Abstract
The fate of pluripotent stem cells is tightly controlled during early embryonic development. Both the derivation and the maintenance of embryonic stem cells (ES cells) in vitro depend on feeder cell-derived growth factors that are largely unidentified. To dissect the mechanisms governing pluripotency, we conducted a screen to identify factors that are produced by mouse embryonic fibroblast STO cells and are required to maintain the pluripotency of ES cells. One of the factors is bone morphogenetic protein 4 (BMP4). Unexpectedly, the major effect of BMP4 on the self-renewal of ES cells is accomplished by means of the inhibition of both extracellular receptor kinase (ERK) and p38 mitogen-activated protein kinase (MAPK) pathways, and inhibitors of ERK and p38 MAPKs mimic the effect of BMP4 on ES cells. Importantly, inhibition of the p38 MAPK pathway by SB203580 overcomes the block in deriving ES cells from blastocysts lacking a functional Alk3, the BMP type IA receptor. These results uncover a paradigm for BMP signaling in the biology of pluripotent stem cells.
Embryonic stem cells (ES cells) are able to form all cell types of the body by following normal embryogenesis (1–3). The pluripotency of ES cells has attracted great attention for their potential use in tissue and cell therapy. However, the molecular and developmental mechanisms controlling pluripotency and differentiation of ES cells are largely unknown, and only a very limited number of genes has so far been shown to affect the fate decisions of inner cell mass (ICM) or ES cells. These genes include Oct4, Fgf4, H2az, Foxd3, Nanog, and Sox2 (4–12).
Growth factors required for ES cell self-renewal are usually provided by feeder cells, or exogenously (13). Leukemia-inhibiting factor (LIF) and its close relatives are the known propluripotency factors for mouse ES cells. It is unclear how many other growth factors or signaling pathways are required for the self-renewal of ES cells. To address these questions, we set out to identify such factors that affect the self-renewal of ES cells. To accomplish this, we isolated and screened sublines of the mouse embryonic fibroblast STO cells for their ability to support the self-renewal of ES cells by using Oct4-GFP as a convenient marker of pluripotency (14–16). By this approach in combination with gene expression profiling, we have identified bone morphogenetic protein 4 (BMP4) as part of the extracellular propluripotency cues. Also, our studies show that BMP4 and LIF have synergistic effect in teratoma formation. Moreover, a number of genes differentially expressed in ES cells cultured with or without exogenous BMP4 have been identified.
One of these differentially expressed genes, X-linked inhibitor of apoptosis (Xiap), is expressed at higher levels in ES cells cultured in the presence of exogenous BMP4 than in its absence. XIAP has been implicated in connecting the type I receptors of BMPs and TGF-βs with the mitogen-activated protein kinase (MAPK) p38 pathway (17–20). Contrary to previous findings in which BMP signaling up-regulates p38 MAP kinase (17, 20, 21), our findings reveal that BMP4 inhibits MAP kinase pathways in ES cells. Moreover, inhibitors of extracellular receptor kinase (ERK) (PD98059) and p38 (SB203580) pathways mimic BMP4 in the self-renewal of ES cells. Importantly, inhibition of p38 by using SB203580 allows the derivation of Alk3 (BmprIa)-/- ES cell lines, which was otherwise not possible. Therefore, these studies by using a combination of biochemical and genetic approaches have identified BMP4 as a propluripotency factor. Inhibition of MAP kinase pathways represents an essential aspect of BMP4/ALK3 signaling in regulating the self-renewal of mouse ES cells.
Materials and Methods
Derivation of ES Cells. Oct4-GFP transgenic mice on a 129/SvEv background were used to obtain embryonic day 3.5 (E3.5) blastocysts that were used to establish ES cell lines with a standard procedure (22). The complete ES cell medium is composed of DMEM (12800-017, GIBCO) supplemented with 4 mM l-glutamine/4.5 g/liter glucose/1.5 g/liter sodium bicarbonate/100 units/ml penicillin/100 μg/ml streptomycin/55 μM 2-mercaptoehtanol/100 μM nonessential amino acid/1,000 units/ml LIF (Chemicon)/20% FBS (HyClone).
Derivation of Sublines of STO Cells and Preparation of Feeder Cells. STO cells were cultured in DMEM supplemented with 4 mM l-glutamine/4.5 g/liter glucose/1.5 g/liter sodium bicarbonate/100 units/ml penicillin/100 μg/ml streptomycin/10% FBS. We transfected ≈5 × 106 STO cells (American Type Culture Collection) with 20 μg of linearized PGK-Neor DNA in 800 μl of PBS by using Gene Pulser II (Bio-Rad) at 250 V and 800 μF. We plated ≈50% of the surviving cells into 960 wells and selected them in the presence of G418 (180 μg/ml) for 14 days. Cells in 300 wells grew up, and 120 sublines with adherent morphology were screened for their ability to support ES cells. COS cells were cultured as described (23). Treatment of feeder cells with mitomycin C (10 μg/ml) and further handling were performed as described (22).
RNA Purification, Microarray Analysis, and RT-PCR. STO cells were cultured to 90–95% confluence in STO medium and then in ES cell medium for 24 h before RNA purification with UltraSpec II RNA isolation kit (Biotecx Laboratories, Houston). For RNA purification from ES cells, ≈2 × 107 cells were plated onto a 100-mm dish containing COS-Bmp4 or Cos-Bmp8bh cells with complete ES cell medium. RNAs were purified 3 days later. Microarray analysis was carried out at the University of Texas Southwestern Medical Center Microarray Core Facility by using U74A V2 chips (Affymetrix, Santa Clara, CA). Total RNAs from STO or ES cells were used to make first-strand cDNAs with avian myeloblastosis virus reverse transcriptase, oligo(dT), and random primers. cDNAs were normalized with Hprt and were then subjected to PCR amplification for other sequences with the different dilutions (×0.1, ×0.5, and ×1.0) to ensure that at least one of the reactions was in relatively linear range of amplification with appropriate cycle numbers. See Table 1, which is published as supporting information on the PNAS web site, for PCR primer sequences.
