Abstract
Glioblastoma (GBM) is the most common and aggressive histologic subtype of brain cancer with poor outcomes and limited treatment options. Here we report the selective overexpression of the protein arginine methyltransferase PRMT5 as a novel candidate theranostic target in this disease. PRMT5 silences the transcription of regulatory genes by catalyzing symmetric di-methylation of arginine residues on histone tails. PRMT5 overexpression in patient-derived primary tumors and cell lines correlated with cell line growth rate and inversely with overall patient survival. Genetic attenuation of PRMT5 led to cell cycle arrest, apoptosis and loss of cell migratory activity. Cell death was p53-independent but caspase-dependent and enhanced with temozolomide, a chemotherapeutic agent used as a present standard of care. Global gene profiling and chromatin immunoprecipitation identified the tumor suppressor ST7 as a key gene silenced by PRMT5. Diminished ST7 expression was associated with reduced patient survival. PRMT5 attenuation limited PRMT5 recruitment to the ST7 promoter, led to restored expression of ST7 and cell growth inhibition. Lastly, PRMT5 attenuation enhanced GBM cell survival in a mouse xenograft model of aggressive GBM. Together, our findings defined PRMT5 as a candidate prognostic factor and therapeutic target in GBM, offering a preclinical justification for targeting PRMT5-driven oncogenic pathways in this deadly disease.
Keywords: PRMT5, glioma, arginine, methylation, epigenetics, cancer
Introduction
High grade astrocytomas are the most common primary central nervous system (CNS) malignancy accounting for nearly 14,000 new cases per year in the United States. While surgery remains the mainstay for the treatment of patients with these CNS tumors, grade III (anaplastic astrocytoma) and grade IV (glioblastoma multiforme, GBM) astrocytomas exhibit a highly invasive clinical behavior that, in most cases, precludes complete surgical resection (1). In contrast to many cancers, the outcome of patients diagnosed with GBM has shown only marginal improvement over the past several decades with a median survival of only fifteen months despite multimodal therapy (2). Recent work has contributed to an improved understanding of the pathophysiology of these high grade malignancies, however, discovery of effective therapies has been limited by the lack of novel targets that are selectively involved in the complex pathogenesis of this disease. Numerous genome-wide studies have demonstrated that GBM possesses a remarkable degree of heterogeneity with regard to genetic mutation, gene expression profiles, and epigenetic modifications (3, 4). This degree of biologic heterogeneity has contributed to the challenge of identifying targets that are critical to the underlying pathogenesis of this aggressive disease.
Post-translational modification of proteins is a common activity involved at virtually all levels of cellular regulation. Enzymes in the protein arginine methyltransferase (PRMT) family represent a group of proteins that are evolutionarily conserved amongst a wide variety of organisms. PRMT enzymes covalently modify both histone and non-histone proteins that are critical to the maintenance of numerous cellular regulatory networks (5–7). The PRMT5 enzyme is a type II arginine methyltransferase that utilizes the co-factor molecule S-adenosyl-L-methionine (SAM) to catalyze the transfer of a methyl group to two of three guanidino nitrogen atoms within the arginine molecule. PRMT5 drives the formation of both ω-NG-monomethyl and ω-NG, NG-symmetric dimethyl arginine residues to affect protein function (8–12). PRMT5-driven methylation of arginine residues leads to symmetric dimethylation of histone proteins H3 (S2Me-H4R3) and H4 (S2Me-H3R8) which in turn alters chromatin structure to promote transcriptional repression (13, 14). A growing number of non histone proteins involved in the control of multiple regulatory networks have also been identified as targets of PRMT5 (6, 15).
We have previously reported that PRMT5 over-expression is involved in the pathogenesis of mantle cell lymphoma and knockdown of PRMT5 expression interfered with growth of transformed B cells (16). While this work contributed information regarding the mechanism of PRMT5 over-expression, the functional consequences of PRMT5 inhibition on maintenance of the malignant phenotype of the transformed cell remain poorly characterized. Here we show that over-expression of PRMT5 is associated with more aggressive disease in patients with GBM. Because this enzyme is intimately involved with numerous processes that are frequently dysregulated in cancer, we sought to determine the consequences of PRMT5 silencing in GBM. We demonstrate that inhibition of PRMT5 over-expression in GBM cells leads to restoration of critical regulatory pathways affecting cell growth, survival, and tumor suppressor activity. Our findings provide new information regarding methods to directly and indirectly affect tumor progression while providing further justification to explore novel approaches to target PRMT5 over-expression in this incurable cancer.
Materials and methods
Cell lines and culture
The human GBM cell lines used in this study are summarized in supplementary table S1. The tet-on inducible P53 line, 2024 (17) cell line was kindly provided by Dr. Erwin Van Meir.
Evaluation of PRMT5 protein expression in primary astrocytoma tumors
Under an IRB-approved protocol, sixty patients with astrocytomas treated at the Ohio State University from January 2003 to October 2007 were identified. Age, gender, race, and previous history of astrocytomas were assessed by reviewing the medical records of these patients. Reports were reviewed to determine tumor grade, Ki67 proliferation index, as well as clinical characteristics of disease. Immunohistochemistry (IHC) was performed using a Ventana Benchmark system (Ventana Medical Systems, Tucson, AZ) and the ultraview universal Fast Red kit, following manufacturer’s recommendations. Optimal conditions for PRMT5 were determined to be 1:125 with antigen retrieval for 30 min using mantle cell lymphoma primary tumor tissues and benign, reactive lymph nodes as the positive and negative controls, respectively. Slides were counterstained with hematoxylin II for 4 min. See supplemental materials for details on calculating PRMT5 expression index.
Small interfering RNA transfection
siRNA and scramble (scr) RNA were constructed by silencer siRNA construction kit by Ambion (Austin, TX). si-PRMT5 or scrRNA were transfected into GBM cells by Lipofectamine 2000 according to manufacturer’s instruction. See supplemental materials for sequences of inhibitory RNAs used in this paper.
Cell cycle, apoptosis, and migration analysis
Cells were harvested after treatment and fixed in 75% EtOH. After digestion with RNase, DNA was stained with propidium iodide (PI) and analyzed with a Beckman Coulter flow cytometer (Brea, CA) and Modfit software (Verity, ME). For apoptosis assays, treated cells were stained with Annexin-V and PI and evaluated by flow cytometry as described (18). Caspase activation followed methods previously described (18). GBM migration was evaluated as previously described (19).
Real-time quantitative RT-PCR, protein detection, and chromatin immunoprecipitation (ChIP) assay
Total RNA was prepared from untreated and treated GBM cells using TRIzol reagent (Invitrogen, Grand Island, NY) according to the manufacturer's instructions. The cDNA was prepared with the MMLV Reverse Transcription Kit (Invitrogen, Grand Island, NY) following the manufacturer's recommendations. Real-time PCR was performed using a TaqMan 2×Universal PCR Master Mix kit per manufacture’s instructions on an Applied Biosystems 7900HT Fast Sequence Detection System (Carlsbad, CA). Immunofluorescence and western blot utilized antibody reagents (supplemental materials) and were performed as described (16). ChIP assays were done using the EZ-Magna ChIP kit (Upstate, Billerica, MA) according to the manufacturer's instructions.
