Abstract
Individual responses to growth hormone (GH) treatment are variable. Short-term generation of insulin-like growth factor-I (IGF-I) is recognized as a potential marker of sensitivity to GH treatment. This prospective, phase IV study used an integrated genomic analysis to identify markers associated with 1-month change in IGF-I (ΔIGF-I) following initiation of recombinant human (r-h)GH therapy in treatment-naïve children with GH deficiency (GHD) (n=166) or Turner syndrome (TS) (n=147). In both GHD and TS, polymorphisms in the cell-cycle regulator CDK4 were associated with 1-month ΔIGF-I (P<0.05). Baseline gene expression was also correlated with 1-month ΔIGF-I in both GHD and TS (r=0.3; P<0.01). In patients with low IGF-I responses, carriage of specific CDK4 alleles was associated with MAPK and glucocorticoid receptor signaling in GHD, and with p53 and Wnt signaling pathways in TS. Understanding the relationship between genomic markers and early changes in IGF-I may allow development of strategies to rapidly individualize r-hGH dose.
Keywords: genetic association, gene expression, growth hormone, genetic marker, insulin-like growth factor-I
Introduction
Insulin-like growth factor-I (IGF-I) is the major mediator of the effect of growth hormone (GH) on body growth and is therefore a biomarker for GH action. As serum IGF-I is GH dependent, its monitoring is part of a standard recombinant human (r-h)GH treatment management, providing a marker for both efficacy and safety, and allowing titration to a personalized r-hGH dose.1, 2, 3, 4, 5, 6, 7, 8 Short-term IGF-I generation under exogenous r-hGH treatment, used mainly as a marker of GH sensitivity, is predictive of growth in children with GH deficiency (GHD) as well as in those with idiopathic short stature.9, 10, 11, 12
Responsiveness to r-hGH treatment varies dramatically both across and within indications.13, 14, 15 Such variability points to a marked range of r-hGH dose required to achieve a target serum IGF-I level and the desired growth effect.1, 5 In addition, response to r-hGH dose attenuates with time across all indications. When r-hGH dose adaptation is based on observed yearly growth results, the opportunity to achieve the best individual growth performance may be lost.13, 14, 15 Current prediction models of growth response to r-hGH treatment have attempted to identify an appropriate individualized r-hGH dose,16, 17, 18 but account for only up to 60% of response variability. In addition to auxological characteristics and GH dose, these prediction models have included biological markers of GH action such as IGF-I, but to date have not taken genomic markers into account.
Quickly establishing an appropriate individualized r-hGH dose by targeting a safe but efficacious IGF-I level would be clinically useful. Furthermore, a small number of patients still experience an inadequate growth response to a sufficient r-hGH dose, justifying treatment discontinuation. Identification of genomic markers that are predictive of an inadequate response to GH therapy could help detect these patients more quickly.
Here we report original data from the PREDICT study (NCT00256126),9, 10 investigating for the first time in a prospective study the association between genomic data (genetic polymorphisms and gene expression) and 1-month change in IGF-I (ΔIGF-I) following initiation of r-hGH treatment in children with GHD or Turner syndrome (TS). These two conditions are associated with significant short stature, and account for ∼50% of r-hGH prescriptions. These data are expected to help rationalize and expedite the identification of a personalized r-hGH dose. In addition, these data not only provide new insights into the genes that regulate early response to r-hGH in GHD or TS, but also point to the involvement of key pathways, including those related to the glucocorticoid receptor (GR).
Patients and methods
Study design
The PREDICT study was a phase IV, multicenter, prospective, open-label, 1-month study to identify genomic markers associated with 1-month IGF-I generation in response to r-hGH therapy in GH treatment-naïve children with GHD or TS. This was an international study that recruited patients from centers in Argentina, Australia, Austria, Canada, Finland, France, Germany, Italy, Korea, Norway, Russia, Spain, Sweden, Taiwan and the United Kingdom.
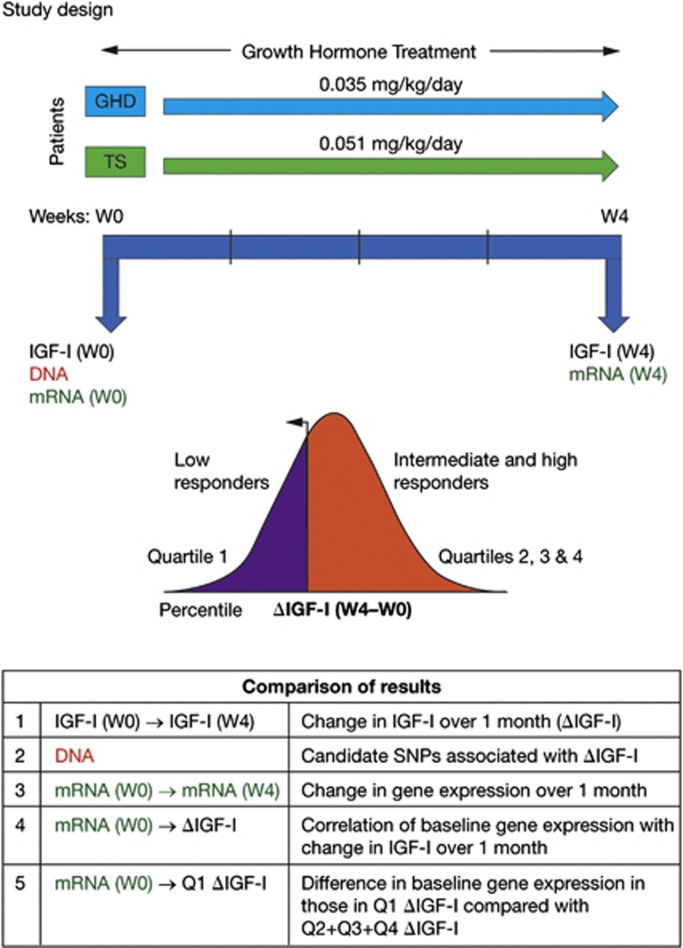
This study involved three stages. First, genes involved in growth and metabolism were identified by literature searches and selected for inclusion, based on advice from a panel of advisors (see Supplementary Table S1 for a list of the candidate genes). Second, genotypes and gene expression data were assessed for their association with 1-month ΔIGF-I. Finally, the predictive potential of the genetic markers (genotypes and baseline gene expression) was evaluated by categorizing the patient population into three groups based on ΔIGF-I over 1 month of treatment: High (top 25%), intermediate (middle 50%) and low (bottom 25%) responders. These patient groups were tested for their association with (a) genetic markers and (b) baseline gene expression profiles. Analysis was carried out separately for children with GHD and for girls with TS (Figure 1).
Figure 1.
Study design and analysis. Growth hormone (GH) was administered daily during 1 month in two different treatment-naïve short pre-pubertal groups at a standard dose: GH deficiency (GHD) or Turner syndrome (TS). Analyses were performed to address specific questions in both GHD and TS. (1) How do serum insulin-like growth factor-I (IGF-I) levels change (ΔIGF-I) over 1 month of recombinant human (r-h)GH therapy (comparing IGF-I before first GH injection (W0) and at week 4 (W4))? (2) Are specific single-nucleotide polymorphisms (SNPs) from a candidate gene list associated with ΔIGF-I (analyzed in DNA extracted from peripheral blood nucleated cells (PBMC) at W0. (3) Which genes display a change in expression over 1 month of GH treatment (analyzed in mRNA from PBMC, W0 versus W4 (full genome))? (4) Does baseline gene expression correlate with 1 month ΔIGF-I (analyzed in mRNA at W0)? (5) Does baseline gene expression differ in lowest quartile of 1-month ΔIGF-I (analyzed in mRNA, comparing W0 Q1 ΔIGF-I with Q2+Q3+Q4 ΔIGF-I)?