DNA Constructs and Transfection. pIRES and pBmp4 were described (23). pBmp8bh was constructed by inserting murine Bmp8b cDNA containing hemagglutinin following the cleavage site into MCS-B of pIRES. Transfection of the above vectors into COS cells was as described (23). The mixture of transfected COS cells with persistent G418 selection was used as feeder cells. CAG-puro was generated by replacing CMV promoter in Puro3 (Clontech) with CAG promoter (24). Transfection of these plasmids into ES cells was carried out with 1 × 107 cells in 800 μl of PBS at 250 V and 600 μF. After transfection, ES cells were selected in the presence of 2 μg/ml puromycin for 14 days to obtain positive clones.
Western Blotting. We plated ≈2 × 106 STO or COS cells into 100-mm plates to grow for 24 h, and we then switched them to 5 ml of medium with 0.5% FBS for 24 h. Conditioned medium was concentrated with Ultrafree-15 centrifugal filter 10KNMWL (Millipore). Equal amounts of concentrates was loaded onto SDS/PAGE gels for size fractionation and Western blot analysis for semiquantitation of BMP4 proteins with an mAb against the mature domain of BMP4 (MAB875, R & D Systems).
Detection of Phospho-MAPKs. Bmp4-/- ES cells were harvested from cultures on STO cells, resuspended in medium containing 10% FBS without LIF, and divided into aliquots (each had 4 × 106 cells in 2 ml of medium). Purified BMP4 proteins (R & D Systems) were added to several aliquots to a final concentration of 50 ng/ml. Cells were harvested 5, 15, 30, and 60 min after BMP4 addition for detection of phospho-ERK, p38, and stress-activated protein kinase for several initial experiments. Western blotting was conducted with reagents obtained from Cell Signaling Technology (Beverly, MA).
MAPK Inhibitors. SB203580 (1 or 2 μM) and PD98059 (12.5 μM) were used as suggested in refs. 9 and 25.
Results
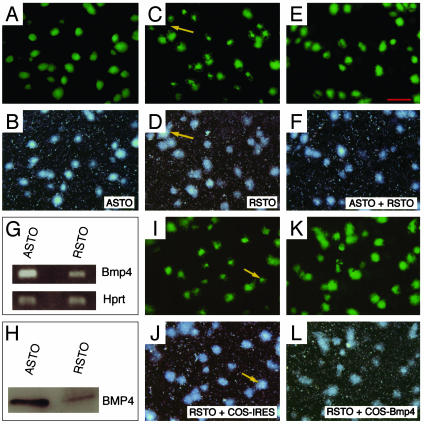
BMP4 as a Candidate Factor That Inhibits ES Cell Differentiation. Oct4-GFP ES cells and STO cell sublines were generated as described in Materials and Methods. We combined 18 STO sublines that supported robust Oct4-GFP expression of ES cells to create a mixture named ES cell-assisting STO (ASTO). One inferior subline was designated as ES cell-resisting STO (RSTO). As shown in Fig. 1 A and B, in low-density culture, every ES cell colony growing on ASTO maintained a robust and uniform Oct4-GFP expression, whereas those on RSTO showed partial loss of Oct4-GFP expression, indicative of differentiation (Fig. 1 C and D). However, ES cells on a mixture of equal number of ASTO and RSTO cells resembled those on ASTO cells (Fig. 1 E and F), indicating that ASTO provided dominant factor(s) inhibiting ES cell differentiation.
Fig. 1.
Identification of BMP4 as one of the STO-produced factors mimicking ASTO activity. We plated ≈2,000 ES cells onto a 35-mm dish containing ASTO or RSTO cells. Photos were taken 5 days after the culture with complete ES cell medium. (A and B) Fluorescent and dark-field photomicrographs of ES cell colonies on ASTO. All colonies on ASTO had robust Oct4-GFP expression. (C and D) Fluorescent and dark-field photomicrographs of ES cell colonies on RSTO. Partial loss of GFP was obvious in many colonies, and a big colony only expressed GFP in a small area (arrow). (E and F) ES cells cultured on an equal mixture of ASTO and RSTO. The Oct4-GFP expression resembled those in A and B. (G) RT-PCR showing ASTO expressed higher levels of Bmp4 than RSTO with Hprt RT-PCR for normalization. (H) Western blotting showing ASTO-conditioned medium containing higher levels of BMP4 than RSTO-conditioned medium (equal loading). (I and J) ES cells cultured on RSTO with COS-IRES-conditioned medium had a similar phenotype as those shown in C and D. (K and L) ES cells cultured on RSTO with COS-Bmp4-conditioned medium showed robust Oct4-GFP expression in almost all colonies. Scale bar in A–F and I–L, 700 μm.
To identify possible factor(s) that inhibits ES cell differentiation, we compared gene expression patterns between ASTO and RSTO by using murine A chips (Affymetrix). Several hundred genes were expressed at higher levels (≥1.5-fold) in ASTO than in RSTO (see Table 2, which is published as supporting information on the PNAS web site). Among these are sequences encoding extracellular proteins, including Igf2, Gas6, Dan, sFrp1, sFrp2, Proliferin, Wnt6, Igfbp4, Vegf, Tnf member 9, Ephrin B1, Scf, and Bmp4. Mutations in Bmp2, Bmp4, and Bmp8b result in either an absence or a reduction in the number of primordial germ cells in embryos after gastrulation (26–29), and primordial germ cells and ES cells are related closely. Therefore, our effort was directed to the possible role of BMP4 in ES cells.