Results
PRMT5 protein is over expressed in GBM cell lines and correlates with proliferation
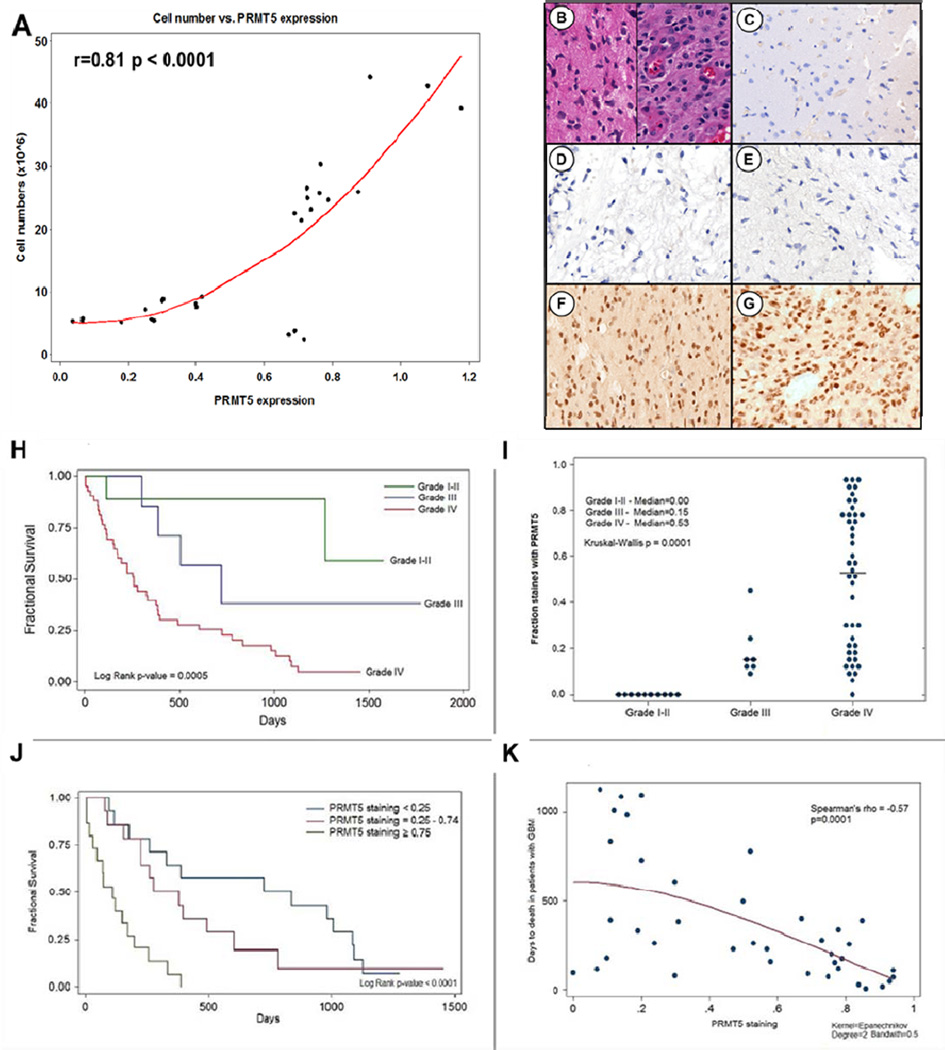
Aberrant expression of PRMT5 protein has been identified in a number of malignancies including mantle cell lymphoma (16) and germ cell tumors (20). To determine whether PRMT5 was over expressed in astrocytomas, we evaluated a panel of cell lines (supplementary Table 1) derived from primary GBM biopsy specimens for PRMT5 protein expression by western blot (supplemental Fig S1). Compared to normal brain tissue (NB) and normal human astrocytes (NHA) that did not express measurable PRMT5 protein, eight astrocytoma-derived cell lines demonstrated abundant levels of PRMT5 protein expression (supplemental Fig S1). Because the various cell lines displayed different growth rates, we examined whether the degree of PRMT5 protein over expression correlated with proliferative index (Fig 1A). Determination of absolute cell numbers was performed in parallel with western blot probing for PRMT5 and β-actin proteins at three different time points (representative blot in supplemental Fig S1). PRMT5 expression (normalized to β-actin control) correlated in a linear fashion with the proliferation of GBM cell lines (r = 0.81; p < 0.0001).
Figure 1. PRMT5 over-expression correlates with human GBM cell proliferation, and inversely correlates with patient survival.
(A) PRMT5 expression correlates with cell growth (r=0.81 with p-value<0.0001). (B) H&E staining of GBM. (D-G) PRMT5 is selectively expressed in high grade (grade III and IV) astrocytomas (F,G), while normal brain (C) and low or intermediate grade astrocytomas (D,E) do not. (H) Kaplan Meier plots by glioma grade. (I) PRMT5 expression index by glioma grade. (J) Kaplan Meier plots of overall survival of GBM patients as a function of PRMT5 protein expression index. (K) Time to death and PRMT5 level in patients who died with GBM. PRMT5 level is continuously associated with time to death (Spearman’s rho = −0.57, p=0.0001).
PRMT5 is over-expressed in primary GBM tumors and correlates with more aggressive disease
The direct correlation between GBM cell line growth and PRMT5 expression suggested that this enzyme may contribute toward the malignant phenotype (Fig 1A). Thus, we next examined PRMT5 expression in primary astrocytoma tumors. High grade astrocytomas that spontaneously develop in a preclinical in vivo murine model allowing for controlled induction of allelic haploinsufficiency at Nf1, P53 and PTEN tumor suppressor gene loci (21) were examined for PRMT5 and Ki67 expression (supplemental Fig S2). While control brain tissue from wild type mice failed to demonstrate any detectable PRMT5 protein expression, all tumor samples taken from tumor bearing Nf1- and P53-deficient mice (Mut3 mice, GFAP-cre; cisNf1−/+; P53−/−) showed over-expression of PRMT5 that coincided with the cellular proliferation signal Ki67. PRMT5 expression was higher in tumor tissue from tumor-bearing Mut3 mice compared to uninvolved brain tissue proximal to tumor or from brain tissue from control mice without CNS tumors of the same genetic background (supplemental Table S3, data not shown).
We next examined PRMT5 protein expression profiles in primary tumor specimens from 60 patients diagnosed with astrocytomas (supplemental Table S2 and Fig 1B–G). To verify that our patient cohort demonstrated clinical behavior consistent with the natural history of this disease (22), we evaluated survival in the context of tumor grade (Fig 1H) and found median survival to correlate with tumor grade as described (23). PRMT5 was over-expressed in grade III and IV tumors with the most prominent expression in a nuclear distribution (Fig 1F, G and I). Normal brain tissue or tumors from patients with grade I (N=10) and grade II astrocytomas (N=7) did not demonstrate any detectable PRMT5 protein (Fig 1C, D, E, and I). The median PRMT5 expression index (definition provided in supplemental materials & methods) seen in grade III tumors was 0.15 (N=7; Inter-quartile range = 0.11–0.25) and for grade IV tumors was 0.53 (N=43, Inter-quartile range = 0.19–0.78). The difference in PRMT5 expression indices between grade III and grade IV tumors was statistically significant (p = 0.01, by standard two-sample Wilcoxon rank-sum test). The differences in PRMT5 expression index observed in grades I/II versus grades III versus IV tumors was statistically significant (p = 0.0001, Kruskal-Wallis equality-of-populations rank test).
Because PRMT5 over-expression was confined to grade III and IV tumors, we excluded patients with grade I and II tumors from our evaluation of other clinical variables related to PRMT5 expression. In patients with grade III and IV tumors, PRMT5 expression index was not significantly associated with gender, race, or prior diagnosis with astrocytoma. In patients with grade IV tumors (GBM), the degree of nuclear PRMT5 expression was associated with age, though the result was of borderline significance (Spearman's correlation coefficient = 0.29, p = 0.057).