This study was conducted in compliance with ethical principles based on the Declaration of Helsinki, the International Conference on Harmonization Tripartite Guideline for Good Clinical Practice and all applicable regulatory requirements.
Patients and treatment
Pre-pubertal children naïve to r-hGH (n=166 with GHD; n=147 with TS) formed the 1-month intention-to-treat cohort. For the genetic analyses, genotype and IGF-I data were available for 160 children with GHD and 139 with TS. Paired expression data for baseline and 1 month of treatment, along with ΔIGF-I, were available for 67 children with GHD and 73 with TS.
The demographic characteristics of the 1-month cohort are illustrated in Table 1. Children with GHD had an established diagnosis based on two different stimulation tests, with a peak serum GH level <10 μg l−1. When children with GHD were categorized according to severity of GHD, based on the peak GH level attained during the stimulation test, 156/166 (94%) children had GH levels ⩽7.0 μg l−1 (77 had levels >4–⩽7 μg l−1 and 79 had levels ⩽4 μg l−1), whereas 13/166 (8%) had GH levels >7–⩽10 μg l−1. Children with GHD associated with etiologies such as central nervous system tumors were excluded. Children born small for gestational age were not excluded. Of the girls with TS, 69/147 (47%) had monosomy X (45 X), consistent with a typical TS population;19 the remainder had partial deletion (46 del[X]), an isochromosome of the long arm of one X chromosome (46 X i[Xq]), or mosaics for 45 X with one or more cell lineages.
Table 1. Baseline characteristics of the study population (intention-to-treat population).
| Characteristic |
GHD |
TS |
||
|---|---|---|---|---|
| Median | Range | Median | Range | |
| Age, years | 9.6 | 1.5, 15.9 | 9.2 | 2.0, 18.7 |
| Height SDS | −2.2 | −6.9, 0.5 | −2.4 | −5.7, 0.4 |
| Weight SDS | −1.4 | −4.1, 6.1 | −1.2 | −3.7, 4.1 |
| BMI SDS | −0.3 | −3.0, 10.3 | 0.4 | −2.2, 6.8 |
| Mid-parental height SDS | −0.7 | −2.9, 1.7 | −0.3 | −2.9, 3.3 |
| Bone age, years | 7.0 | 0.0, 12.5 | 8.0 | 2.0, 15.0 |
| Gestational age, weeks | 40.0 | 27.0, 42.0 | 39.0 | 25.0, 42.0 |
| Birth length, cm | 49.5 | 29.0, 55.0 | 49.0 | 30.0, 55.0 |
| Weight at birth, kg | 3.1 | 0.7, 4.6 | 2.8 | 0.7, 4.1 |
| Baseline IGF-I SDS | −1.8 | −7.810, 1.071 | −1.2 | −6.468, 1.858 |
| Baseline IGFBP3 SDS | −0.2 | −6.568, 2.190 | 0.2 | −3.248, 2.142 |
Abbreviations: BMI, body mass index; GHD, growth hormone deficiency; IGF-I, insulin-like growth factor-I; IGFBP3, IGF-binding protein 3; SDS, s.d. score; TS, Turner syndrome.
r-hGH (Saizen, Merck Serono S.A.—Geneva, Switzerland; 8 mg, containing 0.3% metacresol) was administered subcutaneously, once daily at bedtime using an auto-injector device (one.click, Merck Serono S.A.) for 1 month. Children with GHD received an average r-hGH dose of 0.035 mg kg−1 per day and girls with TS received an average dose of 0.051 mg kg−1 per day.
Serum IGF-I measurement
Serum IGF-I was measured centrally at qLAB (Livingston, Edinburgh, UK) using the DCP chemiluminescent immunoassay (Immulite 2000, Siemens Healthcare Diagnostics , Norwood, MA, USA). IGF-I levels were converted to s.d. scores (SDS) using relevant reference data.20
The study was powered (at 90% P=0.025 one-sided test) to a primary endpoint that would achieve an increase in IGF-I level at 1 month of at least 15%. This critical 15% within-patient change in IGF-I was standardized to SDS changes using gender-matched and median age-related IGF-I standard ranges. A 15% change in IGF-I (μg l−1) corresponded to a 0.35 change in IGF-I SDS.
Genetic analysis
Genotyping was performed on DNA extracted from whole blood using Illumina GoldenGate assays (Illumina, San Diego, CA, USA). In total, 1536 single-nucleotide polymorphisms (SNPs), located on 103 candidate genes, related to: (1) the GH–IGF-I axis; (2) bone and cell growth; and (3) glucose and lipid metabolism, were selected for genotyping. Of these, 1451 SNPs were genotyped successfully. Before analysis, genotyping data were filtered to remove SNPs with low minor allele frequency (<10%), those with a call rate <95%, and those showing significant deviation from the Hardy–Weinberg equilibrium using a Bonferroni correction for 1451 tests (except for X-linked SNPs in girls with TS and boys with GHD). After data cleaning, 1171 SNPs in GHD and 1182 SNPs in TS remained for analysis.
Gene expression analysis
Gene expression profiling was carried out on whole blood collected in PAXgene tubes (PreAnalytiX GmbH, Hombrechtikon, Switzerland) to immediately stabilize the RNA. Children were excluded if they had a chronic infectious disease or an acute/severe illness during the previous 6 months. RNA was extracted centrally by qLAB using the PAXgene 96 blood RNA kit (Qiagen GmbH, Hilden, Germany). Reduction of globin mRNA was undertaken using the Ambion GLOBIN Clear Human Kit (Life Technologies, Carlsbad, CA, USA). Quality of RNA was assessed using a ND-1000 spectrophotometer (Nanodrop, Wilmington, DE, USA) and quantified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Complementary RNA was generated using the Two-Cycle Eukaryotic Target Labeling Kit (Affymetrix, Santa Clara, CA, USA) and a final quality check performed before hybridization to Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays. Arrays were then scanned on an Affymetrix GeneChip 3000 7G scanner and assessed for quality against internal and hybridization controls.
Processing and normalization of the raw gene expression data were performed on GHD and TS samples using a Robust Multi-array Average background correction modified for probe sequence with quantile normalization and median polish (Partek Genomics Suite, version 6.3, St Louis, MO, USA). Confounding effects due to variations in cell populations and outliers were identified by cross validation using principal component analysis and iso-map multidimensional scaling (Qlucore Omics Explorer 2.2, Qlucore, Lund, Sweden). A cut-off was set for the variable with the largest variance (σmax) to remove non-informative probes; this was set at 0.05 (σ/σmax) (Qlucore Omics Explorer 2.2).
Statistical analysis
Continuous analysis
SNPs associated with 1-month ΔIGF-I were identified using the Kruskal–Wallis rank sum test on the following models: genotype, presence of the major allele (dominant model) and presence of the minor allele (recessive model). For non-pseudoautosomal X-linked markers, boys with GHD were analyzed separately from girls with GHD. As a candidate gene rather than a whole genome approach was being used, P-values were adjusted for multiple testing using Bonferroni correction, taking into account the number of linkage disequilibrium blocks present in the gene containing the SNP of interest.
