Abstract
The tissue-specific pattern of mRNA expression can indicate important clues about gene function. High-density oligonucleotide arrays offer the opportunity to examine patterns of gene expression on a genome scale. Toward this end, we have designed custom arrays that interrogate the expression of the vast majority of protein-encoding human and mouse genes and have used them to profile a panel of 79 human and 61 mouse tissues. The resulting data set provides the expression patterns for thousands of predicted genes, as well as known and poorly characterized genes, from mice and humans. We have explored this data set for global trends in gene expression, evaluated commonly used lines of evidence in gene prediction methodologies, and investigated patterns indicative of chromosomal organization of transcription. We describe hundreds of regions of correlated transcription and show that some are subject to both tissue and parental allele-specific expression, suggesting a link between spatial expression and imprinting.
The completion of the human and mouse genome sequences opened an historic era in mammalian biology. One common conclusion from these projects was the determination that mammals have only ≈30,000 protein-encoding genes (1, 2). Yet, despite the apparent tractability of this figure (earlier estimates were much higher), to date all existing research has determined the function of only a fraction of these genes. Currently, only ≈15,000 human and ≈10,000 mouse genes are described in the literature (Medline, www.ncbi.nih.gov/Pubmed). The challenge and opportunity for genomics strategies and techniques are to accelerate the functional annotation of novel genes from the uncharted genome.
High-throughput technologies for biological annotation have the capacity to partially address the discrepancy between the identification of genes and the understanding of their function. For example, proteins have well defined molecular roles encoded in their primary amino acid sequence as domains. Using sequence informatics, these domains can be used as a tool to search the entire genome to find protein family members that likely function in an analogous manner. Gene expression arrays have also been a useful tool for genome-wide studies where changes in gene expression can be associated with physiological or pathophysiological states (3). Recently, other high-throughput techniques such as RNA interference (4) and cDNA overexpression (5) have been developed, further accelerating functional genome annotation. The integration of these diverse strategies is critical to annotation efforts and remains a significant challenge.
Previously, we generated a preliminary description of the human and mouse transcriptome using oligonucleotide arrays that interrogate the expression of ≈10,000 human and ≈7,000 mouse target genes (6). We explored this data set for insights into gene function, transcriptional regulation, disease etiology, and comparative genomics. However, this data set was based on commercially available gene expression arrays and therefore was biased toward previously characterized genes. In this report, we significantly extend this earlier work by determining the expression patterns of previously uncharacterized protein-encoding genes and de novo gene predictions from the mouse and human genome projects. Using custom-designed whole-genome gene expression arrays that target 44,775 human and 36,182 mouse transcripts, we have built a more extensive gene atlas using a panel of RNAs derived from 79 human and 61 mouse tissues. This data set constitutes one of the largest quantitative evaluations of gene expression of the protein-encoding transcriptome to date.
Building on our previous analyses, these expression patterns were examined for global trends in gene expression. We also provide experimental validation of thousands of gene predictions and use these data to determine which of the commonly used types of evidence for gene prediction most accurately correlates with expressed genes. In addition, we used this data set to search for chromosomal regions of correlated transcription (RCTs), which may indicate higher-order mechanisms of transcriptional regulation. Furthermore, we show that some of these tissue-specific coregulated genes are subject to another form of regulation, parental imprinting, and thus that several of these regions are under the control of both tissue- and parental allele-specific expression. Finally, we have made these data publicly available for searching and visualization by keyword, accession number, sequence, expression pattern, and coregulaion at our web site (http://symatlas.gnf.org).
Materials and Methods
Microarray Chip Design. We identified a nonredundant set of target sequences for the human and mouse using the following sources: RefSeq (15,491 human and 12,029 mouse sequences) (7); Celera (49,859 human and 29,331 mouse sequences) (8); Ensembl (33,698 human sequences); and RIKEN (46,299 mouse sequences) (9). First, all sequences were screened with repeat-masker (www.repeatmasker.org) to remove repetitive elements. Next, sequence identity between individual sequences was established by using pairwise blat (10) or blast (11) and sim4 (12). The results from single-linkage clustering were further triaged to produce a final target set of 44,775 human and 36,182 mouse targets with the highest degree of confidence of computational prediction [biasing toward sequences containing Inter-pro domains (13) and away from noncoding RNAs]. Finally, the human sequence set was pruned of all targets already represented on the Affymetrix (Santa Clara, CA) commercially available HG-U133A array, leaving 22,645 target sequences for our custom array. One hundred target sequences from the HG-U133A chip were also included in the GNF1H design for the normalization procedure (see below). The final human and mouse target sets were submitted to the Affymetrix chip design pipeline for fabrication of the GNF1H and GNF1M arrays, respectively.