The heterogeneity in the degree of PRMT5 expression seen in grade IV tumors (Fig 1I) prompted us to examine whether PRMT5 expression index correlated with GBM patient survival. We stratified grade IV tumors into three categories of PRMT5 expression index: (i) ≥0.75; (ii) 0.25 – 0.74; and (iii) < 0.25. Patients with grade IV tumors showing a PRMT5 expression index ≥ 0.75 demonstrated shorter overall survival compared to patients with lower PRMT5 expression indices. Patients with PRMT5 expression index < 0.25 had a median survival of 726 days (Fig 1J, 95% CI = 176 – 1083 days, N=14) compared to 277 days (95% CI = 156 – 604 days, N=14) for those with PRMT5 expression index = 0.25 - 0.74, and 108 days (95% CI = 17 – 173 days, N=15) for those with PRMT5 expression index ≥ 0.75 (Fig 1J, log rank p < 0.0001). Cox proportional hazards were used to model mortality in patients with GBM. In a multivariable model (with age, gender, Ki67 and PRMT5 as effects), PRMT5 expression index (represented as a single continuous variable) was significantly associated with decreased overall survival. In the model a 10% increase in PRMT5 expression index on average was associated with an unadjusted HR = 1.37 (95% CI = 1.18 – 1.57, p < 0.001) and an adjusted HR = 1.40 (95% CI = 1.18 – 1.66, p < 0.001) for mortality. In the model, age, gender, and Ki67 were not significantly associated with mortality. We evaluated the association of PRMT5 expression with time to death graphically in GBM patients who died over follow up (only 4 of 43 individuals, 9.3%, were alive at the end of follow up). We found that PRMT5 expression was strongly and continuously associated with time to death in GBM patients who died during follow up (Fig 1K, Spearman’s rank correlation coefficient = −0.57, p < 0.0001).
Previous GBM tumor history was not significantly associated with PRMT5 expression index or survival in individuals with GBM. Within the 43 patients diagnosed with GBM, 11 had a previous GBM history. We did not observe any difference in survival over follow up for primary GBM or recurrent GBM (Median Survival = 259 days in primary GBM and 254 days in recurrent GBM, Log Rank test = 0.6 on the survival curves). PRMT5 expression index was not significantly associated with prior GBM history, median PRMT5 expression index = 0.56 in primary GBM and 0.31 in recurrent GBM (Wilcoxon rank sum p=0.5).
PRMT5 mRNA in GBM cell lines did not differ from normal human astrocytes (not shown), however, the amount of protein differed markedly between GBM primary tumors, cell lines, normal astrocytes (NHA) and normal brain (Fig 1 and supplemental Fig S1) suggesting that PRMT5 dysregulation occurred at the level of micro-RNA, similar to that observed with mantle cell lymphoma (16). We therefore evaluated PRMT5 expression as a function of survival and found that in the Rembrandt data set for GBM cases (total n=181 cases), no significant correlation between PRMT5 transcript levels and survival (supplemental Fig S1B). We therefore evaluated two microRNAs, miR96 and miR92b, previously shown to be under expressed in mantle cell lymphoma allowing for efficient PRMT5 translation and upregulation of PRMT5 protein levels (16). We utilized nanostring methodology to quantify miR96 (avg = 44 counts) and miR92b (avg = 108 counts) and found both miRs to be under expressed compared to the top 50 detectable miRs (avg counts ranging from 601 to 123,068) for 11 primary GBM RNA samples (supplemental Fig S1C). These observations indicate that miR dysreulation may contribute toward more efficient PRMT5 translation and elevated protein levels.
Consequences of PRMT5 knockdown in GBM cell lines in vitro
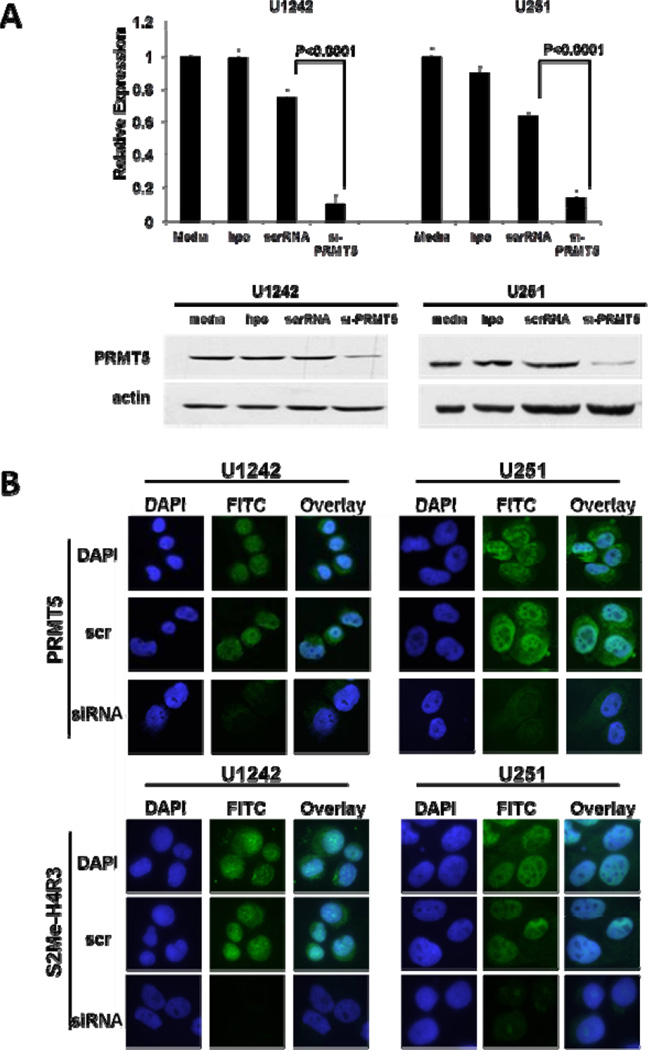
Since expression of PRMT5 correlated with proliferation rate of GBM cells and survival of GBM patients, we next determined the consequences of PRMT5 silencing in vitro. We developed a panel of nine small inhibitor RNA molecules (siRNA, and control scrambled: scrRNA) designed to knock down PRMT5 transcript and protein expression. Significant decreases in mRNA levels (90% and 80% in U1242 and U251, respectively) and in PRMT5 protein expression were achieved with three lead siRNAs (Fig 2A and B, supplemental Fig S3A). Since PRMT5 and PRMT7 have each been shown to possess type II methyltransferase activity, we examined the specificity of our si-PRMT5 reagent. As shown in supplemental Figure S3B, si-PRMT5 did not affect the expression of the other type II PRMT enzyme, PRMT7. More importantly, PRMT5 knockdown by si-PRMT5 led to demethylation of target histone protein arginine residues (H4R3) as determined by immunofluoresence (IF) using an antibody specific for symmetric dimethyl arginine of histone 4 (S2Me-H4R3, Figure 2B). Twenty four hours following siRNA treatment, cytoplasmic and nuclear PRMT5 protein levels were undetectable and this correlated with the loss of the PRMT5-associated epigenetic mark, S2Me-H4R3 (14).
Figure 2. siRNA-mediated silencing of PRMT5 expression.
(A) Human GBM cell lines were treated as indicated and expression of PRMT5 detected by real-time PCR and Western blot. (B) si-PRMT5 reduced expression of PRMT5 and its epigenetic mark S2Me-H4R3 as determined by immunofluresence microscopy.
