Abstract
Most adaptation is thought to occur through the fixation of numerous alleles at many different loci. Consequently, the independent evolution of similar phenotypes is predicted to occur through different genetic mechanisms. The genetic basis of adaptation is still largely unknown, however, and it is unclear whether adaptation to new environments utilizes ubiquitous small-effect polygenic variation or large-effect alleles at a small number of loci. To address this question, we examined the genetic basis of bony armor loss in three freshwater populations of Alaskan threespine stickleback, Gasterosteus aculeatus, that evolved from fully armored anadromous populations in the last 14,000 years. Crosses between complete-armor and low-armor populations revealed that a single Mendelian factor governed the formation of all but the most anterior lateral plates, and another independently segregating factor largely determined pelvic armor. Genetic mapping localized the Mendelian genes to different chromosomal regions, and crosses among these same three widely separated populations showed that both bony plates and pelvic armor failed to fully complement, implicating the same Mendelian armor reduction genes. Thus, rapid and repeated armor loss in Alaskan stickleback populations appears to be occurring through the fixation of large-effect variants in the same genes.
A central tenet of evolutionary theory is that adaptation in the wild, like artificial selection, occurs gradually through the sequential fixation of small-effect variants (1). Consequently, the independent evolution of similar phenotypes is expected to use unique combinations of genes and alleles (2). New populations, however, are often established in novel environments at the edge of an organism's range, and selective pressures faced in these new habitats are often an important causative factor for adaptive radiations (3). Importantly, novel environments may also have immediate disruptive effects on developmental processes that can expose novel genetic variants, some of which may have large effects on evolving phenotypes (4, 5). The importance of genes of major effect is currently the focus of renewed research (6, 7). The role of major effect genes during adaptation, however, is still unclear, as is the frequency with which recurrent phenotypic evolution occurs through changes in the same (8–11) or different (8, 12, 13) genes. In addition, the genetics of adaptation has most often been studied in the laboratory (14), with much less work in natural populations (13). To address these problems, we have taken advantage of a unique natural system, the rapid postglacial diversification of threespine stickleback, Gasterosteus aculeatus (15). Thousands of coastal freshwater populations of stickleback have derived independently from anadromous (sea-run) ancestors. Phenotypically similar throughout their range, anadromous stickleback are protected with bony armor including lateral plates and a robust set of dorsal and pelvic spines (Fig. 1D). In contrast, derived lacustrine populations (Fig. 1 A–C) exhibit extensive recurrent phenotypic diversification, including armor loss (16).
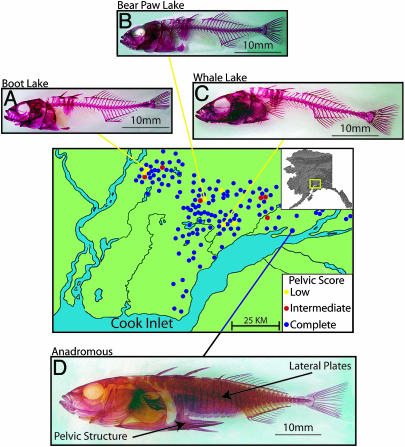
Fig. 1.
Map of the Matanuska-Susitna (Mat-Su) Valley of Alaska with trypsin clearedandAlizarinredstainedsticklebackshowingBootLake(A),BearPawLake(B) and Whale Lake (C) freshwater low-armor populations, and anadromous Rabbit Slough complete-armor ancestor (D). The complete-armor anadromous form has a full set of lateral plates and fully formed pelvic structure (arrows), whereas each freshwater population lacks most of the lateral plates and has either highly reduced (Boot, Bear Paw) or completely absent (Whale) pelvic structures. Blue dots indicate additional populations with mostly complete pelvic structure (mean score of 5.0), red are mostly intermediate (2.0–4.0), and yellow are low (0.0–2.0).
Researchers have long been interested in these armor structures (17–20), and have recently begun to use molecular genetic tools to study stickleback development (21–24). We have built on existing stickleback work by examining the genetic basis of independent armor loss in three freshwater populations in south-central Alaska (Fig. 1 and Table 1), an area that was until recently (9,000–14,000 years B.P.) covered by ice (25). Population genetic analyses indicate that lacustrine populations in this region were derived from a common complete-armor anadromous ancestor that was most likely similar to the present day anadromous population in this region (26). The three armor-reduced freshwater populations we studied are in isolated drainages leading to the sea and are surrounded by populations (some <500 m away) that have complete-pelvic armor (Fig. 1), as would be expected if each population lost its armor independently. The low-armor populations are geographically distant from each other along potential water routes (70–200 km) as well as directly overland (15–35 km), and are separated by numerous geographic barriers (Fig. 1), further supporting their independent armor loss. Although fish from oceanic and lacustrine populations have highly divergent phenotypes and seldom mate in the wild, they can be crossed in the laboratory by using in vitro techniques. We performed complementation tests to ask whether the genetic basis of armor loss was the same in the three different populations, made mapping crosses to follow the segregation of armor phenotypes, and identified the chromosomal location for armor-loss genetic factors.