Categorical analysis
Associated markers were then tested in a further stage of the analysis, in which patients were classified, by quartiles (Q) of 1-month ΔIGF-I, into high (⩾Q3), intermediate (>Q1–<Q3) and low (⩽Q1) responders. Markers were assessed by comparing high responders versus intermediate+low responders and low responders versus intermediate+high responders. All P-values were calculated using Fisher's exact test and adjusted for multiple testing using a Bonferroni correction, taking into account the number of linkage disequilibrium blocks within each candidate gene.
Genes identified as significantly associated with ΔIGF-I were evaluated for their potential as biomarkers for predicting high or low response. In the whole group, including all genotypes, 25% of children would be expected to be high or low IGF-I responders and 75% would be intermediate+low or intermediate+high responders. Marker and/or non-marker genotypes that were associated with ⩾15% change from this distribution were identified. The following criteria were applied: the marker or non-marker genotypes should occur at a frequency within the population of >10% P-values adjusted for multiple testing for both continuous and categorical analyses should be ⩽0.05; positive predictive value should be ⩾40% and/or negative predictive value ⩾90%.
Gene expression profiles
Rank regression analysis was used to correlate change in baseline gene expression with 1-month ΔIGF-I (Figure 1). A paired t-test was used to compare gene expression changes over 1 month and analysis of variance was used to determine the differential baseline gene expression between quartile groups, both tests with age and gender as eliminated factors. False discovery-modified P-values (q) based on hypergeometric tests were also determined. Enriched gene functions and canonical biological pathways were identified with Ingenuity Pathways Analysis software (Ingenuity Systems, Redwood City, CA, USA) using Fisher's exact test.
Network analysis
Analysis of inferred protein:protein interaction (PPI) networks was performed using Ingenuity Pathways Analysis software to increase confidence in the observations of differentially expressed genes by their correlation with biological pathways. Gene expression changes were also mapped to the human interactome (Reactome), and a sub-network of inferred protein interactions was extracted.21 The top 10% of proteins within these networks were scored for network topology (connectivity and bottleneck) and used as markers of essential pathway functions,22, 23 which were identified using the Reactome algorithm24 plug-in for Cytoscape 2.8.225 (Supplementary Figure S2).
Results
IGF-I changes after 1 month of r-hGH treatment
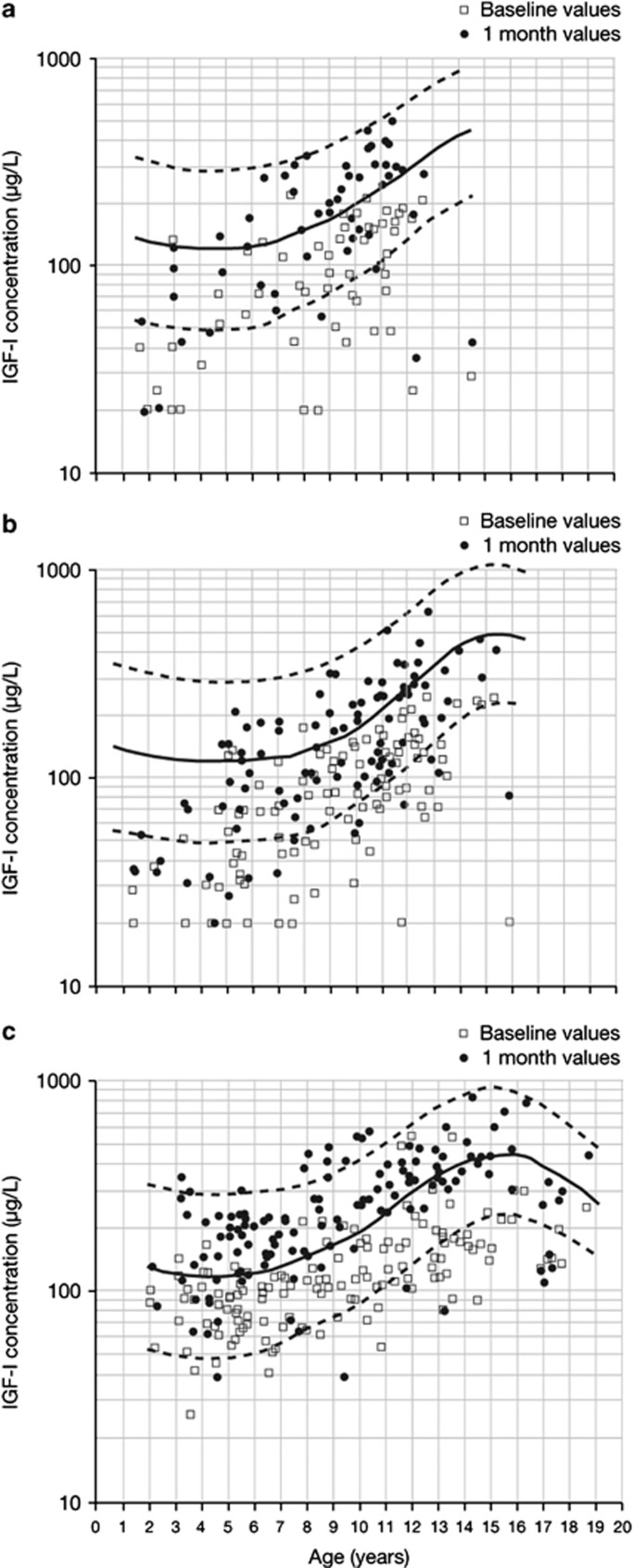
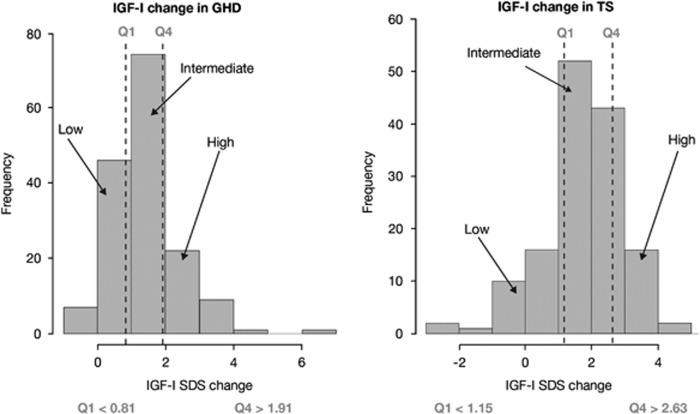
Serum IGF-I levels at baseline and at 1 month in children with GHD or TS are shown in Figure 2. The primary endpoint of an increase of >0.35 in IGF-I SDS from baseline to 1 month was met in 87.0% of children with GHD and in 88.1% of girls with TS. The mean (s.d.; range) 1-month ΔIGF-I SDS was +1.4 (1.0; −0.4, 6.4) in children with GHD and +1.8 (1.2; −2.5, 4.6) in girls with TS (Figure 3). Both 1-month IGF-I (Figure 2) and 1-month ΔIGF-I (Figure 3) show some overlap between GHD and TS.
Figure 2.
Serum insulin-like growth factor-I (IGF-I) (μg l−1) at baseline and 1 month for (a) girls with growth hormone deficiency (GHD), (b) boys with GHD and (c) girls with Turner syndrome (TS). Median growth hormone dose was 0.035 and 0.050 mg kg−1 per day in GHD and TS, respectively. Open squares represent baseline IGF-I and filled circles represent 1-month IGF-I.