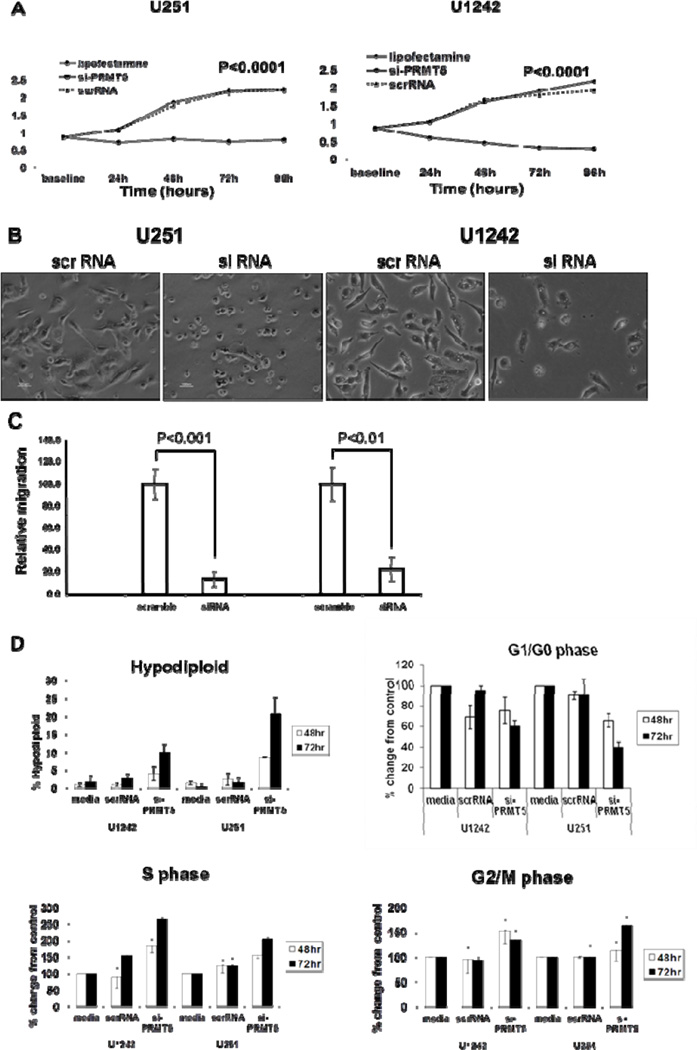
We next determined the effects of PRMT5 depletion on growth properties of GBM cells. As shown in Figure 3A, si-PRMT5 elicited a significant anti-proliferative effect on cell growth (p<0.0001). Invasiveness is a characteristic feature of GBM and a major contributing factor that prevents successful surgical resection leading to the low progression free survival observed in patients with GBM. SiRNA induced loss of PRMT5 expression led to cell morphologic changes in GBM cell lines demonstrating a transition from a cellular morphology with elongated cell shape and dendritic-like projections to a more rounded cellular shape (Fig 3B). To determine if this change in cellular morphology correlated with decreased invasiveness of GBM cells, we evaluated cellular migration by both wound-healing and trans-well migration assays. Treatment of GBM cell lines with lipofectamine or scrRNA controls did not alter cellular migration compared to untreated cells (not shown). Inhibition of PRMT5 over expression with siRNA, however, resulted in marked reduction in migratory activity of both cell lines in both trans-well (Fig 3C) and wound-healing assays (not shown). Altered cellular proliferation, morphology and migratory activity led us to next examine if cell cycle progression was affected with PRMT5 knockdown (Fig 3D). Using parallel IF assays to confirm PRMT5 knockdown and loss of the S2Me-H4R3 epigenetic mark, we were able to confirm a correlation between loss of PRMT5 expression and cell cycle arrest (Fig 3D). Furthermore, reduced expression of PRMT5 led to an overall increase in the number of hypodiploid cells indicating endonucleolytic DNA cleavage associated with cell death (2.05% in scrRNA and 10.80% in si-PRMT5, p<0.001), a decrease in the percentage of cells in G1/G0 (60.2% in scrRNA and 35.5% in si-PRMT5, p<0.0001), and an increased accumulation of cells in S (9.2% in scrRNA and 14.9% in si-PRMT5, p<0.0001) and G2/M (24.3% in scrRNA and 32.7% in si-PRMT5, p=0.0001) phases of the cell cycle (Fig 3D). These results demonstrate that PRMT5 silencing promotes cell cycle arrest and leads to hypodiploid nuclear content consistent with programmed cell death.
Figure 3. Consequences of PRMT5 silencing in GBM in vitro.
(A) Proliferation was evaluated by MTS assay. (B) Change of cell morphology with PRMT5 knockdown. (C) PRMT5 silencing leads to decreased migratory activity of GBM cells measured by transwell assay. (D) Cell cycle analysis by flow cytometry.
Knockdown of PRMT5 leads GBM cells to undergo caspase-dependent apoptosis
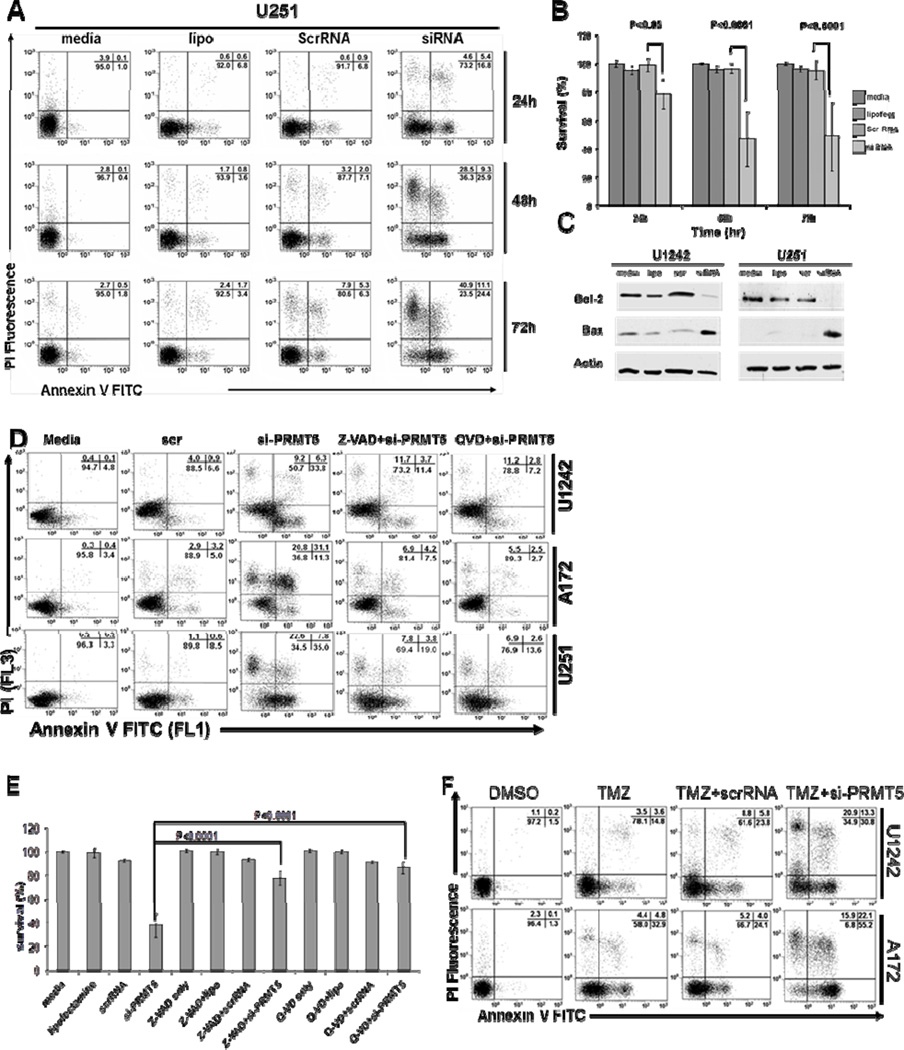
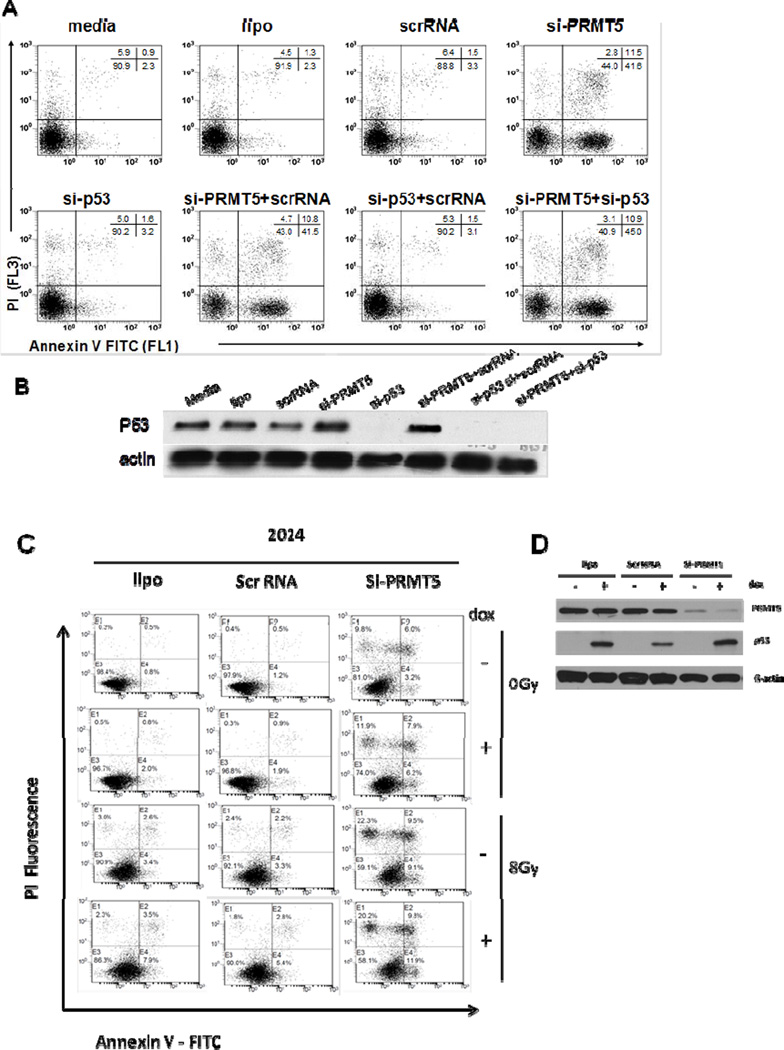
PRMT5 silencing can induce apoptosis in different types of cancer (6, 24). To investigate the effect of PRMT5 knockdown on GBM cell viability, three different cell lines (A172, U1242, and U251) were transfected with either scrRNA or si-PRMT5, and cell viability was determined by annexin-V-FITC/PI staining and flow cytometry. As shown in Figure 4A and 4B, PRMT5 knockdown resulted in the induction of apoptosis in all cell lines compared to scrRNA control (p=0.002 across cell lines and timepoints).