Table 1. Variation in lateral plate number and pelvic score across populations and cross types.
| Lateral plates
|
Pelvic structure
|
||||||
|---|---|---|---|---|---|---|---|
| Type of collection or cross | n | Left | Right | n | Left | Right | |
| Parental Anadromous | Wild | 116 | 33.2 (0.06) | 33.2 (0.07) | 116 | 5.0 (0.00) | 5.0 (0.00) |
| Laboratory | 115 | 33.1 (0.06) | 33.1 (0.06) | 115 | 5.0 (0.00) | 5.0 (0.00) | |
| Parental Bear Paw Lake | Wild | 102 | 3.9 (0.08) | 3.9 (0.08) | 102 | 1.0 (0.08) | 0.7 (0.07) |
| Laboratory | 131 | 3.8 (0.08) | 3.8 (0.08) | 131 | 1.0 (0.09) | 0.5 (0.05) | |
| Parental Boot Lake | Wild | 106 | 5.2 (0.08) | 5.2 (0.08) | 106 | 0.9 (0.10) | 0.9 (0.09) |
| Laboratory | 135 | 4.8 (0.08) | 4.7 (0.07) | 135 | 1.0 (0.08) | 0.8 (0.06) | |
| Parental Whale Lake | Wild | 105 | 2.3 (0.12) | 2.2 (0.12) | 105 | 0.5 (0.05) | 0.2 (0.04) |
| Laboratory | 38 | 4.6 (0.09) | 4.6 (0.12) | 38 | 0.1 (0.03) | 0.0 (0.00) | |
| F1 complementation | Bear Paw-by-Boot | 225 | 5.4 (0.06) | 5.5 (0.06) | 225 | 2.0 (0.12) | 1.1 (0.07) |
| Boot-by-Whale | 57 | 4.9 (0.15) | 4.7 (0.15) | 57 | 0.8 (0.15) | 0.5 (0.11) | |
| F1 mapping | Bear Paw-by-Anadromous | 50 | 32.1 (0.13) | 32.3 (0.15) | 50 | 5.0 (0.00) | 5.0 (0.04) |
| Boot-by-Anadromous | 33 | 31.4 (0.17) | 31.4 (0.23) | 33 | 5.0 (0.00) | 5.0 (0.03) | |
| Whale-by-Anadromous | 42 | 32.2 (0.13) | 32.3 (0.16) | 42 | 5.0 (0.00) | 5.0 (0.02) | |
| F2 mapping | Bear Paw complete armor | 281 | 32.6 (0.07) | 32.6 (0.08) | 324 | 5.0 (0.00) | 5.0 (0.00) |
| Bear Paw low armor | 123 | 6.7 (0.17) | 6.6 (0.19) | 88 | 2.6 (0.18) | 2.2 (0.19) | |
| Boot complete armor | 264 | 32.5 (0.08) | 32.5 (0.07) | 296 | 5.0 (0.00) | 5.0 (0.00) | |
| Boot low armor | 99 | 7.1 (0.23) | 7.2 (0.36) | 70 | 2.7 (0.19) | 2.4 (0.20) | |
| Whale complete armor | 191 | 32.6 (0.10) | 32.5 (0.19) | 207 | 5.0 (0.00) | 5.0 (0.00) | |
| Whale low armor | 81 | 5.7 (0.13) | 5.7 (0.15) | 75 | 2.0 (0.16) | 1.6 (0.15) | |
Mean (SE) numbers of lateral plates and pelvic scores on the left and right side, along with number of individuals sampled (n) for wild-caught and laboratory-bred and reared individuals, crosses between different low-armor populations (F1 complementation), hybrid F1 individuals produced by crossing low-to complete-armor fish (F1 mapping), and the offspring of full-sib crosses between F1 mapping individuals divided between complete and low lateral plate and pelvic classes (F2 mapping). Bold indicates scores within the complete-armor class.