As shown in Fig. 1 G and H, RT-PCR and Western blotting with equal amount of conditioned medium confirmed that BMP4 was indeed expressed at higher levels in ASTO than in RSTO. To test whether BMP4 serves as a factor inhibiting ES cell differentiation, we obtained conditioned medium from COS cells expressing BMP4 (COS-Bmp4) and control COS cells containing parental pIRES vector (COS-IRES), and we then cultured ES cells on RSTO with the conditioned media for 5 days. As shown in Fig. 1 I–L, ES cell colonies grown on RSTO with COS-Bmp4-conditioned medium had little loss of Oct4-GFP. Thus, RSTO cells with sufficient exogenous BMP4 behaved as ASTO cells.
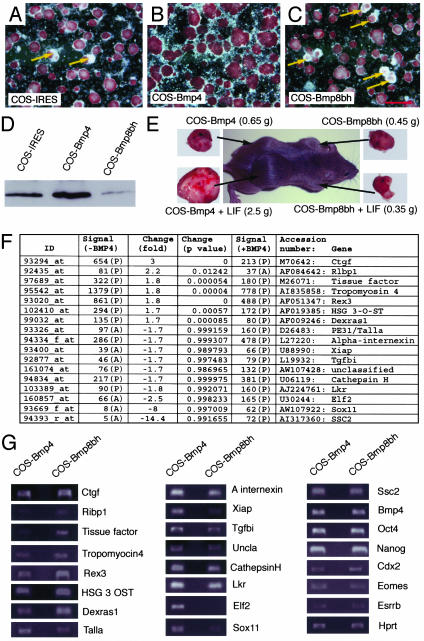
BMP4 and LIF Have Synergistic Effects on the Self-Renewal of ES Cells. To validate that BMP4 serves as a propluripotency factor, we compared alkaline phosphatase (ALP, one of the markers of undifferentiated ES cells) expression in ES cells cultured on COS feeder cells containing different expression vectors in the presence of LIF. These include COS-IRES, COS-Bmp4, and COS-Bmp8bh. As shown in Fig. 2 A–C, 5 days after culture, ES cell colonies on different feeders showed variable loss of ALP (loss of red staining), a sign of differentiation. The effectiveness of different feeder cells in preventing the loss of ALP or Oct4-GFP were summarized in the following order: COS-Bmp4 > COS-IRES > COS-Bmp8bh. These observations correlated well with the levels of BMP4 expression by feeder cells as shown by Western blotting using equal amounts of conditioned medium (Fig. 2D). COS-IRES showed some endogenous BMP4 in COS cells. COS-Bmp4 expressed higher levels of BMP4; however, only very low concentrations of BMP4 were detected in COS-Bmp8bh, suggesting that expression of BMP8BH inhibits the production of mature BMP4.
Fig. 2.
BMP4 prevents the differentiation of ES cells. (A–C) We plated ≈2 × 105 ES cells onto each 35-mm dish with COS-IRES, COS-Bmp4, or COS-Bmp8bh as feeders. After culture for 5 days with complete ES cell medium, cells were fixed in 4% paraformaldehyde/PBS and stained for ALP. (A) ES cells grown on COS-IRES. Loss of ALP was observed in some colonies (arrows). (B) ES cells grown on COS-Bmp4. Little loss of ALP was observed. (C) ES cells grown on COS-Bmp8bh. More colonies lost ALP expression than in A. (D) Western blotting to detect BMP4 proteins in conditioned medium from COS cells. COS-IRES-conditioned medium contained BMP4 corresponding to the endogenous expression. COS-Bmp4 expressed BMP4 at higher levels, whereas COS-Bmp8bh-conditioned medium only contained very low levels of BMP4. (E) Teratoma formation assay. ES cells were cultured on COS-Bmp4 and COS-Bmp8bh with or without LIF for 3 days. We injected ≈2 × 106 ES cells in 50 μl of PBS from these cultures into 129/SvEv males s.c.. Tumors were observed and dissected 4 weeks after injection, with tumor weight labeled for each culture condition. (F) Shown are 17 differentially expressed sequences between ES cells cultured on COS-Bmp4 and COS-Bmp8bh (P < 0.015). (G) Semiquantitative RT-PCR to confirm 17 differentially expressed sequences. Except Ssc2 (no difference), all sequences were shown to be consistent with the microarray data shown in F. RT-PCR also showed that Bmp4, Oct4, and Nanog were expressed in both groups of ES cells at similar levels. After 45 cycles of amplification, markers for trophoblast Cdx2, Eomes, and Esrrb were detected without significant differences between the two samples. Scale bar in A–C, 700 μm.
To further analyze the pluripotency of ES cells cultured with BMP4, we compared the teratoma-forming ability of ES cells cultured on COS-Bmp4 and COS-Bmp8bh cells with or without LIF for 3 days. For these experiments, 2 × 106 ES cells in 50 μl of PBS were injected s.c. into 129/SvEv males. After 4 weeks, three of the four injected mice revealed teratomas in all four groups of ES cells. One of the injected mice had only a small tumor in the COS-Bmp4 plus LIF group after 5 weeks, and no tumors developed from other injected ES cells. Although the sizes of the tumors varied among recipients, the tumor formed from ES cells treated with COS-Bmp4 plus LIF was always the largest (at least four times as big as others) in each recipient (Fig. 2E). These results indicate that BMP4 or LIF alone have only minor effects on pluripotency, whereas in combination, they have synergy on the self-renewal of ES cells. A similar conclusion was reached also by Ying et al. (30).
