression in the variety of developmental processes where Dpp signaling is involved.
Acknowledgments
We thank S. Carroll, J. M. Monje, S. Sotillos, A. Srivastava, and A. Vigano for reagents; H. H. Lee for Mad and Med protocols; L. A. Baena-López for sharing unpublished results; L. Sánchez-Pulido for help in the blast searches; R. Hernandez, A. López, and E. Caminero for technical help; and G. Morata and J. Modolell for critical reading of the manuscript; and especially Antonio García-Bellido and Fotis C. Kafatos for their continuous support. This work was supported by an institutional grant from the Ramón Areces Foundation for Centro de Biología Molecular Severo Ochoa and by Ministerio de Ciencia y Tecnología Grant BMC2000-1191.
Abbreviations: β-gal, β-galactosidase; EMSA, electrophoretic mobility-shift assay.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ578459).
References
- 1.Affolter, M., Marty, T., Vigano, M. A. & Jazwinska, A. (2001) EMBO J. 20, 3298-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Celis, J. F. (2003) BioEssays 25, 443-451. [DOI] [PubMed] [Google Scholar]
- 3.Muller, B., Hartmann, B., Pyrowolakis, G., Affolter, M. & Basler, K. (2003) Cell 113, 221-233. [DOI] [PubMed] [Google Scholar]
- 4.Kirkpatrick, H., Johnson, K. & Laughon, A. (2001) J. Biol. Chem. 276, 18216-18222. [DOI] [PubMed] [Google Scholar]
- 5.Rushlow, C., Colosimo, P. F., Lin, M. C., Xu, M. & Kirov, N. (2001) Genes Dev. 15, 340-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sivasankaran, R., Vigano, M. A., Muller, B., Affolter, M. & Basler, K. (2000) EMBO J. 19, 6162-6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Celis, J. F., Barrio, R. & Kafatos, F. C. (1996) Nature 381, 421-424. [DOI] [PubMed] [Google Scholar]
- 8.de Celis, J. F. & Barrio, R. (2000) Mech. Dev. 91, 31-41. [DOI] [PubMed] [Google Scholar]
- 9.Lecuit, T., Brook, W. J., Ng, M., Calleja, M., Sun, H. & Cohen, S. M. (1996) Nature 381, 387-393. [DOI] [PubMed] [Google Scholar]
- 10.Nellen, D., Burke, R., Struhl, G. & Basler, K. (1996) Cell 85, 357-368. [DOI] [PubMed] [Google Scholar]
- 11.Campbell, G. & Tomlinson, A. (1999) Cell 96, 553-562. [DOI] [PubMed] [Google Scholar]
- 12.Jazwinska, A., Kirov, N., Wieschaus, E., Roth, S. & Rushlow, C. (1999) Cell 96, 563-573. [DOI] [PubMed] [Google Scholar]
- 13.Marty, T., Muller, B., Basler, K. & Affolter, M. (2000) Nat. Cell Biol. 2, 745-749. [DOI] [PubMed] [Google Scholar]
- 14.Baena-Lopez, L. A. & Garcia-Bellido, A. (2003) Development (Cambridge, U.K.) 130, 197-208. [DOI] [PubMed] [Google Scholar]
- 15.Kim, J., Sebring, A., Esch, J. J., Kraus, M. E., Vorwerk, K., Magee, J. & Carroll, S. B. (1996) Nature 382, 133-138. [DOI] [PubMed] [Google Scholar]
- 16.Williams, J. A., Bell, J. B. & Carroll, S. B. (1991) Genes Dev. 5, 2481-2495. [DOI] [PubMed] [Google Scholar]
- 17.Halder, G., Polaczyk, P., Kraus, M. E., Hudson, A., Kim, J., Laughon, A. & Carroll, S. (1998) Genes Dev. 12, 3900-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paumard-Rigal, S., Zider, A., Vaudin, P. & Silber, J. (1998) Dev. Genes Evol. 208, 440-446. [DOI] [PubMed] [Google Scholar]
- 19.Simmonds, A. J., Liu, X., Soanes, K. H., Krause, H. M., Irvine, K. D. & Bell, J. B. (1998) Genes Dev. 12, 3815-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrio, R., de Celis, J. F., Bolshakov, S. & Kafatos, F. C. (1999) Dev. Biol. 215, 33-47. [DOI] [PubMed] [Google Scholar]
- 21.Kühnlein, R. P., Bronner, G., Taubert, H. & Schuh, R. (1997) Mech. Dev. 66, 107-118. [DOI] [PubMed] [Google Scholar]
- 22.Van de Bor, V., Delanoue, R., Cossard, R. & Silber, J. (1999) Cell Death Differ. 6, 557-564. [DOI] [PubMed] [Google Scholar]
- 23.Calleja, M., Moreno, E., Pelaz, S. & Morata, G. (1996) Science 274, 252-255. [DOI] [PubMed] [Google Scholar]
- 24.de Celis, J. F., Barrio, R. & Kafatos, F. C. (1999) Development (Cambridge, U.K.) 126, 2653-2662. [DOI] [PubMed] [Google Scholar]
- 25.Xu, X., Yin, Z., Hudson, J. B., Ferguson, E. L. & Frasch, M. (1998) Genes Dev. 12, 2354-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wharton, K. A., Jr., & Crews, S. T. (1993) Mech. Dev. 40, 141-154. [DOI] [PubMed] [Google Scholar]
- 27.Rubin, G. M. & Spradling, A. C. (1982) Science 218, 348-353. [DOI] [PubMed] [Google Scholar]
- 28.Tanimoto, H., Itoh, S., ten Dijke, P. & Tabata, T. (2000) Mol. Cell 5, 59-71. [DOI] [PubMed] [Google Scholar]
- 29.Guss, K. A., Nelson, C. E., Hudson, A., Kraus, M. E. & Carroll, S. B. (2001) Science 292, 1164-1167. [DOI] [PubMed] [Google Scholar]
- 30.Saller, E. & Bienz, M. (2001) EMBO Rep. 2, 298-305. [DOI] [PMC free article] [PubMed] [Google Scholar]