Figure 3.
One-month change in insulin-like growth factor-I (IGF-I) s.d. score (SDS): distributions in growth hormone deficiency (GHD; left) and Turner syndrome (TS; right), divided into low (⩽Q1), intermediate (>Q1–<Q3) and high (⩾Q3) IGF-I responders. Q, quartile.
Genetic association with IGF-I change
Genetic association with IGF-I change was analyzed using both a continuous and a categorical approach. The continuous analysis allowed assessment of the impact of SNP carriage on ΔIGF-I; the categorical analysis permitted assessment of the association between an SNP and a ‘good' or ‘poor' IGF-I response, therefore allowing the prediction of outcome.
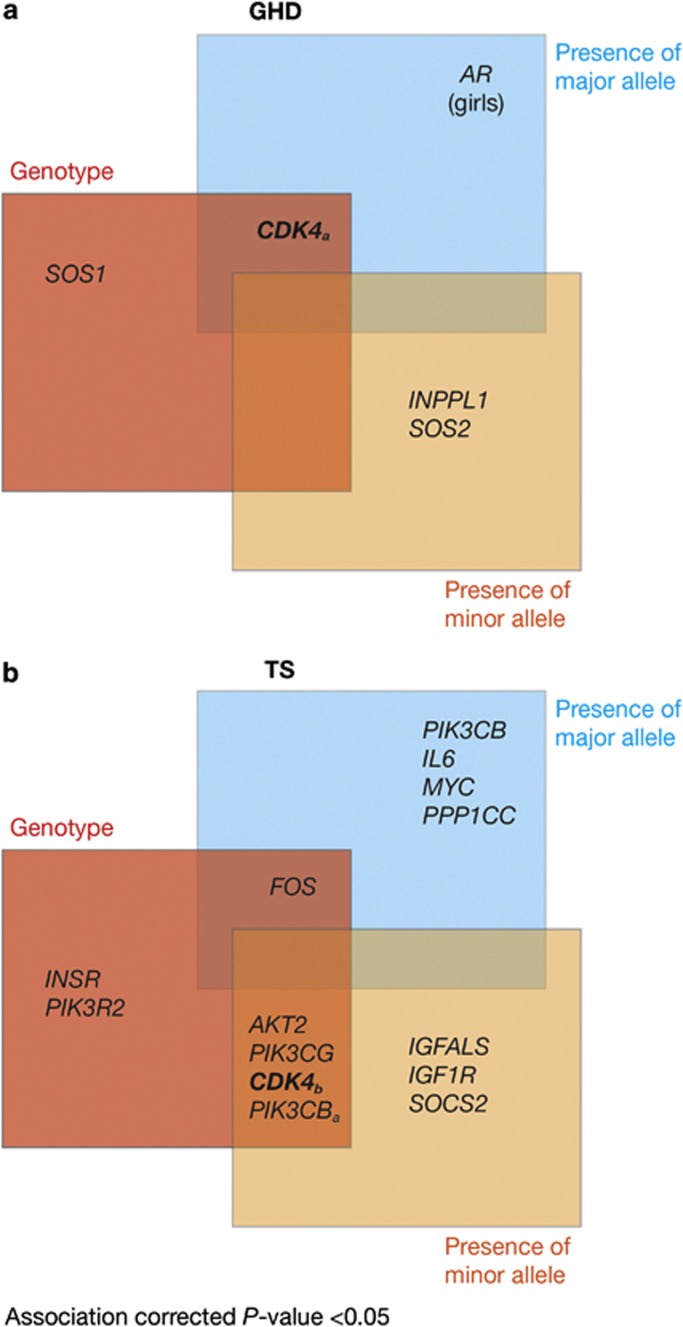
Six SNPs in five different genes for children with GHD and 34 SNPs in 13 separate genes for girls with TS were significantly (P<0.05) associated with 1-month ΔIGF-I SDS (Figure 4). The CDK4 gene was significantly (P<0.05) associated with 1-month ΔIGF-I SDS in both GHD and TS, but the other SNPs differed between the conditions.
Figure 4.
Genes carrying one or more single-nucleotide polymorphism (SNP) associated with 1-month insulin-like growth factor-I (IGF-I) generation in children with (a) growth hormone deficiency (GHD) and (b) Turner syndrome (TS) treated with recombinant human growth hormone. SNPs in the AR gene (rs5918762 and rs5918757) were significant when analyzing GHD girls only.
In children with GHD, SNP rs2270777 within CDK4 met all criteria (Figure 4). The AA genotype was associated with a high IGF-I response. The frequency of this genotype was 22% in the whole study population. For the high versus intermediate+low comparison, 40% of those with the AA genotype had a high response versus an expected 25%. For the low versus intermediate+high comparison, only 9% of those with the AA genotype had a low response, giving a negative predictive value of 91%. Thus, in this population, carriage of the AA genotype was associated with an increased chance (by 1.6-fold) of being a high responder.
For girls with TS, rs2069502 in CDK4 satisfied all criteria (Figure 4). There was a decreased frequency of high responders to r-hGH therapy in carriers of the GG genotype. For the high versus intermediate+low comparison, only 8% of those with the GG genotype had a high response, versus an expected 25%, giving a negative predictive value of 92%. In this population, carriage of the GG genotype was associated with a decreased chance (by 4.7-fold) of being a high responder.
When comparisons were restricted to low (⩽Q1) versus high responders (>Q4), the odds ratios were slightly larger but the P-values were higher because of reduced sample sizes (data not shown).
Gene expression changes over 1 month of treatment with r-hGH
To support the use of gene expression profiling in whole blood as a valid model of GH action with sufficient sensitivity to detect direct and/or indirect GH-induced changes, gene expression in paired samples over 1 month of r-hGH treatment was evaluated. The expression of 5367 and 1850 probe sets in GHD and TS, respectively, with age and gender as confounding factors, changed significantly (t-test, P<0.05), with 552 probe sets overlapping corresponding to 446 genes (Table 2).
Table 2. Change from baseline in gene expression after 1 month of treatment with recombinant human growth hormone.
| Parameter | GHD | TS |
|---|---|---|
| Number of probes detected | 24 707 | 25 313 |
| Upregulated genes, n (%) | 13 242 (53.6) | 13 906 (54.9) |
| Downregulated genes, n (%) | 11 465 (46.4) | 11 407 (45.1) |
| Maximum-fold change | ||
| Upregulated genes | 1.58 | 1.37 |
| Downregulated genes | −1.37 | −1.33 |
| Gene probe sets with a significant expression change from baseline | 5367 | 1850 |
| Unadjusted P-value<0.05 | ||
| Upregulated, n (%) | 3556 (66.3) | 1178 (63.7) |
| Downregulated, n (%) | 1811 (33.7) | 672 (36.3) |
| FDR<0.1 | 1505 | 3 |
| Upregulated, n (%) | 1122 (74.6) | 1 (33.3) |
| Downregulated, n (%) | 383 (25.4) | 2 (66.7) |
Abbreviations: FDR, false discovery rate; GHD, growth hormone deficiency; TS, Turner syndrome.
Gene expression was assessed using the Affymetrix Human Genome U133 Plus 2.0 microarray system. Age-related gene expression probes were removed and the final number of probes detected was further adjusted for probes with low variance (σ/σmax=0.05). The overlap of gene expression probe sets between GHD and TS was 552 (P<0.05), representing 446 genes.