Materials and Methods
Collection, Husbandry, and in Vitro Crosses. Stickleback were collected wild in the Matanuska-Susitna Borough of Alaska from Bear Paw Lake (N 61.6139, W 149.7529), Boot Lake (N 61.7167, W 149.1167), Whale Lake (N 61.5431, W 149.7472), Glenn Highway (Anadromous; N 61.5658, W 149.0486), and Rabbit Slough (Anadromous; N 61.5595, W 149.2583). Some individuals were immediately fixed in either 10% formalin or 100% ethanol after being killed with MS222 (Tricaine Methanesulfonate). Other fish were transported live to the University of Oregon stickleback facility and kept at a density of ≈0.5 fish per liter in water of 5 ppt salinity and 18°C. Juvenile stickleback were fed paramecia, vinegar eels, and newly hatched brine shrimp, and adults were fed a mixture of frozen brine shrimp, blood worms, and dry food (Bio-Oregon, Warrenton, OR). Stickleback were raised in one room under a 10-h light/14-h dark schedule, and were brought into reproductive condition in another room on a 20-h light/4-h dark cycle. For crosses, females were stripped and eggs were fertilized by using macerated testis. Complementation tests in all within- and between-population combinations were performed en masse using 20 females and males from each low-armor population. Mapping crosses were performed by using single mating pairs. Entrails of each fish were removed and stored at -80°C, and the soma was fixed in 10% formalin for phenotypic analysis.
Phentoypic and Statistical Analyses. Fixed fish were cleared by using trypsin and stained with Alizarin red (27). Lateral plates were counted twice on both sides by independent observers, and discrepancies were reexamined by a third observer. The pelvic structure was scored by using a scale modified from ref. 28 based on the presence of the three processes of the supporting structure and the pelvic spine: 0, complete absence of bony structure; 1, a circular remnant of bone without processes; 2, bone with one process, usually the anterior, making an oval vestige; 3, two processes, usually the ascending branch and the anterior process, making an “L” shape; 4, the presence of the anterior and posterior processes along with the ascending branch; and 5, a fully functional pelvis with all processes and a pelvic spine. Statistical analyses were performed by using jmp (v5.01) software (29) with lateral plate counts and pelvic structure scores scaled continuously. Although pelvic scores appear to be categories, they are actually values of a variable (size of the pelvis) with a continuous range from total absence to complete structure. Normality assumptions for each analysis were tested, and when necessary, transformations of the data were performed (30). In many cases, complete normality was not achieved, and we used the scaled variables with the most normal distributions and confined ourselves to ANOVA, which is known to be more robust to deviations from normality than alternative approaches (30).
For all ANOVA models, lateral plate and pelvic scores were included singly as response variables. Because we intentionally chose to examine specific stickleback populations, this factor (population) was treated as a fixed effect in each model as was rearing environment (wild or laboratory). However, because of the stochastic nature of the inheritance of genetic factors, “family” was always considered to be a random effect. As such, analyses that made comparisons only across populations were fixed models, but ANOVAs in which family was nested within population were mixed models. Several ANOVAs were performed to examine lateral plate and pelvic structure variation across crosses and collections. An ANOVA of plate number in complete-armor anadromous fish with one factor (wild or laboratory) was not significant (F1,229 = 1.75; P = 0.1875). An ANOVA of plate number and pelvic structure in low-armor populations with two factors, rearing environment (wild or laboratory) and population (Bear Paw, Boot, or Whale), was significant (plates F5,611 = 129.74; pelvis F5,611 = 17.88) for the main effects (P ≤ 0.0001 for population and class factor in each analysis), as well as the interaction (P ≤ 0.0001) for plates but not pelvis (P = 0.16). An ANOVA compared anadromous to F1 individuals (F3,152 = 18.05, P ≤ 0.0001) as well as parental crosses to Boot-by-Whale (F2,227 = 0.95, P = 0.3903) and Bear Paw-by-Boot complementation crosses (F2,488 = 172.97, P ≤ 0.0001). A nested ANOVA of family within population, partitioned by plate morph, showed significant variation across both factors in the complete (overall model F29,706 = 4.88, P ≤ 0.0001) and low (F29,289 = 1.88, P = 0.005) plate morphs. Similarly, a nested ANOVA of pelvic scores showed significant variation across families (model F28,199 = 3.39, P ≤ 0.0001).
Categorical lateral plate and pelvic morph variables were derived from the distribution of plate and pelvic structures. Fish with a continuous run of >30 plates were categorized as “complete.” Fish that had 10–30 plates, including both anterior plates and plates on the caudal peduncle, were classified as “partial.” Fewer than 20 fish in our entire study exhibited the partial-plate phenotype. Fish with <10 plates that were confined to the anterior pelvic region were categorized as “low.” A pelvic score of 5.0 (fully functional) was classified as “complete” pelvis, and any score <5.0 (nonfunctional pelvis) was “low.” G statistics were used to analyze the frequencies of lateral plate and pelvic morphs (30), and implemented in a computer program written by W.A.C.