Figure 4. PRMT5 Knockdown promotes cell death of human GBM cells.
(A) U251 cells were treated as indicated and cell death assessed flow cytometry. (B) A172, U1242 and U251 were treated as indicated resulting in decreased cell viability of siPRMT5 compared to scrRNA (p=0.002). (C) Western blot for BCL2 and BAX. (D) Caspase inhibitors blocked siPRMT5-induced cell death. (E) Cell death of three human GBM cell lines pretreated with caspase inhibitors. (F) Cells treated with Temozolomide, replated, treated as indicated and cell death determined by flow cytometry.
The BCL2 family of proteins is known to play a central role in the regulation of apoptosis (25). As shown in Figure 4C, si-PRMT5 significantly led to an increase in the BAX/BCL2 ratio associated with activation of downstream caspase enzymes and apoptosis (26). ChIP experiments were performed and failed to show PRMT5 recruitment to the BAX gene promoter (data not shown), suggesting other pathways are involved in the change of BAX/BCL2 following PRMT5 depletion.
As caspase-3 is a key protease associated with DNA fragmentation, we investigated whether apoptosis induced by PRMT5 knockdown was caspase-3-dependent. Flow cytometry analysis of GBM cells stained with an anti-activated-caspase-3 antibody showed increased caspase-3 activation following PRMT5 silencing (supplemental Fig S4A). Pretreatment of GBM cell lines with the pan caspase inhibitors Z-VAD-FMK (100 µM) and Q-VD-OPh (20 µM) led to blockade of caspase-3 activation (supplemental Fig S4A) and prevented apoptosis (Fig 4D and E, p<0.0001), thus consistent with a caspase-dependent mechanism of cell death.
The addition of temozolomide to radiation therapy has improved the survival of patients with GBM and is now considered standard in the management of these patients (27). GBM cells were pre-incubated with 100 µM of temozolomide for 72 hours and transfected with scrRNA or si-PRMT5 (Fig 4F and supplemental Fig S4B). PRMT5 knockdown led to greater than additive induction of programmed cell death when combined with temozolomide.
Apoptosis induced by PRMT5 depletion occurs independent of P53-mutational status
The P53 pathway is commonly deleted or mutated in GBM and associated with more aggressive disease and poor prognosis (28). Our initial observations suggested that induction of GBM cell line cell death following PRMT5 depletion occurred independent of P53 mutational status (Fig 5, A172 P53wt vs. U1242 and U251 P53 mut and Table S1). Recent work has described the mechanism by which PRMT5 modulates the activity of P53 (6, 7). The P53 wild type cell line, A172 (29) was co-transfected with siRNA targeting P53 (si-P53) and si-PRMT5. As shown in Figure 5A and 5B, changes in cell viability due to PRMT5 depletion remained unchanged regardless of P53 expression level. We confirmed these studies using pifithrin-α, an inhibitor of P53 transcriptional activity(Fig S5). Additionally, in P53-null human GBM cells with a doxycycline-inducible P53 expression system (2024 cell line), the loss of cell viability due to PRMT5-depletion was similar despite absence or presence of P53 expression, even after inducing DNA damage with ionizing radiation (Fig 5C and 5D). Furthermore, the cells appeared to be sensitized to radiation-induced cell death upon depletion of PRMT5, regardless of P53 expression (Fig 5C and 5D). We found all cell lines, regardless of P53 mutational status, underwent G2/M cell cycle arrest (Fig 3D and data not shown) and apoptosis following PRMT5 depletion, an important finding when taken in the context that P53 defects are associated with chemo and radio-resistance and portend a poor prognosis in patients with GBM.
Figure 5. si-PRMT5 induced cell death is P53-independent.
(A) P53-wild type cell line, A172, treated as indicated and cell death assessed in triplicate by flow cytometry. (B) A172 cells treated as in (A) and P53 detected by Western blot. (C) The 2024 cell line treated as indicated and irradiated with a X-ray ionizing radiation (8Gy) to introduce DNA damage signal. Cell death was assessed by flow cytometry. (D) Western blot for PRMT5, P53 and β-ACTIN.
PRMT5 inhibition promotes restoration of regulatory gene expression
Several reports have shown that PRMT5 contributes to transcriptional repression of tumor suppressor genes ST7 (16), NM23 (14), RBL2 (p130) (30) and DOC-1 (31). To investigate potential genes targeted by PRMT5 in GBM, we treated cell lines with PRMT5 siRNA or scrRNA control, and carried out transcriptome analysis using Affymetrix cDNA expression arrays (Supplemental Table S4, genes showing >1.5-fold expression with PRMT5 knockdown). Consistent with the findings of Pal et al who reported a similar experiment using a murine cell line, we found 84 genes to become up regulated >1.5-fold compared to only four genes down regulated after PRMT5 knockdown, verifying PRMT5 as a global transcriptional repressor in human GBM cells. We observed a wide variety of genes with functions ranging from tumor suppressor (ST7) to immune modulatory activity (CXCL10, CXCL11, IL6). Similar to data with mantle cell lymphoma cell lines and tumors (16) where we characterized a PRMT5-dependent repressive complex (PRMT5, MBD2, HDAC2), we noted a 1.4-fold increase in ST7 mRNA expression in GBM cell lines treated with si-PRMT5.