Tissue Preparation. Human tissue samples were obtained from several sources: Clinomics Biosciences (Pittsfield, MA), Clontech, AllCells (Berkeley, CA), Clonetics/BioWhittaker (Walk-ersville, MD), AMS Biotechnology (Abingdon, Oxfordshire, U.K.), and the University of California at San Diego. When samples from four or more subjects were available, equal numbers of male and female subjects were used to make two independent pools; when fewer than four samples were available, RNA samples were pooled, and duplicate amplifications were performed for each pool (detailed annotation for human samples is on our web site and in Table 1, which is published as supporting information on the PNAS web site). Adult (10- to 12-week-old) mouse tissue samples were independently generated from two groups of four male and three female C57BL/6 mice by dissection, and tissues were subsequently quickly frozen on dry ice. Tissues were pulverized while frozen, and total RNA was extracted with Trizol (Invitrogen, Carlsbad) by using ≈100 mg of tissue, then further processed by using the RNeasy miniprep kit according to manufacturer's protocols (Qiagen, Chatsworth, CA). The quality of all samples was determined with an Agilent Bioanalyzer (Palo Alto, CA).
Microarray Procedure. Microarray analysis was performed essentially as described (14). Briefly, 5 μg of total RNA was used to synthesize cDNA that was then used as a template to generate biotinylated cRNA. cRNA was fragmented and hybridized to Affymetrix custom or commercially available gene expression arrays. The arrays were then washed and scanned with a laser scanner, and images were analyzed by using the MAS5 algorithm (15). Arrays were normalized by using global median scaling. The human HG-U133A and GNF1H chips, which were hybridized to the same biological sample, were first paired and prenormalized by using the common targets. The condensed data files are available from our web site (http://symatlas.gnf.org) and Gene Expression Omnibus (www.ncbi.nih.gov/geo) (16). Raw CEL files will be provided upon request (http://symatlas.gnf.org).
Identification of RCTs. All target genes were mapped to their corresponding genome assembly (human to National Center for Biotechnology Information Hs34 assembly, mouse to February 2003 Mm30 assembly) by using blat (10). To account for multiple probes interrogating a single gene, target sequences were also compared to UniGene (www.ncbi.nih.gov/UniGene) by using blast. Target sequences that map within 25 kb of each other and to a common UniGene cluster were pooled, and their expression values were averaged and treated as a single target in the RCT analysis. Next, each chromosome was scanned in window sizes of 3–10 adjacent genes. Windows where >50% of all pairwise comparisons of expression pattern showed a Pearson correlation coefficient >0.6 were identified as RCTs. Randomization studies of gene order confirmed the significance of both the overall number of RCTs and the average pairwise correlation of each individual RCT (P < 0.005, correcting for multiple testing). Pairwise sequence similarity within each RCT was assessed by using tblastx (11), where a similarity value is the product of the alignment similarity and the percentage of total sequence length aligned. Synteny between the human and mouse genome assemblies was derived from a published analysis of syntenic anchors (17). For the analysis of evolutionarily conserved RCTs, only the 32 tissues profiled in common between the mouse and human data sets were used. All analyses and visualizations were performed by using r (www.r-project.org).
Imprinting Analysis. Allele-specific probe expression analysis was used to identify genes with an imprinted expression pattern. Two distinct mouse strains, C57BL/6J (B6) and Mus musculus castaneus (CAST/Ei), were bred to produce four independent mouse crosses (male::female): B6::B6, B6::CAST/Ei, CAST/Ei::B6, and CAST/Ei::CAST/Ei. Each litter of embryonic day 14–16 embryos was pooled, and RNA from four to five separate litters was labeled and hybridized to GNF1M arrays. A probe-level analysis was performed to detect naturally occurring polymorphisms between the two strains. Individual probes (but not entire probe sets) showing a significantly different signal between the two homozygous groups were identified as putative polymorphisms in the target gene. Next, the hybridization signal from the two reciprocal crosses was examined for statistically significant differences in signal based on the paternal or maternal allele, as assessed by t test (P < 0.001), indicating a pattern of male or female imprinting.