Differential Gene Expression in ES Cells Cultured on COS-Bmp4 and COS-Bmp8bh. To investigate the mechanisms of BMP signaling and to identify genes regulated by BMP4 in ES cells, gene expression profiling was conducted with mouse A chips (Affymetrix) on ES cells cultured on COS-Bmp4 and COS-Bmp8bh for 3 days. Between the two cell populations, ≈200 sequences were differentially expressed (≥1.5-fold) (see Table 3, which is published as supporting information on the PNAS web site, for the complete microarray data). We chose 17 sequences for further analysis (Fig. 2F). As shown in Fig. 2G, except Ssc2 that showed no difference between the two samples, the differential expression of other 16 sequences were confirmed by RT-PCR. Under these experimental conditions no differences were observed between the two samples in trophoblast markers (Eomes, Esrrb, and Cdx2). This result is contrary to a report (31) that BMP4 causes human ES cells to differentiate into trophoblast.
Among the proteins encoded by these sequences as shown in Fig. 2F, XIAP has been identified as a protein bridging BMP type IA receptor, ALK3, and TAK1 (TGF-β activated kinase 1) through TAB1 (TAK1 binding protein 1) to activate p38 MAPK in Xenopus (17, 32, 33). Therefore, our efforts were directed toward the relationship among BMP4, XIAP, and MAPKs in ES cells.
Overexpression of XIAP Disrupts the Self-Renewal of ES Cells. Although BMP signaling and XIAP are linked to the p38 MAPK pathway during Xenopus axis formation, the relationship between BMP, XIAP, and p38 pathway in ES cells is not known. To address this issue, we transfected a full-length mouse Xiap cDNA in the expression vector CAG-puro into wild-type ES cells. After three rounds of transfection, CAG-Xiap-puro yielded no ES cell colonies containing the transgene, whereas the control CAG-puro vector produced >500 colonies each round. These results suggest that over expression of XIAP disrupts ES cell self-renewal. This notion is supported further by a tetracycline-regulated expression of XIAP (Tet-on) in four independent ES cell lines. ES cell colonies were fewer and smaller when 1 μg/ml doxycyline was added to the culture than those without doxycycline (see Fig. 6, which is published as supporting information on the PNAS web site).
The evidence that XIAP transduces BMP signaling by upregulating p38 MAPK was obtained by means of its interaction with ALK3. It is not clear whether other type I BMP receptor(s) interacts with XIAP to elicit similar responses. We, therefore, conducted another round of transfection experiments into both wild-type and Alk3-/- ES cells (their derivation is explained below). Consistent with the above findings, no or few colonies were obtained in wild-type ES cells. Interestingly, >500 colonies were obtained in Alk3-/- ES cells, indicating that XIAP-induced disruption of ES cell self-renewal is ALK3-dependent. Therefore, these genetic data strongly suggest that ALK3 is the only type I BMP receptor in ES cells that interacts with XIAP to cause ES cell differentiation.
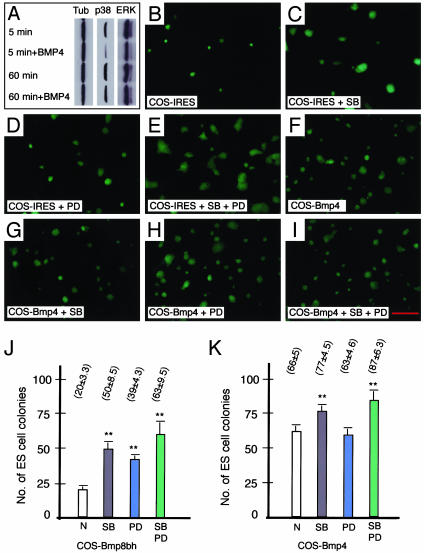
BMP4 Inhibits Both ERK and p38 MAPKs in ES Cells. If BMP4 stimulates p38 MAPK through XIAP, the consequence is not major because the net effect of BMP4 on the self-renewal of ES cells is opposite to that of XIAP. To investigate further the influence of BMP4 on MAPKs, we examined the activities of three classes of MAPKs in ES cells cultured with or without exogenous BMP4 under feeder cell-free conditions. Bmp4-/- ES cells were used in this experiment to eliminate the influence of endogenous BMP4. As shown in Fig. 3A, the active p38 and ERK MAPKs (phospho forms) in ES cells were reduced shortly (5 min) after the addition of BMP4 compared with the control. However, the phosphorylation of ERK recovered completely at 60 min, whereas the phosphorylation of p38 only recovered partially. Similar recoveries were observed at 15 and 30 min, suggesting that a regulatory mechanism is in place to balance MAPK activities rapidly in ES cells. Finally, the stress-activated protein kinase appeared to be unaffected by BMP4 (data not shown).
Fig. 3.
Relationship of BMP4 signaling and MAPK pathways. (A) BMP4 inhibits p38 and ERK MAPKs. A Bmp4-/- ES cell line was used for these studies. The levels of phospho-p38 and ERK were reduced significantly 5 min after BMP4 addition (50 ng/ml). A complete recovery for ERK and partial recovery for p38 were observed at 60 min. α-Tubulin was used as a loading control. (B) ES cells cultured on COS-IRES. We plated ≈5 × 104 ES cells on a 35-mm dish that contained COS-IRES feeder cells. ES cells were cultured for 3 days and then photos were taken to show colonies. (C) ES cells cultured on COS-IRES with p38 inhibitor SB203580 (1 μM). Colonies were larger than those in B. (D) ES cells cultured on COS-IRES with MEK inhibitor PD98059 (12.5 μM). Colonies were similar to those in C.(E) ES cells cultured on COS-IRES with both PD98059 and SB203580. (F) ES cells cultured on COS-Bmp4. (G) ES cells cultured on COS-Bmp4 plus SB203580. (H) ES cells cultured on COS-Bmp4 with PD98059. (I) ES cells cultured on COS-Bmp4 plus both inhibitors. (J) Effects of MAPK inhibitors on colony formation of ES cells. We plated 500 ES cells on COS-Bmp8bh feeder cells in each of the 35-mm dish and cultured in ES cell medium with or without MAPK inhibitors. Colonies were counted 5 days later. Each group was in triplicate. (K) Colony formation experiments as described for J were conducted with COS-Bmp4 as feeders. *, P < 0.01. Scale bar in B–I, 700 μm.