Genetic mapping DNA was extracted from individuals in a single family from a Bear Paw-by-Anadromous cross using standard phenol-chloroform techniques (31). Bulked segregant analysis (32) was performed by using bulks of DNA from 10 individuals of each of the four combinations of plate and pelvis phenotype classes. This provided 20 individuals in each of the complete and low bulks. Primers for microsatellite markers from previous studies (22, 33–35) were used for genotyping (31). If genotypic differences appeared between the bulks, we then ran the same markers, as well as flanking markers, on the entire panel of F2 fish. Mapping data were analyzed by using the program map manager qtx (36). Distances between markers were compared to the map of Peichel et al. (22) and were generally in agreement. Once Mendelian lateral plate and pelvic loci were localized in the primary cross, we also mapped them in two additional crosses (Boot- and Whale-by-Anadromous), providing mapping data for each of the three low-armor populations in our study.
Results and Discussion
In wild populations, the complete-armor anadromous and low-armor lacustrine populations differ in lateral plate number and pelvic score, and this population-level difference is maintained in fish crossed and reared in the laboratory (Table 1 and Figs. 1 and 2). In Alaska, complete-armor anadromous stickleback averaged 33 plates per side regardless of whether they were wild-caught or laboratory-reared, and low-armor wild and laboratory fish exhibited zero to nine plates per side (Table 1 and Fig. 2). In stickleback from the three freshwater lakes in our study, fish from Whale Lake had two to three fewer plates than Bear Paw Lake or Boot Lake, and laboratory-reared fish had one to two more plates than wild fish (Table 1 and Fig. 2), indicating two genetically differentiated lateral plate classes in the parental populations that correspond respectively to the complete and low lateral plate morphs previously recognized in stickleback (19). Anadromous fish also had higher pelvic scores in both wild-caught and lab-reared fish (all score of 5.0; Table 1 and Fig. 2), and the freshwater populations had lower scores equivalent to either absence of bone (0.0), or the presence of a small bony vestige without spines or processes (Fig. 3). Whale Lake fish had lower scores (0.0–0.5) than stickleback from Bear Paw or Boot Lakes (0.5–1.0), and pelvic scores for lab-reared fish were lower than wild-caught fish. Similar to lateral plates, two major classes of pelvic structure exist in the parental populations: complete and low. In summary, variation in armor phenotypes between oceanic and freshwater stickleback populations has a strong genetic component as evidenced by the persistence of these phenotypes under common laboratory conditions.
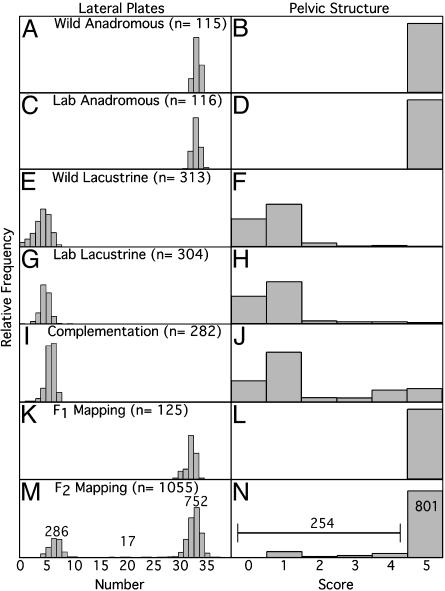
Fig. 2.
Distribution of number of lateral plate and pelvic structure scores in wild-caught complete-armor anadromous (A and B, Wild Anadromous), laboratory-bred and reared anadromous (C and D, Lab Anadromous), low-armor freshwater parental populations (E and F, Wild Lacustrine; ∼100 individuals each pooled from Bear Paw, Boot, and Whale Lakes), lab-reared fish from intrapopulation low-armor crosses (G and H, Lab Lacustrine; pool of ∼100 individuals from each population), F1 complementation hybrid (I and J, Complementation; pool of offspring from Bear Paw-by-Boot and Boot-by-Whale crosses), F1 mapping hybrids (K and L, F1 Mapping; pool of all F1 fish from crosses between anadromous-by-each low-armor population), and F2 mapping hybrid (M and N,F2 Mapping; pool of F2 fish from full-sib crosses of F1 Mapping individuals from anadromous-by-each low-armor parent). The lateral plate number and pelvic scores are the average of the left and right sides of the fish. Numbers above data in the F2 Mapping panels (M and N) are the counts of the complete to low lateral plate and pelvic structure classes respectively, indicating an approximate 3:1 ratio for each trait.
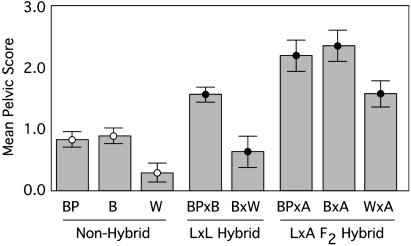
Fig. 3.