Results and Discussion
The tissue-specific RNA expression pattern of a gene can indicate important clues to its physiological function. To build an extensive atlas of tissue-specific gene expression, we created custom arrays that interrogate the expression of known and predicted protein-encoding genes from the mouse and human genomes. The design process used a nonredundant set of known genes and gene predictions compiled from Refseq, Celera, Ensembl (for human), and RIKEN (for mouse). For our GNF1H custom human array, we further removed gene targets that were already represented on the commercially available HG-U133A array from Affymetrix. Finally, we biased the final selection toward gene predictions with likely protein-coding regions. In total, the U133A/GNF1H chipset interrogates 44,775 probe sets, and our custom-designed GNF1M mouse array interrogates 36,182 probe sets. As of the most current annotation in January 2004, these correspond to 33,698 and 33,825 unique human and mouse genes, respectively, after accounting for multiple probe sets interrogating single genes and split transcripts.
Using these whole-genome gene expression arrays, we measured the expression of an extensive set of transcripts and transcript predictions on a single technology platform across a diverse panel of 79 human and 61 mouse tissues. This gene atlas represents the normal transcriptome and allowed us to examine global trends in gene expression. Classical reassociation kinetics (Rot) has been used to assess global trends in gene expression at a population level (18). The analysis of our data set expands this knowledge by examining transcript expression across a large number of tissues at the individual transcript level. We find that 52% (16,454) and 59% (17,924) of target genes are detected in at least one tissue in the human and mouse, respectively (Fig. 4A, which is published as supporting information on the PNAS web site). The average number of transcripts expressed in a single tissue was ≈8,200 (mouse). These observations generally concur with previous findings derived from Rot analyses, which indicate that ≈10,000–15,000 mRNAs are expressed in a given tissue at ≈1–10 copies per cell, and that 90% of these are common between two tissues (19). However, although Rot analysis suggests that the majority of transcripts are present in many or all tissues, our data show that <1% of human target sequences are ubiquitously expressed. Approximately 3% of mouse target sequences are detected in all samples profiled, although this number will certainly decline as the number of samples increases. Not surprisingly, the expression of these ubiquitously expressed housekeeping genes is ≈30-fold higher than for all genes in the data set (Fig. 4B).
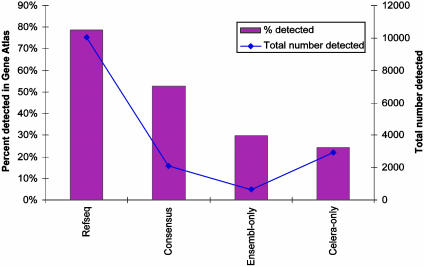
Another valuable use of this data set is characterization of novel predicted genes derived from the mouse and human genome projects (1, 2). Many of these exist solely as in silico predictions, and therefore evidence of their expression can serve as validation of these predictions. Furthermore, determining the expression pattern of an uncharacterized gene can indicate the appropriate tissue(s) from which the transcript can be cloned, as well as provide a base layer of physiological annotation. Gene prediction is an inexact art, where distinct methods and researchers often produce largely nonoverlapping sets of gene predictions (20). For the human data, we subdivided the transcripts into four classes based on annotation information at the time of design: known genes found in Refseq, genes predicted independently by two groups (Celera and Ensembl), singleton predictions found by the Ensembl group only, and singleton predictions found by the Celera group only. As expected, the set of known genes (Refseq) has the highest rate of detection in our data set, because 79% have detectable expression in at least one sample (Fig. 1). Because all Refseq genes are known to be expressed, this suggests that our methodologies and current tissue libraries have a minimum false-negative rate of ≈21% in detection of expression. This can certainly be improved with the profiling of additional tissues and cell types. Consensus gene predictions have a higher rate of detectable expression (53%) than either singleton gene predictions offered by Ensembl or Celera only (30% and 24%, respectively) (Fig. 1). Although the Ensembl-only group had a slightly higher rate of detection, a greater total number of Celera-only predictions was detected (2,918 Celera vs. 618 Ensembl predictions). Analogous results are seen in the mouse data set, in which Refseq genes had a higher rate of detection than gene predictions by Celera (79% vs. 46%). The differences among these three classes are also reflected in the quantitative measures of gene expression. On average, human Refseq genes are expressed at a level 2-fold higher than consensus predictions, which in turn are 66% higher than singleton predictions (P<<0.001; data not shown). This observation likely reflects a historical bias in the biology of studying highly abundant proteins. In total, we find evidence of expression for 5,641 (31.2%) human and 2,629 (46.2%) mouse gene predictions through detection of their transcribed mRNA product in at least one tissue. In addition, we describe the expression pattern for 9,708 mouse RIKEN-derived genes, many of which lack significant expression annotation. It is important to note that the gene predictions for which we do not observe detectable expression are not necessarily incorrect, because the appropriate tissue(s) for a given gene may have not been profiled, the gene may be present in a small number of copies (e.g., in a small subset of cells within a tissue), or the probe set may not properly interrogate the expression of the gene (e.g., UTRs, split transcripts, or missing or mistaken terminal exons). Despite these caveats, this data set provides the expression pattern of thousands of gene predictions and poorly characterized transcripts from the mouse and human genome projects, offering the opportunity to study the function of these genes in their most relevant tissues.