To establish the proportion of differentially expressed genes that were potentially regulated directly by GH, we compared the data by orthology with mouse genes identified as having STAT5 bound to putative regulatory elements by chromatin immunoprecipitation sequencing (ChIP:seq).26 In GHD and TS, the proportion of genes potentially directly regulated by GH through STAT5 was similar (18.2% and 18.3%, respectively).
Table 3 shows the top ten canonical biological pathways associated with the changes in gene expression that occurred over 1 month of r-hGH treatment (q<0.05); glucocorticoid signaling was represented in both GHD and TS analyses, but other pathways differed between the conditions. IGF-I signaling was ranked 18th in GHD and 118th in TS (Figure 5). These observations were supported by network analysis showing the involvement of PPI networks related to ‘growth' mechanisms, cellular assembly and organization in GHD (P<1 × 10−33), and carbohydrate metabolism and protein synthesis in TS (P<1 × 10−44).
Table 3. Biological pathways associated with gene expression changes over 1 month.
| Canonical pathways | −log(B-H P-value) |
|---|---|
| (a) GHD | |
| Protein ubiquitination pathway | 13.80 |
| Estrogen receptor signaling | 3.54 |
| Glucocorticoid receptor signaling | 3.51 |
| Huntington's disease signaling | 2.66 |
| Actin nucleation by ARP–WASP complex | 2.56 |
| Oxidative phosphorylation | 2.56 |
| Mitochondrial dysfunction | 2.37 |
| NRF2-mediated oxidative stress response | 2.36 |
| Aldosterone signaling in epithelial cells | 2.10 |
| Purine metabolism | 2.09 |
| (b) TS | |
| Rac signaling | 2.86 |
| Integrin signaling | 2.50 |
| CD28 signaling in T-helper cells | 2.50 |
| Inositol phosphate metabolism | 2.23 |
| Glucocorticoid receptor signaling | 2.03 |
| Signaling by rho family GTPases | 2.03 |
| Role of pattern recognition receptors in recognition of bacteria and viruses | 1.99 |
| Huntington's disease signaling | 1.92 |
| RAN signaling | 1.79 |
| PI3K signaling in B lymphocytes | 1.79 |
The top 10 canonical pathways from (a) growth hormone deficiency (GHD) and (b) Turner syndrome (TS) associated with gene expression changes over 1 month, as defined by Ingenuity Knowledge Base, are shown. Fisher's exact test with a P-value corrected for false discovery rate by the Benjamini–Hochberg (B-H P-value) method.
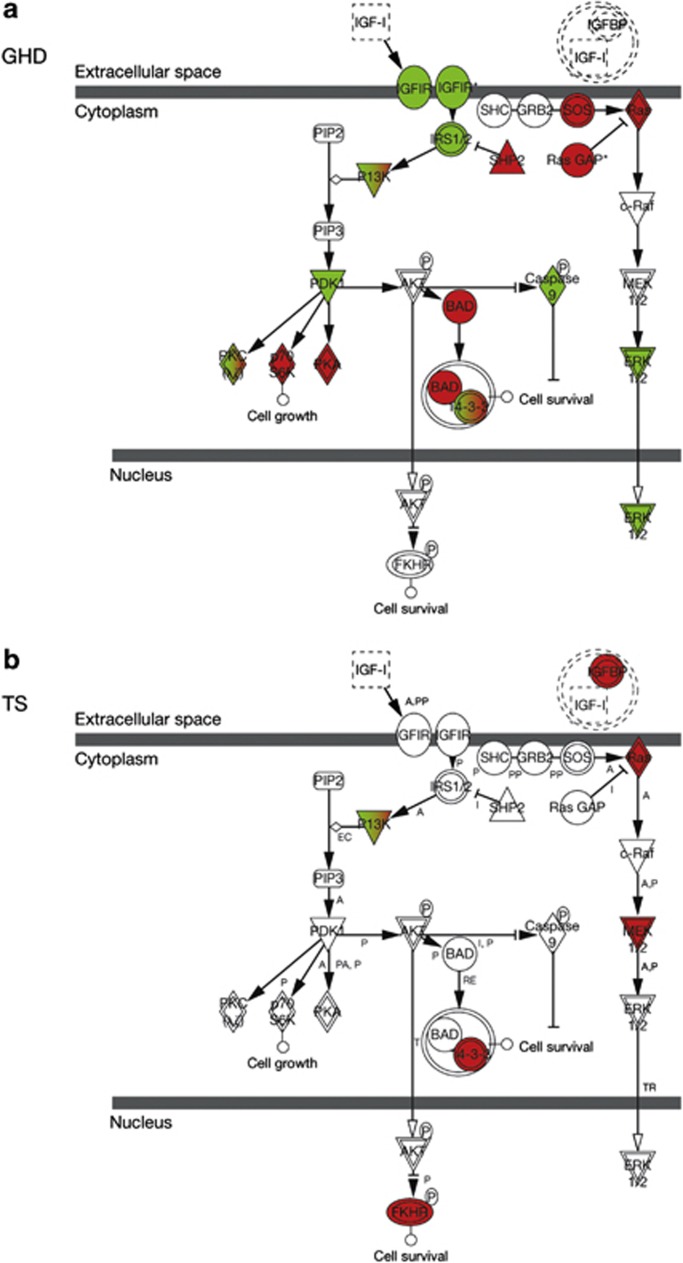
Figure 5.
Insulin-like growth factor-I (IGF-I) signaling pathways: change in gene expression over 1 month of recombinant human growth hormone (r-hGH) therapy in (a) growth hormone deficiency (GHD) and (b) Turner syndrome (TS), as defined by Ingenuity Knowledge Base. Red indicates overexpressed genes and green suppressed genes. Gene expression within the IGF-I signaling pathway differs between GHD and TS in both number and nature of genes modulated.
Baseline gene expression profiles associated with IGF-I response
To evaluate whether gene expression could have a role in predicting response to r-hGH, the relationship between profiles at baseline (treatment-naïve patients) and change in serum IGF-I was assessed by rank regression analysis. In children with GHD (n=89) and in girls with TS (n=87), 536 and 59 baseline gene expression probe sets, respectively, correlated with 1-month ΔIGF-I SDS (r=0.3; P<0.01).
Baseline gene expression differences in lowest quartile of 1-month ΔIGF-I
In addition, the change in serum IGF-I over 1 month of treatment in low IGF-I responders (⩽Q1), when compared with intermediate and high responders, was significantly associated with differential expression of genes at baseline (treatment-naïve patients) in 1092 and 713 probe sets in GHD and TS, respectively (analysis of variance, P<0.05), with 24 overlapping probe sets accounting for 21 known genes. In these low IGF-I responders, canonical pathway analysis highlighted IGF-I and GR in GHD (q<0.14 and q<0.0062, respectively), but not in TS (q<0.48 and q<0.31) (Supplementary Table S2 and Supplementary Figure S1). Inferred network analysis using these genes demonstrated the involvement of growth and development biological functions (P<1 × 10−30).
Integration of genetic and gene expression data
Network analysis places the genes for which changes in expression were observed in the context of a model of all known PPIs, termed the interactome. The properties of these inferred networks can then be examined and correlated with biological function (Supplementary Figure S2).