Comparison of the mean (SE) pelvic scores across intrapopulation (Non-Hybrid) crosses of bear paw (BP), boot (B), and whale (W), low-by-low complementation (L × L Hybrid) crosses of Bear Paw-by-Boot (BP × B) and Boot-by-Whale (B × W), and the low-pelvic class of the low-by-anadromous F2 hybrids (L × A F2 Hybrid) whose parents were the product of Bear Paw-by-Anadromous (BP × A), Boot-by-Anadromous (BP × A), and Whale-by-Anadromous (BP × A) crosses. Only individuals from the low-pelvic class (scores < 5) were used to calculate the statistics for the L × AF2 hybrid families presented here.
Previous studies have shown that, in stickleback, three naturally occurring lateral plate morphs (complete, partial, and low) have a simple genetic basis in some crosses, and more complex in others, prompting the formulation of at least six different one- or two-locus genetic models in addition to polygenic models (19). To determine whether the genetic basis of armor loss in the Alaskan populations generally matches any of the previously presented genetic models, we mated each of the three lacustrine populations to the complete-armor anadromous ancestral population in a series of single-pair matings. Heterozygous F1 animals showed a complete lateral plate and complete-pelvis phenotype that was similar to the anadromous parent, indicating that the full armor phenotypes were specified by dominant alleles. F1 full siblings were mated to produce a total of 1,055 individuals in 30 F2 families. F2 animals had either the complete (752) or the low lateral plate phenotype (286), with few (17) showing a partial phenotype (Fig. 2M). All families showed similar ratios that were not statistically different from 3:1 (see Table 2, which is published as supporting information on the PNAS web site), indicating a Mendelian basis for much of the armor loss. Our findings of a simple 3:1 Mendelian ratio match only one other study of lateral plate morphs, in which Avise (17) hypothesized the evolution of modifier loci that stabilized the dominance relationship. Alternatively, in young lacustrine stickleback populations, such as in Alaska, the “ground state” for the morph locus may be complete dominance. Analysis of the pelvic structure in the same mapping families showed a pattern similar to lateral plates, with F1 families exhibiting the parental complete-pelvic phenotype (Table 1 and Fig. 2), and all F2 families showing a similar 3:1 proportion of complete (score, 5.0) to low (score, ≤5.0; Table 2) classes, indicating that much of the pelvic loss can also be accounted for by a Mendelian factor. Although the plate and pelvis phenotypes both exhibited a Mendelian basis, each phenotype is caused by a different genetic factor. A ratio of ≈9:3:3:1 for the plate and pelvic phenotypic combinations occurred in the F2 generation (Fig. 4 and ref. 37).
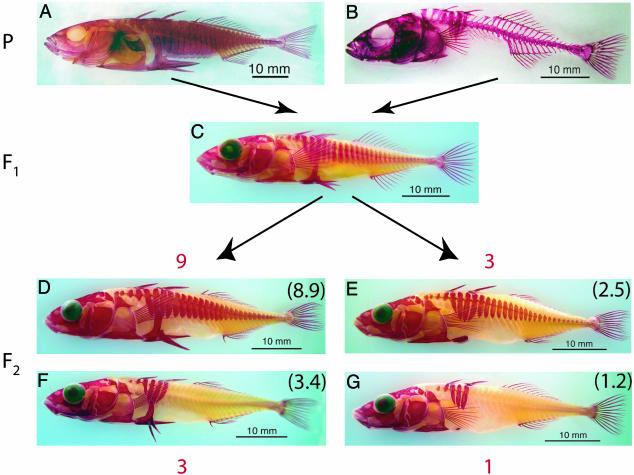
Fig. 4.
Representative phenotypes of the parental complete armor (A), parental low armor (B), F1 mapping hybrid (C), and F2 mapping hybrid (D–G) generations. The major axes of variation in the F2 intercross generation indicate the segregation of armor loss as a 9:3:3:1 dihybrid Mendelian ratio (red; observed ratio in black) of the parental armor classes. (D) The complete-armor phenotype of the F2 generation. (G) The low-armor phenotype. (E and F) The complete-plate/low-pelvic structure and complete-pelvic/low-plate recombinant phenotypes, respectively.
The three low-armor freshwater populations are geographically isolated from one another and surrounded by numerous complete-pelvic populations (Fig. 1), indicating that the loss of armor occurred independently. To test whether the same armor-loss genetic factors were involved in all three populations, each of the three low-armor populations was mated to both of the others. No increase in lateral plates was seen in Boot-by-Whale complementation crosses over the parental control crosses, and the Bear Paw-by-Boot complementation crosses had on average only one to two more plates than either of the parentals. The distribution is similar to the within-population low-armor crosses (Fig. 2 and Table 1), and does not approach the number of complete-plate phenotypes (increases of 25–35 plates) that would have been expected had the major Mendelian loci complemented. Pelvic phenotype failed to complement in the Boot-by-Whale cross, but a significant increase in the pelvic score was seen in the Bear Paw-by-Boot complementation crosses (Figs. 2 and 3) with the mean pelvic score of the hybrids (1.54) significantly higher than either parent (0.76 and 0.84 for Bear Paw and Boot, respectively). Similar to lateral plates, however, the number of complete phenotypes (score of 5.0) is much less than expected if the major Mendelian loci were complementing (Figs. 2 and 3). Therefore, complementation of the Mendelian lateral plate and pelvic loci is not occurring in the crosses between low-armor populations, implicating the same genes of major effect in each population.