Fig. 1.
Validation of gene predictions in humans. Gene targets on the GNF1H array were divided into four categories: contained in Refseq, predicted by both gene prediction efforts considered (“Consensus”), and predicted by only one group (“Ensembl-only” and “Celera-only”). On the left axis (solid bars), rates of validation are shown, where detectable expression in at least one tissue is taken as evidence of the validity of a gene prediction. The right axis (blue line) indicates the total number of validated genes per group.
Given the differing methods and subsequent results from gene prediction efforts, we next investigated which characteristics of a predicted transcript were better indicators of its detectable expression. In the methodology used by Celera, the following lines of evidence were considered in their gene prediction algorithm: “conservation between mouse and human genomic DNA, similarity to human [and] rodent transcripts (ESTs and cDNAs), and similarity of the translation of human genomic DNA to known proteins” (1). Using the detectable expression of a gene product as validation of the prediction, we created receiver operating characteristic curves for each line of evidence that plot the true positive rate as a function of the false positive rate. The area under the curve (AUC) measures the strength of the predictor; a perfect predictor would have AUC = 1, and a random factor would have AUC = 0.5. When comparing the predictor strength among the three lines of evidence above in the human data set, we find that although no single line of evidence is universally predictive of expression, EST evidence has the most predictive value (AUC = 0.77) (Fig. 5, which is published as supporting information on the PNAS web site), an observation likely linked to the fact that highly expressed genes are more likely to be represented in EST databases. Protein homology support and sequence similarity between human and mouse genomic sequences both had a lesser impact on the validation of gene predictions (AUC of 0.66 and 0.65, respectively). The availability of additional mammalian genome sequences should increase the power of sequence conservation in gene prediction. Somewhat surprisingly, simply the length of the transcript prediction was also a reasonable predictor of detection in our data set (AUC = 0.68), suggesting that incomplete transcript predictions were significant factors in the nonobservation of many gene targets.
We and others have used gene expression information, genome sequence, and de novo motif discovery tools to search for enhancer elements that direct tissue-specific gene expression (21, 22). In contrast to enhancers that generally direct the expression of a single gene, locus control regions (LCR) are characterized by their ability to promote the expression of multiple genes at a single locus. To date, only a handful of LCRs have been reported (23). Recently, Spellman and Rubin (24) used Drosophila gene expression arrays to identify ≈200 clusters of adjacent and similarly expressed genes and suggest that these patterns are most consistent with regulation of chromatin structure. Others (25–27) have also performed similar analyses in humans, Caenorhabditis elegans, and yeast on more limited sets of experimental conditions.