CDK4 was found to carry SNPs associated with IGF-I response in both GHD and TS (Figure 4). To examine the biological function of the CDK4 SNPs, we generated inferred biological pathways using baseline gene expression significantly (P<0.05) associated with carriage of different CDK4 alleles in low IGF-I responders. Parameters of network topology in these inferred pathways were calculated as markers of essential function.22, 23 In children with GHD, changes were identified in MAPK- and GRB2/SOS-mediated signaling pathways (e.g., IGF-I) associated with carriage of the A allele (GRB2 decreased expression −1.2-fold, SOS1 increased expression +1.2-fold). The GR signaling pathway associated with carriage of the G allele of the rs2270777 CDK4 SNP showing increased GR co-regulator activity (e.g., NCOA3, DRIP205, SMARCA4 and CBP increased expression +1.2-fold) (Table 4, Supplementary Figure S3). In girls with TS, p53 signaling was associated with carriage of the G allele of the rs2069502 CDK4 SNP (including a +1.5-fold increase in MDM2 expression, implying a repression of P53 function,27 and a +1.5-fold increase in ATM expression, implying that DNA damage increased activation of P53.28 Wnt signaling was associated with carriage of the A allele of the rs2069502 CDK4 SNP (including a −1.5-fold change in expression of β-catenin, a primary modulator of WNT signaling and a concomitant −1.4-fold change in expression of TCF4, a developmental transcription factor that is a downstream target of WNT (Table 4, Supplementary Figure S4). These data demonstrate both disease and individual genetic modulation in response to r-hGH treatment.
Table 4. Interactions of two different CDK4 SNPs (rs2270777 and rs2069502) carried by a single allele with genes, for which differential baseline gene expression is associated with low 1-month change in IGF-I SDS (⩽Q1 versus Q2–Q4).
|
Signaling pathways |
GHD: canonical pathways |
TS: canonical pathways |
|---|---|---|
| Carriage of rs2270777 1769 probe sets (P<0.05) Allele A | Carriage of rs2069502 473 probe sets (P<0.05) Allele A | |
| p53 | — | — |
| Wnt | — | q=1.3 × 10–5 |
| GR | — | — |
| IGF-I (MAPK) | q=1.0 × 10–5 | — |
|
Carriage of rs2270777 1112 probe sets (P<0.05)
Allele G |
Carriage of rs2069502 1194 probe sets (P<0.05)
Allele G |
|
| p53 | — | q=6.8 × 10–6 |
| Wnt | — | — |
| GR | q=1.2 × 10–5 | — |
| IGF-I (MAPK) | — | — |
Abbreviations: GHD, growth hormone deficiency; GR, glucocorticoid receptor; IGF-I, insulin-like growth factor-I; MAPK, mitogen-activated protien kinase; Q, quartile; SDS, s.d. score; SNP, single-nucleotide polymorphism; TS, Turner syndrome.
Number of gene expression probe sets associated with carriage of SNP alleles in baseline gene expression (analysis of variance, P<0.05). False discovery-modified P-values (q) based on hypergeometric tests are shown. In low IGF-I responders to growth hormone treatment, changes in several genes differ within four signaling pathways, IGF-I (via MAPK/GRB2/SOS, which are major pathways mediating IGF-I signaling), p53, Wnt and GR, depending on the carriage of the specified CDK4 SNP allele.
Discussion
To date, the PREDICT study is the largest prospective study carried out to evaluate the association between genomic markers and change in serum IGF-I after 1 month of r-hGH therapy in children with two different conditions, GHD and TS.
Serum IGF-I is commonly used to monitor GH treatment and adapt r-hGH dose. In both GHD and TS, we have observed high variability among children in serum IGF-I levels, both at baseline and after 1 month of r-hGH treatment, as well as in the individual ΔIGF-I (Figure 1). In children with GHD, this variability in early IGF-I changes has been shown to relate to long-term changes in growth.1, 3, 4 These early IGF-I changes offer an opportunity to make rapid adjustments to r-hGH dose. The feasibility of reducing response variability to GH treatment by IGF-I titration has recently been shown.7 As individual r-hGH dose finding requires time when based on observed growth response, and because the first year of treatment is the most sensitive to r-hGH dose, the capacity to predict early IGF-I generation would have clinical utility. For example, those predicted to have a low early IGF-I response could start treatment on a higher r-hGH dose and have their r-hGH dose adapted based on levels of serum IGF-I assessed on a monthly basis, for a few months, following initiation of treatment.14, 15, 16, 17
We have taken a genomic approach to evaluate whether polymorphisms in genes related to growth or metabolism and gene expression profiles are associated with early changes in serum IGF-I. The SNP approach is limited by the selection of candidate genes, with our focus in this study being on growth- and metabolism-regulating genes. However, the gene expression data provide a whole genome view, and integration of the two data sets provides additional confidence in the associations observed. We recognize the inherent difficulties of controlling for statistical false positives in data sets from patients with complex diseases, reflecting the heterogeneous nature of the conditions studied.29 Therefore, we have also used the analysis of PPI networks inferred from the data30 and co-expression analysis24 to increase confidence that the association with specific biological pathways is genuine.31
Here, IGF-I generation at 1 month was found to be associated with polymorphisms in 5 genes in GHD and 13 genes in TS. Most of these genes are not involved in classic GH/IGF-I pathways. CDK4, a member of the early cell-cycle gene family and not known to be directly involved in response to GH, is the only common gene associated with IGF-I generation in the two cohorts of children. It is recognized that girls with TS received a higher dose of r-hGH than children with GHD, doses that were determined by standard clinical practice. Despite the dose discrepancy, the 1-month ΔIGF-I can be seen to overlap between GHD and TS, likely reflecting differences in sensitivity to r-hGH, yet the polymorphisms associated with this change in serum IGF-I differ. This could be related to the underlying diagnosis, but it is not possible to rule out an association influenced by r-hGH dose. Nevertheless, our strategy was to identify polymorphisms in each condition independently when r-hGH doses commonly employed for these conditions were used. Our findings indicate that it is not possible to generalize about genetic association with change in serum IGF-I across r-hGH-treated conditions, and each condition (e.g., short children born small for gestational age, those with Prader Willi syndrome or those with chronic renal failure) needs to be evaluated at the dose used in clinical practice.
While validation in an independent cohort is required, the PREDICT study has potentially identified genetic markers that may have clinical utility in predicting early IGF-I generation and that could contribute to individualization of r-hGH dosing. In addition, we have shown that 1 month of r-hGH treatment is associated with changes in the expression of a large set of genes involved in regulating processes such as the cell cycle, cellular movement and function and carbohydrate metabolism. These observations give further insights into the genetic modulation of GH action and point to similarities and differences between GHD and TS. Not surprisingly, these changes involve the IGF-I pathway (Figure 5). These changes in gene expression could reflect both direct and indirect GH action; we have shown in both GHD and TS that 18% of the modulated genes could be mediated directly through STAT5 activation. However, the identification of other canonical pathways represented among the genes whose expression is altered by r-hGH emphasizes the complexity of the potential mediators of GH action in addition to IGF-I (Table 3). One potentially important observation is the consistent finding of GH treatment-related changes in gene expression involving the GR pathway in both GHD and TS. Glucocorticoid excess, including that associated with therapy, strongly inhibits growth and alters metabolism as well as body composition.32 The relationship between GH, cortisol and insulin is key to the regulation of body growth, and bone, fat, protein and carbohydrate metabolism, confirming GH as a metabolic hormone as much as a growth hormone.33, 34, 35, 36 These data contribute to identifying gene interactions and networks that may be helpful in improving our understanding of GH action (although extrapolating our observations from whole blood to other tissues requires a cautious approach). This work may also bring new insights into diseases that alter body composition, such as obesity, type 2 diabetes, adult GHD, acromegaly and glucocorticoid excess.32, 33, 34, 35, 36, 37, 38 Additionally, these findings highlight other potential gene targets not examined in our current candidate gene approach that could be examined for their association with GH-induced responses. As this study extends to long-term follow-up with analysis of genetic association on annual growth responses, we have the opportunity of looking at genetic influence on much longer-term responses to r-hGH.