Despite the absence of complementation of the major-effect loci, some quantitative complementation of minor loci is apparent, indicating a partitioning of genetic variance affecting these traits across populations. In addition, ANOVAs with family nested within population, partitioned by Mendelian plate and pelvic class, showed significant variation across both factors in the complete- and low-plate and pelvic classes. Therefore, genetic variation in lateral plate and pelvic modifier loci is partitioned across families within populations, as well as across freshwater populations (Fig. 3). This partitioning of genetic variance is similar to that found for microsatellite markers in stickleback populations in south-central Alaska, in which only ≈20% of the ancestral variation was partitioned across populations, whereas 80% of the variation still existed within populations (26). The increase in pelvic score in crosses between Bear Paw Lake and Boot Lake fish supports the conclusion that these populations have different distributions of variants at minor loci (Fig. 3). In addition, because the F2 low-armor class (homozygous recessive Mendelian locus genotype) pelvic scores are significantly greater than the low-armor parental scores (Fig. 3), much of this additional minor genetic variation is probably present in the anadromous ancestor, once again mirroring neutral population genetic data.
For one F2 mapping family (family 1, Bear Paw-by-Anadromous, Table 2), we used bulk segregant analysis (32) to randomly screen through the stickleback genome and localize the Mendelian lateral plate and pelvic loci to chromosomal regions of a linkage map produced by Peichel et al. (22). Because this map is constructed with microsatellite markers, it is an excellent tool to compare results from independent studies of the genetic basis of stickleback phenotypic variation. We found strong linkage to markers in two regions of the genome (see Fig. 5). The pelvic structure locus maps to the end of linkage group (LG) VII, whereas the lateral plate morph maps to LG XVIII. These regions are distinct from QTLs identified as affecting armor in stickleback from British Columbia (22), showing that plate and pelvic classes, as compared to number of plates and length of spine within classes that were examined in British Columbia, are different traits affected by alleles segregating at independent loci. To confirm the conclusions of our complementation results, we mapped the plate and pelvic loci in crosses in Boot-by-Anadromous and Whale-by-Anadromous F2 families (families 13 and 21, Table 2). As expected, the lateral plate locus maps to the same position in each family, as does the pelvic locus, supporting the conclusion from the complementation crosses that the same genetic loci regulate lateral plate and pelvic armor loss in all three populations.
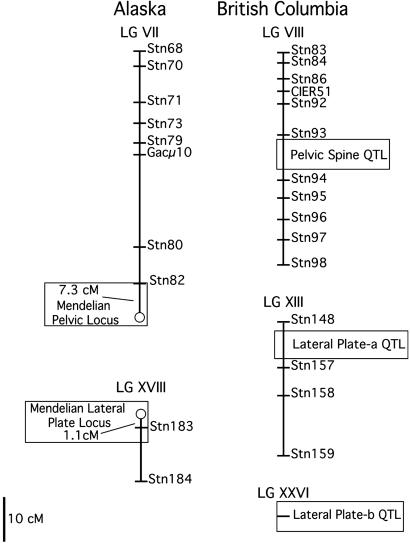
Fig. 5.
Positions of Alaskan Mendelian lateral plate (Mendelian Lateral Plate Locus) and pelvic (Mendelian Pelvic Locus) loci on the stickleback linkage map (22). Alaskan Mendelian plate and pelvic loci localized to linkage group (LG) VII and LGXVIII, respectively. The Alaskan lateral plate locus maps 1.1 centi-Morgans (cM) away from Stn183, and the pelvic phenotype locus maps 7.3 cM from marker Stn82. Open boxes on the right side of the figure indicate the position of QTLs important for lateral plate and pelvic spine variation in a cross between low-plate, complete-pelvic stickleback species from British Columbia (22).