To identify potential loci in our data set, the expression of which may be controlled in a locus-dependent manner, we mapped the transcripts represented on our gene expression arrays to genome assemblies and scanned each chromosome for windows of genes with correlated expression patterns. We called these sites RCTs as a general term encompassing LCRs and correlated expression achieved through gene duplication. It is important to note that detection of these RCTs is heavily influenced by comparison algorithms, normalization procedures, and underlying data. In particular, the inclusion of several purified immune cell populations in our human sample set skewed the normalization procedure and led to an increase in RCTs whose expression is enriched in these samples. In total, we identified 156 and 108 RCTs in human and mouse, respectively (descriptions of all RCTs are available for download from http://symatlas.gnf.org). Tissues with very specific clusters of genes such as those in the immune system, liver, testis, and placenta had more RCTs than other tissues in both the mouse and human data sets. Mechanistically, expression of these RCTs is likely mediated through either common promoter elements (resulting from gene duplication) or through higher-order gene regulation such as site-specific chromatin remodeling. To separate these two possibilities, we identified likely paralogs using tblastx, a local six-frame translated nucleotide-to-nucleotide alignment algorithm (11). RCTs whose genes share significant sequence similarity in their coding sequences are likely to be products of gene duplication, whereas dissimilar genes may result from an LCR or other higher-order transcriptional regulation.
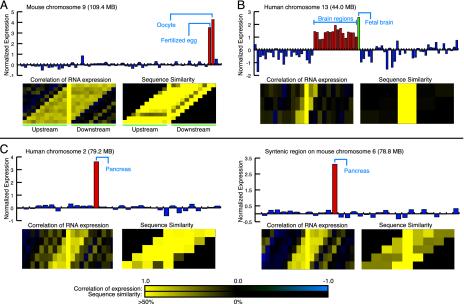
As expected, we found RCTs with both related and unrelated genes. Fig. 2A illustrates an example of an RCT driven by gene duplication. This cluster of genes on mouse chromosome 9 represents a family of 11 uncharacterized F-box and WD40 repeat containing proteins that are specifically expressed in fertilized eggs and oocytes. Because of their high degree of sequence similarity, we hypothesize that their correlated expression pattern is a result of duplicated regulatory elements present in their structural genes, and that these genes may play an important role in the specialized protein expression of oocytes. In contrast, we also note a cluster of three genes with no apparent sequence similarity on human chromosome 13 that are highly enriched in samples derived from brain tissues, particularly the fetal brain sample (Fig. 2B). The genes in this cluster are neurobeachin, an uncharacterized mRNA, and doublecortin- and calmodulin kinase-like 1 protein (DCAMKL1). It is appealing to hypothesize that the correlated expression patterns of these genes and their colocalization at a chromosomal locus indicate a common role in a neurological process or network. Because these genes do not share sequence similarity, this region may also contain a previously unrecognized LCR or strong regional enhancer. Overall, 97 (62%) and 78 (72%) of the human and mouse RCTs identified have an average pairwise sequence similarity of <20% and do not encode related genes.
Fig. 2.
RCT. (A) An RCT was identified on mouse chromosome 9, consisting of 11 genes that share a highly conserved expression pattern. (Upper) The y axis is average normalized expression value, the x axis contains the 61 different tissues, and red bars are fertilized egg and oocytes. The correlation plot (Lower Left) visualizes the pairwise correlation coefficients. Each row represents a gene, ordered vertically according to their position on the chromosome. The center yellow vertical strip represents autocorrelation (R = 1); positions to the right of center represent correlation of the gene to its downstream neighbors, whereas positions to the left represent correlation to the upstream neighbors. Yellow indicates high correlation; blue indicates low correlation (scale at bottom). The sequence similarity plot (using tblastx, Lower Right) has the same structure as the correlation plot, except pairwise sequence similarity is shown. In this RCT with high expression levels in fertilized eggs and oocytes, the genes share a high degree of sequence similarity, likely indicating they are all members of a single gene family and the result of one or more gene duplication events. (B) An example RCT is identified on human chromosome 13, which contains three genes with highly correlated expression (red bars are brain regions, green bar is fetal brain). In contrast to the first example, these genes share very little pairwise sequence similarity. (C) An evolutionarily conserved RCT is shown from human chromosome 2 (Left) and the syntenic region on mouse chromosome 6 (Right). These RCTs share a pancreas-enriched expression pattern (red bar), as well as significant sequence similarity.