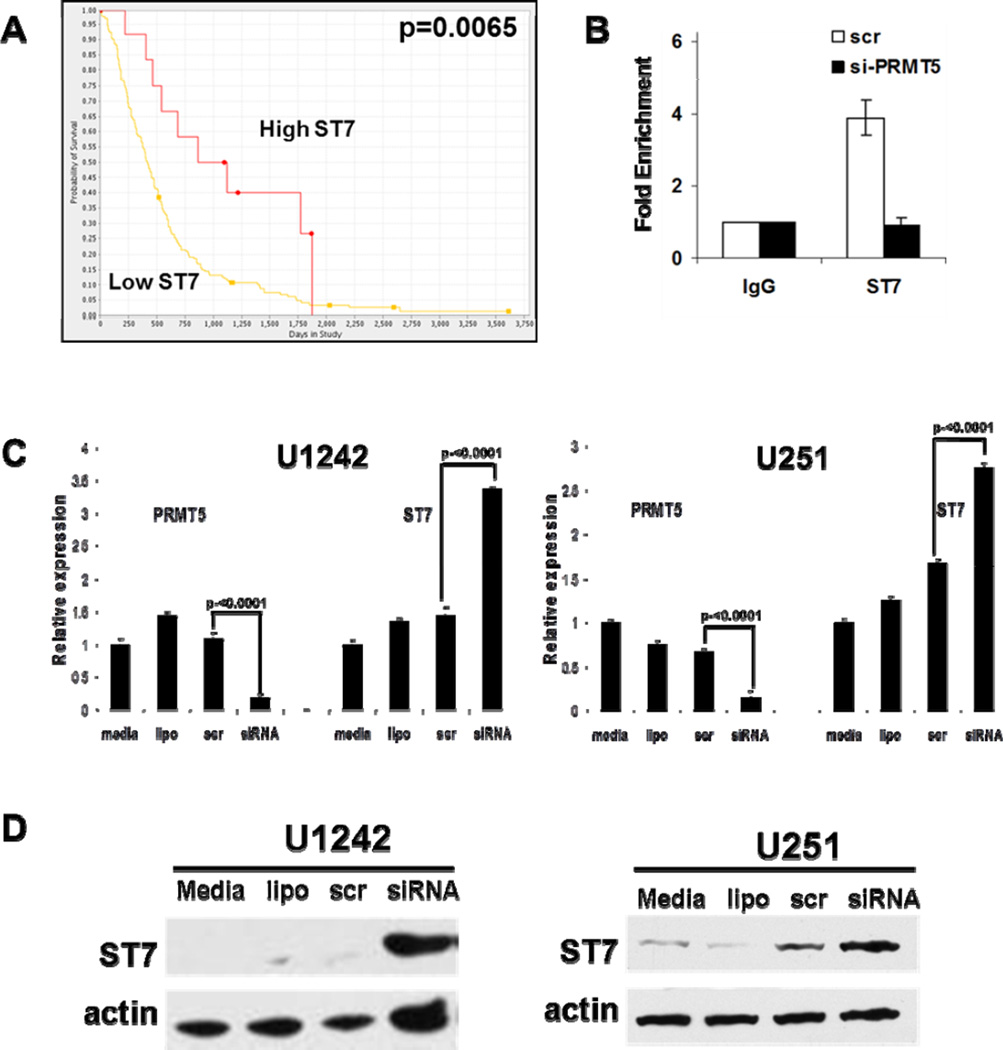
Based on this cell line in vitro data suggesting that ST7 is a functional downstream target of PRMT5, we sought to further examine ST7 expression levels as they related to clinical outcomes in GBM. We used the Rembrandt data set to analyze microarray gene expression data to associated survival data. Kaplan-Meier analysis was performed on all glioma patients (grade I-IV, Supplementary Figure S6) and GBM only (Fig 6A). Both analyses demonstrated that ST7 expression correlated with statistically significant differences in overall survival in glioma patients. Specifically, two-fold up-regulation of ST7 was associated with a statistically significant increase in overall survival. This supports our hypothesis that lower ST7 expression levels attributed to increased levels of PRMT5 and transcriptional silencing of ST7 correlate with a more aggressive and worse clinical outcome. It also supports the notion that ST7 contributes to the pathogenesis in supporting the malignant behavior of GBM.
Figure 6. ST7 is a PRMT5 target gene.
(A) Kaplan-Meier plot for GBM cases with differential ST7 gene expression (Low ST7 n=169; High ST7 n=12). (B) ChIP assays using IgG control or anti-PRMT5 antibody performed on chromatin from U251 or U1242 treated as indicated. Fold-enrichment with each antibody was calculated relative to the IgG control (in triplicate). The data were normalized by the input DNA (Delta Ct=Ct(IP)-Ct(input)) and one-way ANOVA was used to analyze the normalized data for each cell line. (C) Expression of ST7 is up-regulated by si-PRMT5 determined by by real-time PCR and (D) Western blot.
Given that ST7 repression in GBM tumors associated with significant reduction in survival (Fig 6A) and the role played by ST7 in maintenance of extracellular matrix and modulation of genes that prevent metastasis (32), we examined the functional consequences of PRMT5 inhibition and modulation of this anti-cancer target gene. To verify that PRMT5 was physically recruited to the promoters of these genes and associated with a co-repressor complex, we isolated chromatin preparations and performed ChIP studies. GBM cell lines were grown in the presence of si-PRMT5 (validated with 3 separate siRNA constructs as shown in supplemental Fig S3), scrRNA or left untreated, and 24 hours later cross-linked chromatin was prepared for ChIP assays. Using antibodies specific for PRMT5 and non-reactive antibody control and primers specific for regions within target gene promoters, we confirmed PRMT5 recruitment to the ST7 promoter (Fig 6B). When PRMT5 levels were knocked down, enrichment of PRMT5, MBD2, DNMT3a and HDAC2 on ST7 promoter was markedly diminished consistent with our prior findings (14, 16, 32; Fig 6B, and data not shown). Loss of PRMT5 recruitment on target promoters led to enhanced transcriptional (Fig 6C) and translational activity (Fig 6D). Furthermore, we noted that over-expression of ST7 in three GBM cell lines (A172, U1242, U251) was able to induce cell death and reduce cell proliferation (supplemental Fig S7), indicating that ST7 played a role as a downstream effector of PRMT5 dysregulation.