Conclusions
Because these Mendelian armor-loss genes have the same penetrance in all 30 F2 hybrid families from three geographically isolated populations, our study indicates that large genetic steps that happen recurrently may be important during the rapid postglacial diversification of stickleback. Each F2 mapping family in our study was a combination of recently derived freshwater and ancestral genomes (26) whose hybridization in the laboratory most likely recapitulated genetic combinations that existed in natural Alaskan stickleback populations as they evolved the low-armor phenotype over the last 14,000 years. The rapid evolution of low-armor freshwater phenotypes from complete-armor oceanic phenotypes leads to three testable hypotheses for recurrent armor reduction in these populations. (i) Low-armor populations became isolated and their independent evolution included the fixation of different armor-loss alleles that arose through independent mutations after the invasion of fresh water (demographic and mutational independence). (ii) Low-armor populations were isolated and evolved independently but fixed the same ancestral armor-loss allele that was inherited from the oceanic ancestor (demographic independence but mutational nonindependence). (iii) Low-armor populations are all derived from a single lacustrine population that fixed the low-armor allele and subsequently founded each of the other low-armor populations through a complex pattern of gene flow (demographic and mutational nonindependence). We hypothesize that, because the freshwater populations are so young and low-armor populations are so isolated, the recessive low-armor alleles were probably present at low frequency in a polymorphic anadromous ancestor, maintained passively because of large population sizes or actively because of selection (i.e., hypothesis ii). Tests of these hypotheses will require population genetic analyses of numerous neutral loci to establish the level of demographic independence of the populations, as well as genealogical analyses of the DNA sequence of the causative regions to establish independence of the mutational events.
It is as yet unclear how frequently genes of major effect, as in these stickleback results and those of other recent studies (38, 39), provide the genetic basis of evolving traits (6, 7). The loss of structures, for example, may have a simpler genetic basis than those involving the gain of new functions. In addition, despite the role of major loci in stickleback armor loss, minor loci clearly play a part in the fine-tuning of phenotypes independently in each population; for example, compare the dorsal-ventral extent of plates in the anadromous parent to the F1 or complete-plate F2 (Fig. 4). In fact, despite the smaller phenotypic roles of minor loci, they may play major evolutionary roles in the divergence of stickleback populations by uncovering the phenotypic effect of Mendelian alleles in some geographic locales or populations, but not others. Developmental systems affected by minor loci may bias the genetic variation that is uncovered when populations are exposed to similar environmental conditions, potentially providing an explanation for the parallel genetic basis for stickleback armor loss in different Alaskan populations (40). Testing this hypothesis requires not only the identification of the molecular genetic bases of both armor maintenance and loss in geographically isolated populations, but also necessitates an intricate understanding of the environmental and population genetic contexts of gene expression in stickleback from ancestral and derived populations. The extensive work on stickleback behavior, ecology, and evolution (15), the emergent role of stickleback as a model for developmental embryology (24), and the global distribution of ancestral and recurrently derived populations make threespine stickleback an ideal system for this task.
Supplementary Material
Acknowledgments
We thank F. von Hippel for use of laboratory space; C. Hulslander for conceptual, field, and laboratory assistance; S. Bassham, S. Foster, D. Kingsley, K. McGuigan, and C. Peichel for discussions; two anonymous reviewers for helpful comments; B. Trevarrow and the University of Oregon zebrafish facility for rearing assistance; and S. Foster, J. Baker, and their students for helping to develop the Alaskan stickleback system. This material is based on work supported by National Science Foundation Grants IBN 023639 and EAR9870037 (to M.A.B.) and National Institutes of Health Grants R01RR10715 and 5F32GM020892 (to J.H.P. and W.A.C., respectively). National Science Foundation Integrative Graduate Education and Research Traineeship Program DGE 9972830 supported undergraduates D. Bradley, R. Loda, A. Ngo, J. O' Brien, M. Robinson, and M. Rothgary, who helped phenotype and screen genetic libraries.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fisher, R. A. (1930) The Genetical Theory of Natural Selection (Clarendon, Oxford).
- 2.Stern, D. L. (2000) Evolution (Lawrence, Kans.) 54, 1079-1091. [DOI] [PubMed] [Google Scholar]
- 3.Schluter, D. (2000) The Ecology of Adaptive Radiations (Oxford Univ. Press, Oxford).
- 4.Waddington, C. H. (1952) Nature 169, 278. [DOI] [PubMed] [Google Scholar]
- 5.Stearns, S. C. (2002) Proc. Natl. Acad. Sci. USA 99, 10229-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orr, H. A. & Coyne, J. A. (1992) Am. Nat. 140, 725-742. [DOI] [PubMed] [Google Scholar]
- 7.Orr, H. A. (2001) Trends Ecol. Evol. 16, 343-350. [DOI] [PubMed] [Google Scholar]
- 8.Wichman, H. A., Badgett, M. R., Scott, L. A., Boulianne, C. M. & Bull, J. J. (1999) Science 285, 422-424. [DOI] [PubMed] [Google Scholar]
- 9.Calboli, F. C. F., Kennington, W. J. & Partridge, L. (2003) Evolution (Lawrence, Kans.) 57, 2653-2658. [DOI] [PubMed] [Google Scholar]
- 10.Gompel, N. & Carroll, S. B. (2003) Nature 424, 931-935. [DOI] [PubMed] [Google Scholar]
- 11.Sucena, E., Delon, I., Jones, I., Payre, F. & Stern, D. L. (2003) Nature 424, 935-938. [DOI] [PubMed] [Google Scholar]
- 12.Wittkopp, P. J., Williams, B. L., Selegue, J. E. & Carroll, S. B. (2003) Proc. Natl. Acad. Sci. USA 100, 1808-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoekstra, H. E. & Nachman, M. W. (2003) Mol. Ecol. 12, 1185-1194. [DOI] [PubMed] [Google Scholar]
- 14.Elena, S. F. & Lenski, R. E. (2003) Nat. Rev. Genet. 4, 457-469. [DOI] [PubMed] [Google Scholar]
- 15.Bell, M. A. & Foster, S. A. (1994) The Evolutionary Biology of the Threespine Stickleback (Oxford Univ. Press, Oxford).