We next examined both the mouse and human data for RCTs that were identified in both data sets and are likely evolutionarily conserved. The majority of the RCTs were not found in both human and mouse, in many cases because the orthologs or syntenic regions have not yet been defined or the patterns were not conserved. However, in some cases, the apparent lack of conservation likely reflects physiological differences between the two organisms. For example, we observed RCTs with expression enriched in the olfactory bulb present in the mouse but not the human data set. Nevertheless, several RCTs were conserved, including a cluster of pancreas-specific genes mapping to human chromosome 2 and its syntenic region on mouse chromosome 6 (Fig. 2C). The human cluster is comprised of five genes, including pancreatitis-associated proteins (PAP), three regenerating islet-derived proteins (REG1A, REG1B, and REGL), and one protein of unknown function (LPPM429). The mouse cluster contains the ortholog to PAP, four isoforms of regenerating islet-derived proteins, and islet neogenesis-associated protein-related protein. The conservation of this RCT in human and mouse suggests that these genes perform analogous and important roles in both of these mammals.
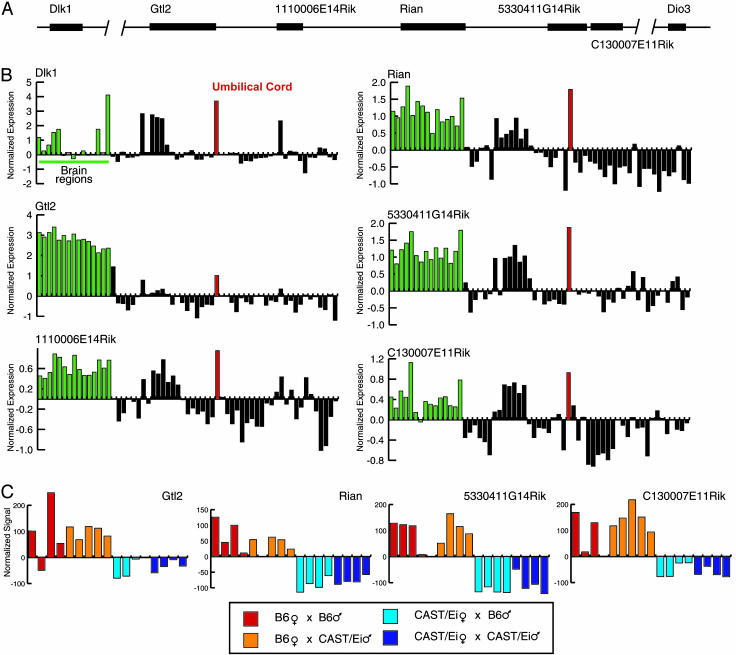
After mapping all target genes to their respective genome assemblies, we noted a region of mouse chromosome 7 (130 Mb) that contained several genes previously shown to be imprinted (28–30), three of which (H19, Igf2, and Cdkn1c) shared a pattern of enriched expression in placenta, umbilical cord, and embryonic tissues. We also noted another pair of adjacent genes (Zim1 and Peg3) elsewhere on chromosome 7 (6 Mb) that shared this tissue-specific expression pattern, and whose expression has been shown to be imprinted (31). Prompted by these observations, we examined our set of RCTs for other imprinted genes that were clustered in a single locus. On mouse chromosome 12 (103 Mb), we observed an RCT that consists of six adjacent genes, all with enriched expression in brain regions and umbilical cord (Fig. 3 A and B). Recently, several groups showed that two genes in this locus, Dlk1 and Gtl2, are imprinted (reviewed in ref. 32). Later, it was also shown that another gene at this locus, Rian, and several adjacent tandemly repeated C/D small nucleolar RNA genes are also imprinted (33, 34). Furthermore, although we do not have a probe set on our array that reliably detects its expression, Dio3 is located proximal to this locus and has also shown to exhibit genomic imprinting (35). The imprinting status of the three remaining R IKEN clones at this locus (1110006E14Rik, 5330411G14Rik, and C130007E11Rik) is not known, although they share the brain- and umbilical cord-enriched expression characteristic of all of the genes in the RCT.
Fig. 3.
Six genes on mouse chromosome 12 share a distinctive pattern of expression. (A) A genomic view of this region (not to scale). Locations of the genes on the mouse genome assembly: Dlk (103.508 Mb), Gtl2 (103.593 Mb), 1110006E14Rik (103.646 Mb), Rian (103.696 Mb), 5330411G14Rik (103.788 Mb), C130007E11Rik (103.798 Mb), and Dio3 (104.328 Mb). (B) These genes share enriched expression in brain regions (green bars) and umbilical cord (red bar). The y axes indicate normalized expression values, whereas each bar along the x axis indicates a sample profiled in our data set. (C) Three of these genes (Dlk1, Gtl2, and Rian) have been previously reported to be imprinted. Using our allele-specific probe expression analysis approach (see text), we confirmed the imprinted regulation of Gtl2 and Rian and report two previously undescribed imprinted transcripts at this locus (5330411G14Rik and C130007E11Rik). The y axes indicate the normalized signal intensity for individual probes on the array, and each bar represents a pooled sample from a cross indicated by color (see key).