MAPK Inhibitors Mimic BMP4 in Supporting ES Cells. Inactivation of either BMP type I receptor Alk3 or type II receptor BmprII causes mouse embryos to die without gastrulation (34, 35). Furthermore, efforts to derive Alk3-/- ES cells were unsuccessful (36, 37). Although SMAD proteins are considered to be the major signal transducers of TGF-βs and BMPs, ES cells can be derived readily from blastocysts lacking SMAD4 (the common partner of all SMADs), indicating that the highest activity of SMAD1/5/8 is not essential for the self-renewal of ES cells (38). To test whether inhibition of MAPKs is a crucial aspect of BMP signaling in pluripotency, we cultured ES cells on COS feeder cells with or without MEK inhibitor (upstream of ERK) PD98059 or p38 inhibitor SB203580. Growing ES cells on COS-IRES with LIF had only a minor effect on the proliferation of ES cells, whereas adding PD98059, SB203580, or both improved ES cell self-renewal dramatically (Fig. 3 B–E). However, these inhibitors only revealed some minor effects on ES cells cultured on COS-Bmp4 (Fig. 3 F–I). These results were further corroborated by colony formation assays. ES cells cultured on COS-Bmp8bh had a 2- to 3-fold increase in colony numbers when SB203580, PD98059, or both were included in the culture medium (Fig. 3J). By contrast, under similar conditions these inhibitors had minor or no effect on colony numbers when ES cells were cultured on COS-Bmp4, suggesting that BMP4 was already inhibiting these pathways (Fig. 3K).
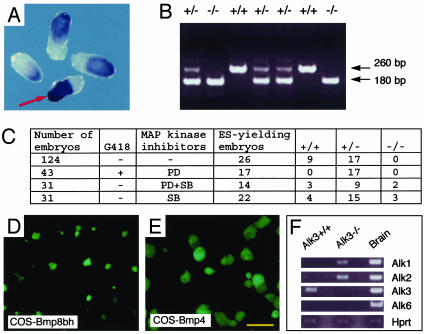
Inhibition of p38 Pathway Allows the Establishment of Alk3-/- ES Cells. Alk3 is the only type I BMP receptor detectable in the pluripotent ICM and early epiblast (34). Although Alk3-/- mutants survive to E6.5–7.0, they do not form mesoderm and fail to gastrulate. Before their demise these embryos have a burst of high levels of Oct4 expression in the epiblast (Fig. 4A), one of the first signs of differentiation for pluripotent cells (8, 39). Furthermore, after numerous attempts with 124 blastocysts from Alk3+/- crosses, Alk3-/- ES cells were not obtained, validating the importance of BMP signaling through ALK3 in maintaining pluripotency (Fig. 4C).
Fig. 4.
Derivation of Alk3-/- ES cells by inhibition of p38 MAPK. (A) Wholemount in situ hybridization of Alk3 mutant embryos at E6.0–6.5 with Oct4 antisense RNA probes. Only one embryo was Alk3-/- and showed a much higher level of Oct4 expression and wrinkled edges of the epiblast (arrow). The rest of the embryos were either wild type or Alk3+/-.(B) Genotype results for ES cells containing the Alk3 null allele in C.(C) Inhibition of p38 MAPK pathway rescued Alk3-/- ES cells. Culturing 124 blastocysts from Alk3+/- crosses in complete ES cell medium only yielded ES cells from wild type (n = 9) or heterozygotes (n = 17). With ERK pathway inhibitor PD98059 and G418 (180 μg/ml) in culture medium, 17 of 43 blastocysts gave rise to ES cells and all were Alk3+/-. With a combination of PD98059 and SB203580 in the medium, the following 14 of 31 blastocysts yielded ES cells: three wild type, nine heterozygotes, and two homozygotes. To test whether inhibition of p38 pathway alone was sufficient or not to derive Alk3-/- ES cells, 31 blastocysts were cultured in the presence of SB203580. Among 22 ES-cell-yielding blastocysts, 3 were Alk3-/- (4 wild-type and 15 heterozygotes). (D) Alk3-/- ES cells cultured on COS-Bmp8bh. We plated ≈1 × 104 ES cells onto COS-Bmp8bh and cultured them in ES cell medium for 4 days. ES cell colonies were rather small. (E) Alk3-/- ES cells cultured on COS-Bmp4. ES cell colonies were much larger than those shown in D, indicating that Alk3-/- ES cells are still responsive to BMP4 and other BMP type I receptors compensate for the loss of Alk3. (F) Expression of putative BMP type I receptors in wild-type and Alk3-/- ES cells by RT-PCR. Brain cDNA was used as a control. Alk3 was the major type I BMP receptor in wild-type ES cells, whereas both Alk1 and Alk2 were present in Alk3-/- mutant ES cells. Scale bar in A, D, and E, 700 μm.