- 16.Bell, M. A. (2001) Genetica 112, 445-461. [PubMed] [Google Scholar]
- 17.Avise, J. C. (1976) Genet. Res. 27, 33-46. [Google Scholar]
- 18.Bell, M. A. (1981) Evolution (Lawrence, Kans.) 35, 67-74. [Google Scholar]
- 19.Bańbura, J. & Bakker, T. C. M. (1995) Behaviour 132, 15-16. [Google Scholar]
- 20.Reimchen, T. & Nosil, P. (2001) Can. J. Zool. 79, 533-539. [Google Scholar]
- 21.Ahn, D. & Gibson, G. (1999) Dev. Genes Evol. 209, 482-494. [DOI] [PubMed] [Google Scholar]
- 22.Peichel, C., Nereng, K., Ohgi, K., Cole, B., Colosimo, P., Buerkle, C. A., Schluter, D. & Kingsley, D. M. (2001) Nature 414, 901-905. [DOI] [PubMed] [Google Scholar]
- 23.Cole, N. J., Tanaka, M., Prescott, A. & Tickle, C. (2003) Curr. Biol. 13, R951-R952. [DOI] [PubMed] [Google Scholar]
- 24.Cresko, W. A., Yan, Y. L., Baltrus, D. A., Amores, A., Singer, A., Rodriguez-Mari, A. & Postlethwait, J. H. (2003) Dev. Dyn. 228, 480-489. [DOI] [PubMed] [Google Scholar]
- 25.Reger, R. D. & Pinney, D. S. (1996) in Adventures Through Time: Readings in the Anthropology of Cook Inlet, Alaska, eds. Davis, N. Y & Davis, W. E. (Cook Inlet Historical Soc., Anchorage, AK).
- 26.Cresko, W. A. (2000) Ph.D. thesis (Clark Univ., Worcester, MA).
- 27.Potthoff, T. (1984) in Ontogeny and Systematics of Fish, ed. Moser, H. G. (Allen Press, Lawrence, KS), Vol. 1, pp. 35-37. [Google Scholar]
- 28.Bell, M. A., Ortí, G., Walker, J. A. & Koenings, J. P. (1993) Evolution (Lawrence, Kans.) 47, 906-914. [DOI] [PubMed] [Google Scholar]
- 29.SAS Institute (2002) JMP (SAS Institute, Cary, NC), Version 5.01.
- 30.Sokal, R. R. & Rohlf, F. J. (1995) Biometry (Freeman, New York), 3rd Ed.
- 31.Sambrook, J., Fritsche, E. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 32.Postlethwait, J., Johnson, S., Midson, C. N., Talbot, W. S., Gates, M., Ballenger, E. W., Africa, D., Andrews, R., Carl, T., Eisen, J. S., et al. (1994) Science 264, 699-703. [DOI] [PubMed] [Google Scholar]
- 33.Rico, C., Zadworny, D., Kuhnlein, U. & Fitzgerald, G. J. (1993) Mol. Ecol. 2, 271-272. [DOI] [PubMed] [Google Scholar]
- 34.Taylor, E. B. (1998) Mol. Ecol. 7, 930-931. [Google Scholar]
- 35.Largiader, C. R., Fries, V., Kobler, B. & Bakker, T. C. M. (1999) Mol. Ecol. 8, 342-344. [PubMed] [Google Scholar]
- 36.Manly, K. F., Cudmore, R. H., Jr., & Meer, J. M. (2001) Mamm. Genome 12, 930-932. [DOI] [PubMed] [Google Scholar]
- 37.Mendel, G. (1863) Experiments in Plant Hybridization (Cambridge Univ. Press, Cambridge, U.K.), reprinted 1958.
- 38.Ffrench-Constant, R. H. (1994) Insect Biochem. Mol. Biol. 24, 335-345. [DOI] [PubMed] [Google Scholar]
- 39.Sucena, E. & Stern, D. L. (2000) Proc. Natl. Acad. Sci. USA 97, 4530-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West-Eberhard, M. J. (2003) Developmental Plasticity and Evolution (Oxford Univ. Press, Oxford).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.