In both GHD and TS, we have identified a baseline gene expression signature in low IGF-I responders. Together with the SNPs associated with IGF-I generation, these data highlight the value of assessing individual genetic backgrounds, and may ultimately provide key new information for the rapid individualization of r-hGH dose.5 Importantly, the decision to discontinue GH in very poor responders could also be made sooner. The genomic markers identified in this study extend the interest of pharmacogenomics applied to GH treatment.39
In summary, genetic polymorphisms of candidate genes, as well as baseline gene expression, have been associated with ΔIGF-I SDS over the first month of r-hGH treatment in children with GHD or TS. The integration of DNA with mRNA data has provided insight into key pathways associated with poor IGF-I generation. Moreover, individual genotypes in those with poor IGF-I responses can be related to specific genetic pathways. This work is an important step in the development of a personalized GH treatment strategy to rapidly individualize r-hGH dose.
Acknowledgments
This study was supported by Merck Serono S.A.—Geneva, Switzerland. We thank MaiLee Wong and Laura McDonagh of Caudex Medical (supported by Merck Serono S.A. — Geneva, Switzerland) for their assistance in the preparation of this manuscript.
Appendix
PREDICT investigator group: Argentina: A Belgorosky, Buenos Aires. Australia: G Ambler, Westmead. Austria: K Kapelari, Innsbruck. Canada: C Deal, Montreal; J Hamilton, Toronto. Finland: J Jääskeläinen, Kuopio. France: Y Brusquet, Puyricard; P Chatelain, Lyon; M Colle, Bordeaux; R Coutant, Angers; Y Le Bouc, Paris; R Reynaud, Marseille; J-P Salles, Toulouse; J Weill, Lille. Germany: R Pfäffle, Leipzig; M Ranke, Tübingen; G Binder, Tübingen. Italy: M Bozzola, Pavia; F Buzi, Brescia; M Cappa, Rome; A Cicognani, Bologna; M Maghnie, Genova; L Tato, Verona; F Antoniazzi, Verona. Korea: DH Kim, Seoul; SW Yang, Seoul; HW Yoo, Seoul. Norway: E Vangsøy Hansen, Bergen; D Veimo, Bodø. Russia: E Bashnina, St Petersburg; V Peterkova, Moscow; J Skorodok, St Petersburg; L Sultanova, Kazan. Spain: A Carrascosa, Barcelona; A Ferrandez Longas, Zaragoza; R Gracia Bouthellier, Madrid; JP Lopez Siguero, Malaga; S Quinteiro, Las Palmas de Gran Canaria; MD Rodriguez-Arnao, Madrid; A Rodriguez Sanchez, Madrid. Sweden: J Dahlgren, Göteborg; L Hagenäs, Stockholm. Taiwan: JW Hou, Taoyuan; TJ Wang, Kaohsiung County. UK: P Clayton, Manchester; C Kelnar, Edinburgh.
PCh and PCl have received research support and honoraria as speakers and consultants from Merck Serono. AS has received honoraria as a speaker from Merck Serono. LT has received research support and honoraria as a speaker from Merck Serono. HWY has received honoraria and MDR-A has received research support from Merck Serono. MZ, GDC, BD and AC are employees of Merck Serono S.A. — Geneva, Switzerland. JZ is an employee of Merck KGaA, Darmstadt, Germany. JR has received consultation fees from Merck Serono. JS and GRA declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
Supplementary Material
References
- Park P, Cohen P. The role of insulin-like growth factor I monitoring in growth hormone-treated children. Horm Res. 2004;62 (Suppl 1:59–65. doi: 10.1159/000080760. [DOI] [PubMed] [Google Scholar]
- [No authors listed].. Critical evaluation of the safety of recombinant human growth hormone administration: statement from the Growth Hormone Research Society J Clin Endocrinol Metab 2001861868–1870. [DOI] [PubMed] [Google Scholar]
- Cohen P, Bright GM, Rogol AD, Kappelgaard AM, Rosenfeld RG. Effects of dose and gender on the growth and growth factor response to GH in GH-deficient children: implications for efficacy and safety. J Clin Endocrinol Metab. 2002;87:90–98. doi: 10.1210/jcem.87.1.8150. [DOI] [PubMed] [Google Scholar]
- Cohen P, Germak J, Rogol AD, Weng W, Kappelgaard AM, Rosenfeld RG. Variable degree of growth hormone (GH) and insulin-like growth factor (IGF) sensitivity in children with idiopathic short stature compared with GH-deficient patients: evidence from an IGF-based dosing study of short children. J Clin Endocrinol Metab. 2010;95:2089–2098. doi: 10.1210/jc.2009-2139. [DOI] [PubMed] [Google Scholar]
- Cohen P, Rogol AD, Howard CP, Bright GM, Kappelgaard AM, Rosenfeld RG. Insulin growth factor-based dosing of growth hormone therapy in children: a randomized, controlled study. J Clin Endocrinol Metab. 2007;92:2480–2486. doi: 10.1210/jc.2007-0204. [DOI] [PubMed] [Google Scholar]
- Copeland KC, Underwood LE, Van Wyk JJ. Induction of immunoreactive somatomedin C human serum by growth hormone: dose-response relationships and effect on chromatographic profiles. J Clin Endocrinol Metab. 1980;50:690–697. doi: 10.1210/jcem-50-4-690. [DOI] [PubMed] [Google Scholar]
- Kristrom B, Aronson AS, Dahlgren J, Gustafsson J, Halldin M, Ivarsson SA, et al. Growth hormone (GH) dosing during catch-up growth guided by individual responsiveness decreases growth response variability in prepubertal children with GH deficiency or idiopathic short stature. J Clin Endocrinol Metab. 2009;94:483–490. doi: 10.1210/jc.2008-1503. [DOI] [PubMed] [Google Scholar]
- Ranke MB, Schweizer R, Elmlinger MW, Weber K, Binder G, Schwarze CP, et al. Relevance of IGF-I, IGFBP-3, and IGFBP-2 measurements during GH treatment of GH-deficient and non-GH-deficient children and adolescents. Horm Res. 2001;55:115–124. doi: 10.1159/000049982. [DOI] [PubMed] [Google Scholar]
- Clayton P, Chatelain P, Yoo HW, Rodriguez-Arnao MD, Salles JP, Lopez-Siguero JP, et al. Single nucleotide polymorphisms (SNPs) associated with, and predictive of, growth response after 1 and 2 years on growth hormone (GH) therapy in girls with Turner syndrome (TS) Horm Res Paediatr 20117697(abstract P1-d1-308).21778758 [Google Scholar]
- Chatelain P, Clayton P, Quinteiro S, Colle M, Cicognani A, Weill J, et al. Single nucleotide polymorphisms (SNPs) associated with, and predictive of, growth response after 1 and 2 years on growth hormone (GH) therapy in children with GH deficiency (GHD) Horm Res Paediatr 201176101(abstract P1-d2-319). [Google Scholar]
- Buckway CK, Guevara-Aguirre J, Pratt KL, Burren CP, Rosenfeld RG. The IGF-I generation test revisited: a marker of GH sensitivity. J Clin Endocrinol Metab. 2001;86:5176–5183. doi: 10.1210/jcem.86.11.8019. [DOI] [PubMed] [Google Scholar]