As described above, BMP signaling inhibits both ERK and p38 MAPK pathways, and inhibitors of either pathways support the self-renewal of ES cells in the absence of exogenous BMP4. If inhibition of ERK and/or p38 pathways is the most important aspect of BMP signaling in ES cells, it should be possible to derive ES cell lines from Alk3-/- embryos by adding PD98059 and/or SB203580. Our first trial of 43 blastocysts with PD98059 and G418 (to eliminate wild-type embryos) only yielded ES cells from 17 heterozygous embryos. Thus, PD98059 alone appeared ineffective. The second trial of 31 blastocysts with PD98059 and SB203580 yielded ES cells from 14 embryos, including 2 Alk3-/- mutants, suggesting either the combination of two inhibitors or SB203580 alone was effective. The third trial of 31 blastocysts with SB203580 alone obtained ES cells from 22 embryos, including 3 Alk3-/- mutants. Therefore, SB203580 alone was effective. When established, these ES cells were able to selfrenew in the absence of SB203580. Furthermore, Alk3-/- ES cells contributed to the whole epiblast in chimeras, but they failed to gastrulate just as Alk3-/- embryos did, indicating that Alk3-/- ES cells do not behave differently than their in vivo counterparts (data not shown). Moreover, these mutant ES cells were also responsive to exogenous BMP4 (Fig. 4 D and E), which is very likely because of the up-regulation of other putative type I BMP receptors, Alk1 and/or Alk2, after several passages, to compensate for the loss of Alk3 (Fig. 4F).
Discussion
The function of Bmp4 appears complex during early mouse embryogenesis. Our previous epiblast culture studies showed that BMP4 and BMP8B acted synergistically to induce primordial germ cells (23). However, epiblasts from Bmp4-/- embryos could not be induced by BMP4 and BMP8B to form primordial germ cells, suggesting that Bmp4-/- epiblasts had lost pluripotency by E6.0. Therefore, based on these results, we proposed that BMP4 had an earlier role at blastocyst and/or early epiblast stages by maintaining pluripotency (23). This notion is validated by the current article and by Ying et al. (30).
MAPK pathways are on the crossroad of signal transduction of many mitogens including LIF, BMPs, and FGFs (40, 41). It is likely that the pluripotency of ES-like cells results from a delicate balance of MAPK activity influenced by many growth factors in vivo and in vitro. A slight increase in MAPK activity can potentially tip the balance to direct ES cells toward a differentiation path, whereas even minor inhibition of MAPK pathways may be sufficient to refrain ES cells from differentiation. The reduction in ERK and p38 phosphorylation after BMP4 addition are likely due to increased activity of dual-specificity (threonine/tyrosine) and/or tyrosine-specific MAPK phosphatases, which are rapidly modulated by positive and negative feedback regulations (42). One of these phosphatases, hematopoietic protein tyrosine phosphatase (HePTP), is expressed in ES cells (data not shown). HePTP was shown to negatively regulate both ERK and p38 MAPKs in T cells (43). The cAMP-dependent protein kinase (PKA) inhibits, whereas protein phosphatase 1 and 2A stimulate, the activity of HePTP in T cells. The effect of BMP4 signaling on any of these components in ES cells should be further investigated.
Ying et al. (30) reported that BMP4 suppresses ES cell differentiation by induction of Id genes. At present, it is not clear whether such up-regulation results solely from activated SMAD proteins or from a combined SMAD activation and inhibition of MAPKs. Ying et al. (30) also tested BMP4 on MAPKs in ES cells and did not find the same effect as reported here. They further examined the effect of 30 μM SB203580 on ES cells, and no positive effect was observed. It is worth noting that the IC50 of SB203580 on p38 MAPK is 0.5 μM according to Davies et al. (44), and a concentration of 30 μM should have affected other kinases. On the other hand, 1 or 2 μM SB203580 was used in this study, and its positive effect on ES cells was supported further by the derivation of ES cell lines from Alk3-/- embryos.
Alk3-/- and wild-type blastocysts appear to be indistinguishable morphologically. However, upon culture for several days in vitro by using conventional methods, the ICM of Alk3-/- mutants failed to expand and no ES cell colonies could be formed. This result does not mean that Alk3 is not necessary for ICM generation. Because Alk3 is rather ubiquitously expressed including in ovary and oocytes, maternal ALK3 may allow the Alk3-/- embryos to form the epiblast. As soon as the maternal proteins are exhausted in early embryonic cells, they will lose pluripotency unless a downstream major target is altered to mimic BMP4/ALK3 signaling, in this case, by means of the inhibition of p38 MAPK by SB203580. Therefore, the significance of p38 MAPK inhibition by BMP4 is demonstrated biochemically and genetically and should be the major effect of BMP4/ALK3 signaling. As shown by Ying et al. (30), SMAD proteins and Id genes play roles in BMP4 signaling during ES cell self-renewal; however, it has not yet been demonstrated that activation of any of these genes is able to rescue Alk3-/- ES cells. Furthermore, the fact that ES cells can be readily derived from Smad4-/- blastocysts casts some doubts about the functions of SMAD proteins in ES cell self-renewal (38).
If other BMP receptors did not compensate for the loss of ALK3, Alk3-/- ES cells would quickly differentiate upon withdrawal of SB203580. The continued self-renewal of Alk3-/- ES cells after SB203580 withdrawal and their responsiveness to BMP4 suggest an up-regulation of another type I BMP receptor. According to previous studies, the most likely candidate type I receptor should be Alk6 (45) because only ALK3 and ALK6 have been shown to bind BMP4. Surprisingly, Alk6 was not up-regulated in Alk3-/- ES cells. In contrast, an up-regulation of both Alk1 and Alk2 was observed (Fig. 4F), suggesting that ALK1 and/or ALK2 proteins are able to serve as BMP4 type I receptors in the absence of ALK3.