PRMT5 inhibition leads to enhanced survival in a preclinical GBM xenograft model
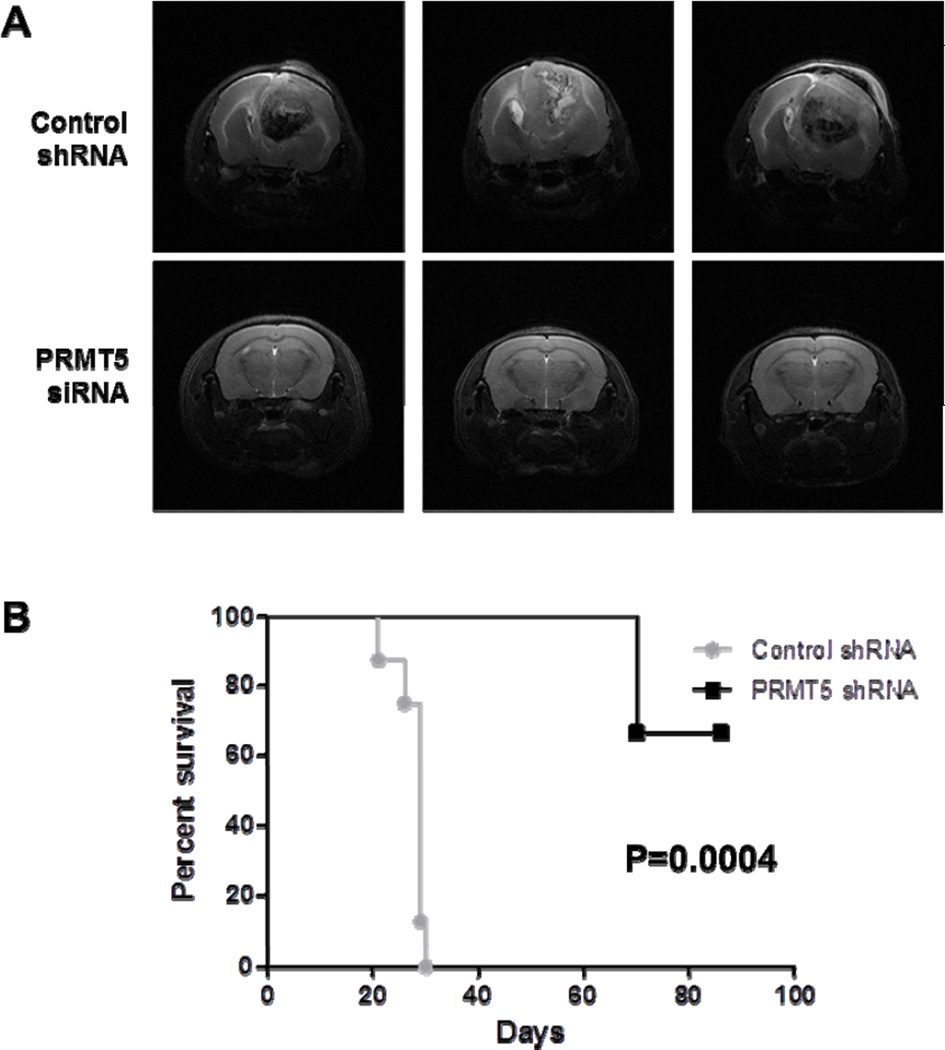
To further examine whether PRMT5 protein over expression is relevant to GBM pathogenesis, we studied the consequences of PRMT5 inhibition in an aggressive in vivo GBM xenograft model. Nude mice underwent intracranial implantation of Gli36 delta cells that were treated either with scrRNA or si-PRMT5 (equivalent cell number and viability was validated prior to engraftment, n=8/group). Mice engrafted with Gli36 cells treated with a single exposure to PRMT5 shRNA group were shown to have prolonged survival (P = 0.0004, Fig 7B) compared to mice in control shRNA group. Tumor burden was tracked by clinical assessment and by MRI at day 28 (Fig 7A). These in vivo data provide further support of the relevance PRMT5 plays in supporting the malignant phenotype of GBM.
Figure 7. PRMT5 knockdown improves survival in vivo.
(A) Representative brain MRI images taken on day 28 of mice in control and siPRMT5 treated groups. (B) Kaplan Meier plot of animals in control and siPRMT5 treated groups (n=8/grp).
Discussion
High grade astrocytomas are the most common primary brain tumors and are associated with a poor prognosis despite aggressive surgical and medical management (23). Over the last several decades, we have observed improved outcome and survival for patients with a wide variety of cancers, however, the minimal improvement in survival of patients with GBM illustrates the need to discover novel targets to develop new therapeutic approaches. In addition to classic gene mutations, inactivation of regulatory and tumor suppressor genes by epigenetic repression is frequently observed in these tumors. Histone deacetylase inhibitors have been shown to possess anti-tumor activity in malignant astrocytomas (33), clinical trials investigating these agents have been disappointing, highlighting the need to identify other promising epigenetic targets.
The PRMT family of enzymes represents a group of proteins that are highly conserved in nature and involved with a wide variety of developmental and biological processes (34). PRMT5, a type II arginine methyltransferase, silences the transcription of genes by symmetric di-methylation of arginine residues on histone proteins and works more efficiently when associated with other co-repressor enzymes such as HDAC and MBD proteins (13, 35). Recent studies have shown PRMT5 to be a major pro-survival factor regulating eIF4E expression and P53 function (7). Furthermore, PRMT5 depletion sensitizes human colon cancer, lung cancer, and fibrosarcoma cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) without affecting TRAIL resistance in non-transformed cells (24). Here we show that PRMT5 is selectively over-expressed in high grade astrocytomas and that the degree of PRMT5 expression correlated with cell growth and patient survival. Our work examining potential mechanisms of PRMT5 over expression point toward micro RNA dysregulation and we are currently investigating how PRMT5 translation is efficiently supported in GBM.
Our findings showing a correlation between PRMT5 over-expression and poor clinical outcome of patients with GBM are intriguing with regard to potential utility as a prognostic factor to identify patients with more aggressive disease. While several abnormalities affecting epigenetic processes have been described in GBM tumors including expression of DNMT enzymes (36), and methyl-CpG binding domain proteins (37), the prognostic relevance of these markers remains unclear. Identifying biomarkers like PRMT5 that are of prognostic and biologic significance will assist in developing novel approaches to this disease. Future prospective analysis will be essential to determine the true prognostic value of PRMT5 over-expression in GBM patients.
Apart from epigenetic modifications, PRMT5 has been shown to mediate arginine methylation of P53 to regulate its function (7). Since P53 has been shown to be highly relevant as a critical tumor suppressor for both primary and secondary subtypes of GBM (38), we explored the role of P53 in cell death triggered by PRMT5 knock down. We observed equivalent degrees of cell death induced by PRMT5 silencing in both P53 wild-type, mutant and deleted GBM cells. Work by several groups has demonstrated that PRMT5 directly targets P53 function by methylating R333, R335 and R337, modifications that negatively affects P53 recruitment to target gene promoters during genotoxic stress and cell death. Scoumanne et al (7) showed that PRMT5 knockdown led to decreased P53 stability, reduced expression of the P53 target gene CDKN1A, and decrease of breast cancer cell proliferation. Other work by Jansson et al (6) demonstrated that the effect of PRMT5 depletion with siRNA on DNA damage-induced apoptosis was more significant in P53 wild type compared to P53 null colorectal carcinoma cell lines. Our results suggest that cell death induced by PRMT5 silencing occurs independent of P53 mutational status indicating that the relevance of arginine methylation of P53 may differ between tumor types and, perhaps, PRMT5 might serve as an equally attractive target in GBM tumors where P53 is often mutated or deleted and associated with a poor prognosis.
Changes observed in the GBM transcriptome following PRMT5 depletion helped us identify PRMT5 as a master transcriptional repressor in this disease. We identified the tumor suppressor gene ST7 as a potential target as it became derepressed with PRMT5 inhibition. Consistent with these results, ChIP experiments identified the ST7 promoter to be direct targets of PRMT5. The human ST7 gene was first recognized as a candidate tumor suppressor based on its chromosomal location (7q31.1), a site of frequent loss of heterozygosity in transformed cells (39) and its reduced expression in cancer (40). Here we show that reduced ST7 expression correlates with poor outcome in patients with GBM. ST7 protein activity is also involved with extracellular matrix remodeling in several solid tumor models and may mediate tumor suppression by altering the tumor microenvironment and affecting tumor invasion (32). The invasive nature of GBM tumors precludes complete surgical resection in the majority of patients with this disease. Thus, development of experimental therapeutic strategies to target PRMT5 over-expression promoting restoration of tumor suppressors that negatively regulate malignant cell invasion and microenvironment may affect metastatic potential of this disease in vivo. We are currently examining the direct role ST7 plays in GBM invasiveness.
Previous work has shown that PRMT5 associates with SWI/SNF chromatin remodeling complexes along with other co-repressor molecules like HDAC2 (35), DNMT3a (13) and MBD proteins (31). Biochemical assays have demonstrated that PRMT5 activity on target H4R3 and H3R8 histone arginine residues is markedly enhanced when lysine residues become deacetylated by HDAC2 (35). Our results showed that inhibition of PRMT5 leads to loss of recruitment at target gene promoters (ST7 and, CXCL10 and CXCL11, data not shown). Importantly, our prior work and current data suggest that by inhibiting PRMT5 over-expression alone, additional co-repressor proteins contributing to anti cancer gene silencing may also be affected, providing rationale to further investigate the anti-tumor activity of combination strategies incorporating agents that target the epigenome (HDAC inhibitors, hypomethylating agents) with PRMT5 inhibitors.