- Rudman D, Kutner MH, Blackston RD, Cushman RA, Bain RP, Patterson JH. Children with normal-variant short stature: treatment with human growth hormone for six months. N Engl J Med. 1981;305:123–131. doi: 10.1056/NEJM198107163050302. [DOI] [PubMed] [Google Scholar]
- van Teunenbroek A, de Muinck Keizer-Schrama SM, Stijnen T, Jansen M, Otten BJ, Delemarre-van de Waal HA, et al. Yearly stepwise increments of the growth hormone dose results in a better growth response after four years in girls with Turner syndrome. Dutch Working Group on Growth Hormone. J Clin Endocrinol Metab. 1996;81:4013–4021. doi: 10.1210/jcem.81.11.8923853. [DOI] [PubMed] [Google Scholar]
- Bakker B, Frane J, Anhalt H, Lippe B, Rosenfeld R. Height velocity targets from the National Cooperative Growth Study for first-year growth hormone responses in short children. J Clin Endocrinol Metab. 2008;93:352–357. doi: 10.1210/jc.2007-1581. [DOI] [PubMed] [Google Scholar]
- Ranke MB, Lindberg A. Observed and predicted growth responses in prepubertal children with growth disorders: guidance of growth hormone treatment by empirical variables. J Clin Endocrinol Metab. 2010;95:1229–1237. doi: 10.1210/jc.2009-1471. [DOI] [PubMed] [Google Scholar]
- Ranke MB, Lindberg A, Chatelain P, Wilton P, Cutfield W, Albertsson-Wikland K, et al. Derivation and validation of a mathematical model for predicting the response to exogenous recombinant human growth hormone (GH) in prepubertal children with idiopathic GH deficiency. KIGS International Board. Kabi Pharmacia International Growth Study. J Clin Endocrinol Metab. 1999;84:1174–1183. doi: 10.1210/jcem.84.4.5634. [DOI] [PubMed] [Google Scholar]
- Ranke MB, Lindberg A, Chatelain P, Wilton P, Cutfield W, Albertsson-Wikland K, et al. Prediction of long-term response to recombinant human growth hormone in Turner syndrome: development and validation of mathematical models. KIGS International Board. Kabi International Growth Study. J Clin Endocrinol Metab. 2000;85:4212–4218. doi: 10.1210/jcem.85.11.6976. [DOI] [PubMed] [Google Scholar]
- Wikland KA, Kristrom B, Rosberg S, Svensson B, Nierop AF. Validated multivariate models predicting the growth response to GH treatment in individual short children with a broad range in GH secretion capacities. Pediatr Res. 2000;48:475–484. doi: 10.1203/00006450-200010000-00010. [DOI] [PubMed] [Google Scholar]
- Sybert VP, McCauley E. Turner's syndrome. N Engl J Med. 2004;351:1227–1238. doi: 10.1056/NEJMra030360. [DOI] [PubMed] [Google Scholar]
- Elmlinger MW, Kuhnel W, Weber MM, Ranke MB. Reference ranges for two automated chemiluminescent assays for serum insulin-like growth factor I (IGF-I) and IGF-binding protein 3 (IGFBP-3) Clin Chem Lab Med. 2004;42:654–664. doi: 10.1515/CCLM.2004.112. [DOI] [PubMed] [Google Scholar]
- Haw R, Hermjakob H, D'Eustachio P, Stein L. Reactome pathway analysis to enrich biological discovery in proteomics data sets. Proteomics. 2011;11:3598–3613. doi: 10.1002/pmic.201100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol. 2007;3:e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhao Z. A comparative study of cancer proteins in the human protein-protein interaction network. BMC Genomics. 2010;11 (Suppl 3:S5. doi: 10.1186/1471-2164-11-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu BM, Kang K, Yu JH, Chen W, Smith HE, Lee D, et al. Genome-wide analyses reveal the extent of opportunistic STAT5 binding that does not yield transcriptional activation of neighboring genes. Nucleic Acids Res. 2012;40:4461–4472. doi: 10.1093/nar/gks056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesugi M, Verdine GL. The alpha-helical FXXPhiPhi motif in p53: TAF interaction and discrimination by MDM2. Proc Natl Acad Sci USA. 1999;96:14801–14806. doi: 10.1073/pnas.96.26.14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Guo M, Ouyang H, Li X, Cordon-Cardo C, Kurimasa A, et al. The catalytic subunit of DNA-dependent protein kinase selectively regulates p53-dependent apoptosis but not cell-cycle arrest. Proc Natl Acad Sci USA. 2000;97:1584–1588. doi: 10.1073/pnas.97.4.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Pavelka N. When one and one gives more than two: challenges and opportunities of integrative omics. Front Genet. 2011;2:105. doi: 10.3389/fgene.2011.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci USA. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Z. Mechanisms of steroid impairment of growth. Horm Res. 2002;58 (Suppl 1:33–38. doi: 10.1159/000064764. [DOI] [PubMed] [Google Scholar]
- van Rossum EF, Voorhoeve PG, te Velde SJ, Koper JW, Delemarre-van de Waal HA, Kemper HC, et al. The ER22/23EK polymorphism in the glucocorticoid receptor gene is associated with a beneficial body composition and muscle strength in young adults. J Clin Endocrinol Metab. 2004;89:4004–4009. doi: 10.1210/jc.2003-031422. [DOI] [PubMed] [Google Scholar]
- Finken MJ, Meulenbelt I, Dekker FW, Frolich M, Walther FJ, Romijn JA, et al. Abdominal fat accumulation in adults born preterm exposed antenatally to maternal glucocorticoid treatment is dependent on glucocorticoid receptor gene variation. J Clin Endocrinol Metab. 2011;96:E1650–E1655. doi: 10.1210/jc.2011-0288. [DOI] [PubMed] [Google Scholar]
- Binnerts A, Deurenberg P, Swart GR, Wilson JH, Lamberts SW. Body composition in growth hormone-deficient adults. Am J Clin Nutr. 1992;55:918–923. doi: 10.1093/ajcn/55.5.918. [DOI] [PubMed] [Google Scholar]
- Masuzaki H, Paterson J, Shinyama H, Morton NM, Mullins JJ, Seckl JR, et al. A transgenic model of visceral obesity and the metabolic syndrome. Science. 2001;294:2166–2170. doi: 10.1126/science.1066285. [DOI] [PubMed] [Google Scholar]
- Mehls O, Himmele R, Homme M, Kiepe D, Klaus G. The interaction of glucocorticoids with the growth hormone-insulin-like growth factor axis and its effects on growth plate chondrocytes and bone cells. J Pediatr Endocrinol Metab. 2001;14 (Suppl 6:1475–1482. [PubMed] [Google Scholar]
- Agha A, Monson JP. Modulation of glucocorticoid metabolism by the growth hormone—IGF-1 axis. Clin Endocrinol (Oxf) 2007;66:459–465. doi: 10.1111/j.1365-2265.2007.02763.x. [DOI] [PubMed] [Google Scholar]
- Rosenfeld RG. Editorial: the pharmacogenomics of human growth. J Clin Endocrinol Metab. 2006;91:795–796. doi: 10.1210/jc.2006-0026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.