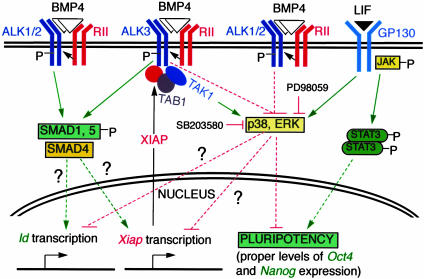
The signaling networks that regulate ES cell biology are complex, and our knowledge of these regulations is rather primitive at present. As summarized in a working model in Fig. 5, XIAP serves as one of the feedback inhibitors of BMP4 signaling in ES cells. However, it is not clear how BMP4 signaling accomplishes the up-regulation of XIAP and the inhibition of ERK and p38 MAPKs. These questions should be addressed in future studies.
Fig. 5.
Schematic representation of BMP4 and LIF signaling in pluripotency. Upon dimeric BMP4 binding to the receptor complex, type II BMP receptor (RII) phosphorylates and activates type IA receptor (ALK3). Activated ALK3 causes the inactivation (dephosphorylation) of p38 and ERK MAPKs by unknown mechanisms. This inactivation relieves the negative effects of MAPKs on pluripotency (the proper levels of Oct4 and Nanog expression), as confirmed further by using MAPK inhibitors SB203580 and PD98059. As a result of the activation of ALK3, transcription of Xiap is up-regulated either by means of the inhibition of p38/ERK or activation of SMAD1/5/8 pathway. XIAP protein in the cytoplasm interacts with ALK3 and TAB1 to result in the activation of TAK1 by ALK3, consequently, a recovery of p38 activity. In the absence of ALK3, ALK1, or ALK2 proteins act as BMP4 type I receptor to maintain the pluripotency of ES cells. XIAP does not cause the loss of pluripotency in the absence of ALK3, presumably because of its inability to interact with ALK1 or ALK2. Lastly, the synergistic effect of BMP4 and LIF is likely attributed to the ability of BMP4 to antagonize the stimulation of ERK by LIF, inhibit p38 MAPK activity, and up-regulate Id genes and to the ability of LIF to activate JAK-STAT pathway. Solid lines indicate a direct effect.
Supplementary Material
Acknowledgments
We thank Drs. Richard Behringer, Melanie Cobb, David Garbers, Robert Hammer, Brigid Hogan, and Timothy Megraw for helpful suggestions on the study and critical comments on the manuscript; Drs. Yasuhisa Matsui and Hans Scholer for Oct4-GFP mice; and Dr. Sarah Comerford and Nikolaus Schultz for advice on gene expression profiling. This work was supported in part by National Institute of Child Health and Human Development Grants HD36218 and HD39154 (to G.-Q.Z.) and the Cecil H. and Ida Green Center for Reproductive Biology Sciences.
Abbreviations: ES cell, embryonic stem cell; En, embryonic day n; BMP, bone morphogenetic protein; ICM, inner cell mass; LIF, leukemia-inhibiting factor; MAPK, mitogen-activated protein kinase; ERK, extracellular receptor kinase; ASTO, ES cell-assisting STO; RSTO, ES cell-resisting STO; ALP, alkaline phosphatase.
References
- 1.Martin, G. R. (1981) Proc. Natl. Acad. Sci. USA 78, 7634-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans, M. J. & Kaufman, M. H. (1981) Nature 292, 154-156. [DOI] [PubMed] [Google Scholar]
- 3.Bradley, A., Evans, M., Kaufman, M. H. & Robertson, E. (1984) Nature 309, 255-256. [DOI] [PubMed] [Google Scholar]
- 4.Nichols, J., Zevnik, B., Anastassiadis, K., Niwa, H., Klewe-Nebenius, D., Chambers, I., Scholer, H. & Smith, A. (1998) Cell 95, 379-391. [DOI] [PubMed] [Google Scholar]
- 5.Hanna, L. A., Foreman, R. K., Tarasenko, I. A., Kessler, D. S. & Labosky, P. A. (2002) Genes Dev. 16, 2650-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faast, R., Thonglairoam, V., Schulz, T. C., Beall, J., Wells, J. R., Taylor, H., Matthaei, K., Rathjen, P. D., Tremethick, D. J. & Lyons, I. (2001) Curr. Biol. 11, 1183-1187. [DOI] [PubMed] [Google Scholar]
- 7.Feldman, B., Poueymirou, W., Papaioannou, V. E., DeChiara, T. M. & Goldfarb, M. (1995) Science 267, 246-249. [DOI] [PubMed] [Google Scholar]
- 8.Niwa, H., Miyazaki, J. & Smith, A. G. (2000) Nat. Genet. 24, 372-376. [DOI] [PubMed] [Google Scholar]
- 9.Burdon, T., Stracey, C., Chambers, I., Nichols, J. & Smith, A. (1999) Dev. Biol. 210, 30-43. [DOI] [PubMed] [Google Scholar]
- 10.Mitsui, K., Tokuzawa, Y., Itoh, H., Segawa, K., Murakami, M., Takahashi, K., Maruyama, M., Maeda, M. & Yamanaka, S. (2003) Cell 113, 631-642. [DOI] [PubMed] [Google Scholar]
- 11.Chambers, I., Colby, D., Robertson, M., Nichols, J., Lee, S., Tweedie, S. & Smith, A. (2003) Cell 113, 643-655. [DOI] [PubMed] [Google Scholar]
- 12.Avilion, A. A., Nicolis, S. K., Pevny, L. H., Perez, L., Vivian, N. & Lovell-Badge, R. (2003) Genes Dev. 17, 126-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith, A. G. (2001) Annu. Rev. Cell Dev. Biol. 17, 435-462. [DOI] [PubMed] [Google Scholar]