Finally, PRMT inhibition was shown to prolong survival in an aggressive preclinical mouse GBM model. Collectively, our data suggests that PRMT5 over-expression represents an unfavorable prognostic marker and an attractive therapeutic target for GBM. Experimental therapeutic strategies aimed at inhibiting the effects of PRMT5 over-expression in cancer might lead to a better understanding of how to directly and indirectly affect GBM tumor progression. These findings further justify our current efforts to explore novel approaches to target PRMT5 activity in GBM and other cancers where this enzyme is dysregulated.
Supplementary Material
Acknowledgements
This work was supported by grants from 1R21NS071346-01 (RAB and CL), The Ohio Cancer Research Associates (RAB), V Foundation (RAB), 2009 AACR-National Brain Tumor Society Fellowship, in Memory of Bonnie Brooks (FY) and the European Society of Hematology/American Society of Hematology (LA). Fengting Yan is a 2013 recipient of the American Association for Cancer Research-Millennium Scholar in Training Award.
References
- 1.Walker MD, Green SB, Byar DP, Alexander E, Jr, Batzdorf U, Brooks WH, et al. Randomized comparisons of radiotherapy and nitrosoureas for the treatment of malignant glioma after surgery. N Engl J Med. 1980;303:1323–1329. doi: 10.1056/NEJM198012043032303. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Gilbert MR, Chakravarti A. Chemoradiotherapy in malignant glioma: standard of care and future directions. J Clin Oncol. 2007;25:4127–4136. doi: 10.1200/JCO.2007.11.8554. [DOI] [PubMed] [Google Scholar]
- 3.Karberg S. Switching on epigenetic therapy. Cell. 2009;139:1029–1031. doi: 10.1016/j.cell.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 4.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aggarwal P, Vaites LP, Kim JK, Mellert H, Gurung B, Nakagawa H, et al. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell. 2010;18:329–340. doi: 10.1016/j.ccr.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 7.Scoumanne A, Zhang J, Chen X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 2009;37:4965–4976. doi: 10.1093/nar/gkp516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 2002;3:641–645. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majumder S, Alinari L, Roy S, Miller T, Datta J, Sif S, et al. Methylation of histone H3 and H4 by PRMT5 regulates ribosomal RNA gene transcription. J Cell Biochem. 2010;109:553–563. doi: 10.1002/jcb.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meister G, Fischer U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 2002;21:5853–5863. doi: 10.1093/emboj/cdf585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rank G, Cerruti L, Simpson RJ, Moritz RL, Jane SM, Zhao Q. Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression. Blood. 2010;116:1585–1592. doi: 10.1182/blood-2009-10-251116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren J, Wang Y, Liang Y, Zhang Y, Bao S, Xu Z. Methylation of ribosomal protein S10 by protein-arginine methyltransferase 5 regulates ribosome biogenesis. J Biol Chem. 2010;285:12695–12705. doi: 10.1074/jbc.M110.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, et al. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276:32971–32976. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 16.Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26:3558–3569. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albertoni M, Shaw PH, Nozaki M, Godard S, Tenan M, Hamou MF, et al. Anoxia induces macrophage inhibitory cytokine-1 (MIC-1) in glioblastoma cells independently of p53 and HIF-1. Oncogene. 2002;21:4212–4219. doi: 10.1038/sj.onc.1205610. [DOI] [PubMed] [Google Scholar]
- 18.Alinari L, White VL, Earl CT, Ryan TP, Johnston JS, Dalton JT, et al. Combination bortezomib and rituximab treatment affects multiple survival and death pathways to promote apoptosis in mantle cell lymphoma. MAbs. 2009;1:31–40. doi: 10.4161/mabs.1.1.7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nowicki MO, Dmitrieva N, Stein AM, Cutter JL, Godlewski J, Saeki Y, et al. Lithium inhibits invasion of glioma cells; possible involvement of glycogen synthase kinase-3. Neuro Oncol. 2008;10:690–699. doi: 10.1215/15228517-2008-041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckert D, Biermann K, Nettersheim D, Gillis AJ, Steger K, Jack HM, et al. Expression of BLIMP1/PRMT5 and concurrent histone H2A/H4 arginine 3 dimethylation in fetal germ cells, CIS/IGCNU and germ cell tumors. BMC Dev Biol. 2008;8:106. doi: 10.1186/1471-213X-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon CH, Zhao D, Chen J, Alcantara S, Li Y, Burns DK, et al. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res. 2008;68:3286–3294. doi: 10.1158/0008-5472.CAN-07-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 23.Wen PY, Kesari S. Malignant Gliomas in Adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka H, Hoshikawa Y, Oh-hara T, Koike S, Naito M, Noda T, et al. PRMT5, a novel TRAIL receptor-binding protein, inhibits TRAIL-induced apoptosis via nuclear factor-kappaB activation. Mol Cancer Res. 2009;7:557–569. doi: 10.1158/1541-7786.MCR-08-0197. [DOI] [PubMed] [Google Scholar]
- 25.Brunelle JK, Letai A. Control of mitochondrial apoptosis by the Bcl-2 family. J Cell Sci. 2009;122:437–441. doi: 10.1242/jcs.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- 27.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 28.Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 29.Van Meir EG, Kikuchi T, Tada M, Li H, Diserens AC, Wojcik BE, et al. Analysis of the p53 gene and its expression in human glioblastoma cells. Cancer Res. 1994;54:649–652. [PubMed] [Google Scholar]
- 30.Wang L, Pal S, Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol. 2008;28:6262–6277. doi: 10.1128/MCB.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, et al. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hooi CF, Blancher C, Qiu W, Revet IM, Williams LH, Ciavarella ML, et al. ST7-mediated suppression of tumorigenicity of prostate cancer cells is characterized by remodeling of the extracellular matrix. Oncogene. 2006;25:3924–3933. doi: 10.1038/sj.onc.1209418. [DOI] [PubMed] [Google Scholar]
- 33.Entin-Meer M, Yang X, VandenBerg SR, Lamborn KR, Nudelman A, Rephaeli A, et al. In vivo efficacy of a novel histone deacetylase inhibitor in combination with radiation for the treatment of gliomas. Neuro Oncol. 2007;9:82–88. doi: 10.1215/15228517-2006-032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, Pestka S. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther. 2007;113:50–87. doi: 10.1016/j.pharmthera.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, et al. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol. 2003;23:7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajendran G, Shanmuganandam K, Bendre A, Mujumdar D, Goel A, Shiras A. Epigenetic regulation of DNA methyltransferases: DNMT1 and DNMT3B in gliomas. J Neurooncol. 2011 doi: 10.1007/s11060-010-0520-2. [DOI] [PubMed] [Google Scholar]
- 37.Schlegel J, Guneysu S, Mennel HD. Expression of the genes of methyl-binding domain proteins in human gliomas. Oncol Rep. 2002;9:393–395. [PubMed] [Google Scholar]
- 38.Fukushima T, Favereaux A, Huang H, Shimizu T, Yonekawa Y, Nakazato Y, et al. Genetic alterations in primary glioblastomas in Japan. J Neuropathol Exp Neurol. 2006;65:12–18. doi: 10.1097/01.jnen.0000196132.66464.96. [DOI] [PubMed] [Google Scholar]
- 39.Zenklusen JC, Conti CJ, Green ED. Mutational and functional analyses reveal that ST7 is a highly conserved tumor-suppressor gene on human chromosome 7q31. Nat Genet. 2001;27:392–398. doi: 10.1038/86891. [DOI] [PubMed] [Google Scholar]
- 40.Charong N, Patmasiriwat P, Zenklusen JC. Localization and characterization of ST7 in cancer. J Cancer Res Clin Oncol. 2011;137:89–97. doi: 10.1007/s00432-010-0863-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.