- 14.Yoshimizu, T., Sugiyama, N., De Felice, M., Yeom, Y. I., Ohbo, K., Masuko, K., Obinata, M., Abe, K., Scholer, H. R. & Matsui, Y. (1999) Dev. Growth Differ. 41, 675-684. [DOI] [PubMed] [Google Scholar]
- 15.Anderson, R., Copeland, T. K., Scholer, H., Heasman, J. & Wylie, C. (2000) Mech. Dev. 91, 61-68. [DOI] [PubMed] [Google Scholar]
- 16.Scholer, H. R., Balling, R., Hatzopoulos, A. K., Suzuki, N. & Gruss, P. (1989) EMBO J. 8, 2551-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi, K., Nagai, S., Ninomiya-Tsuji, J., Nishita, M., Tamai, K., Irie, K., Ueno, N., Nishida, E., Shibuya, H. & Matsumoto, K. (1999) EMBO J. 18, 179-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birkey Reffey, S., Wurthner, J. U., Parks, W. T., Roberts, A. B. & Duckett, C. S. (2001) J. Biol. Chem. 276, 26542-26549. [DOI] [PubMed] [Google Scholar]
- 19.Kimura, N., Matsuo, R., Shibuya, H., Nakashima, K. & Taga, T. (2000) J. Biol. Chem. 275, 17647-17652. [DOI] [PubMed] [Google Scholar]
- 20.von Bubnoff, A. & Cho, K. W. (2001) Dev. Biol. 239, 1-14. [DOI] [PubMed] [Google Scholar]
- 21.Iwasaki, S., Iguchi, M., Watanabe, K., Hoshino, R., Tsujimoto, M. & Kohno, M. (1999) J. Biol. Chem. 274, 26503-26510. [DOI] [PubMed] [Google Scholar]
- 22.Hogan, B., Beddington, R., Constantini, F. & Lacy, E. (1994) Manipulating the Mouse Embryo: A Laboratory Manua (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 23.Ying, Y., Qi, X. & Zhao, G. Q. (2001) Proc. Natl. Acad. Sci. USA 98, 7858-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niwa, H., Burdon, T., Chambers, I. & Smith, A. (1998) Genes Dev. 12, 2048-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, X. Z. & Ron, D. (1996) Science 272, 1347-1349. [DOI] [PubMed] [Google Scholar]
- 26.Ying, Y., Liu, X. M., Marble, A., Lawson, K. A. & Zhao, G. Q. (2000) Mol. Endocrinol. 14, 1053-1063. [DOI] [PubMed] [Google Scholar]
- 27.Ying, Y. & Zhao, G. Q. (2001) Dev. Biol. 232, 484-492. [DOI] [PubMed] [Google Scholar]
- 28.Lawson, K. A., Dunn, N. R., Roelen, B. A., Zeinstra, L. M., Davis, A. M., Wright, C. V., Korving, J. P. & Hogan, B. L. (1999) Genes Dev. 13, 424-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao, G. Q. & Garbers, D. L. (2002) Dev. Cell 2, 537-547. [DOI] [PubMed] [Google Scholar]
- 30.Ying, Q. L., Nichols, J., Chambers, I. & Smith, A. (2003) Cell 115, 281-292. [DOI] [PubMed] [Google Scholar]
- 31.Xu, R. H., Chen, X., Li, D. S., Li, R., Addicks, G. C., Glennon, C., Zwaka, T. P. & Thomson, J. A. (2002) Nat. Biotechnol. 20, 1261-1264. [DOI] [PubMed] [Google Scholar]
- 32.Shibuya, H., Yamaguchi, K., Shirakabe, K., Tonegawa, A., Gotoh, Y., Ueno, N., Irie, K., Nishida, E. & Matsumoto, K. (1996) Science 272, 1179-1182. [DOI] [PubMed] [Google Scholar]
- 33.Shibuya, H., Iwata, H., Masuyama, N., Gotoh, Y., Yamaguchi, K., Irie, K., Matsumoto, K., Nishida, E. & Ueno, N. (1998) EMBO J. 17, 1019-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishina, Y., Suzuki, A., Ueno, N. & Behringer, R. R. (1995) Genes Dev. 9, 3027-3037. [DOI] [PubMed] [Google Scholar]
- 35.Beppu, H., Kawabata, M., Hamamoto, T., Chytil, A., Minowa, O., Noda, T. & Miyazono, K. (2000) Dev. Biol. 221, 249-258. [DOI] [PubMed] [Google Scholar]
- 36.Mishina, Y. (2003) Front Biosci. 8, D855-D869. [DOI] [PubMed] [Google Scholar]
- 37.Zhao, G. Q. (2003) Genesis 35, 43-56. [DOI] [PubMed] [Google Scholar]
- 38.Sirard, C., de la Pompa, J. L., Elia, A., Itie, A., Mirtsos, C., Cheung, A., Hahn, S., Wakeham, A., Schwartz, L., Kern, S. E., Rossant, J. & Mak, T. W. (1998) Genes Dev. 12, 107-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmieri, S. L., Peter, W., Hess, H. & Scholer, H. R. (1994) Dev. Biol. 166, 259-267. [DOI] [PubMed] [Google Scholar]
- 40.Cobb, M. H. (1999) Prog. Biophys. Mol. Biol. 71, 479-500. [DOI] [PubMed] [Google Scholar]
- 41.Kyriakis, J. M. & Avruch, J. (2001) Physiol. Rev. 81, 807-869. [DOI] [PubMed] [Google Scholar]
- 42.Keyse, S. M. (2000) Curr. Opin. Cell Biol. 12, 186-192. [DOI] [PubMed] [Google Scholar]
- 43.Nika, K., Huynh, H., Williams, S., Paul, S., Bottini, N., Tasken, K., Lombroso, P. J. & Mustelin, T. (2003) Biochem. J. 378, 335-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies, S. P., Reddy, H., Caivano, M. & Cohen, P. (2000) Biochem. J. 351, 95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi, Y. & Massague, J. (2003) Cell 113, 685-700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.