To investigate whether these three genes were also imprinted, we used two distinct mouse strains, C57BL/6J (B6) and M. m. castaneus (CAST/Ei), to set up four independent mouse crosses (male::female): B6::B6, B6::CAST/Ei, CAST/Ei::B6, and CAST/Ei::CAST/Ei. Four independent litters of pooled embryonic day 14–16 embryos were dissected, and RNA expression was analyzed by allele-specific probe expression analysis, which allows us to determine whether the transcript is expressed exclusively or preferentially from either the paternal or maternal allele. This analysis reconfirmed the imprinted expression of Gtl2 and Rian (Fig. 3C). Because no probes could distinguish between the B6 and CAST/Ei forms of Dlk1, we were unable to reconfirm its imprinted expression. Two of the uncharacterized RIKEN genes at this locus, 5330411G14Rik and C130007E11Rik, showed expression from the maternal allele only, further expanding the number of known imprinted genes at this locus (Fig. 3C). Because these cDNAs are within 10 kb of one another, it is possible they are derived from the same structural gene. The third gene (1110006E14Rik), like Dlk1, did not contain a probe capable of ascertaining its imprinting status. During the preparation of this manuscript, another gene in this locus sharing the 3′-end of C130007E11Rik was also shown to be imprinted (36). In sum, the allele-specific probe expression analysis method has identified another two imprinted transcripts at this locus. Furthermore, based on the observation that well-characterized imprinted loci on mouse chromosomes 7 and 12 share a common pattern of gene expression in our data, we speculate that the LCR machinery that regulates the parental expression of these genes may also influence their tissue-specific expression pattern.
Conclusion
Here we report an extensive compendium of gene expression of the protein-encoding transcriptomes of the mouse and humans. Further augmentation by additional samples, including region-specific dissections using laser capture microdissection or even cell type-specific gene expression, will undoubtedly increase the utility of these resources. We have investigated this data set for global signatures in tissue-specific gene regulation, expression characteristics of de novo predicted transcripts, and chromosomal RCTs. The identification of several known imprinted loci in our tissue-specific RCT list suggests that these regulatory mechanisms that direct tissue- or parental allele-specific expression may be intertwined. Consistent with this observation, we were able to identify two previously undescribed transcripts that were imprinted on mouse chromosome 12 based on the observation that they share a tissue-specific expression pattern with their neighbors.
With the sequencing phase of the human and mouse genome projects nearly complete, and with the rapid progress in the sequencing of other mammalian genomes, we are now poised to develop and exploit a variety of methods to ascertain the function of the thousands of recently described genes. In this regard, the genome-scale RNA expression data described herein provide a framework for the functional annotation process. By making the underlying data available on our web site (http://symatlas.gnf.org) and through the Gene Expression Omnibus (www.ncbi.nih.gov/geo), we anticipate that this study will aid researchers throughout the global research community to reap the harvests of the human and mouse genome projects.
Supplementary Material
Acknowledgments
We thank the following individuals for providing human RNA samples: Gino Van Heeke, Novartis (bronchial epithelial cells); Graeme Bilbe, Novartis (fetal thyroid); Clifford Shults, University of California at San Diego (whole blood); Bill Sugden, University of Wisconsin, Madison (721 B-lymphoblasts); Joseph D Buxbaum, Mt. Sinai School of Medicine, New York (prefrontal cortex). We also thank Ines Hoffmann and Satchin Panda for preparation of mouse embryonic samples and Peter Dimitrov, Christian Zmasek, and Michael Heuer for technical expertise. This work was supported by the Novartis Research Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RCT, regions of correlated transcription; AUC, area under the curve; LCR, locus control regions.
References
- 1.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- 2.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 3.Su, A. I., Welsh, J. B., Sapinoso, L. M., Kern, S. G., Dimitrov, P., Lapp, H., Schultz, P. G., Powell, S. M., Moskaluk, C. A., Frierson, H. F., Jr., et al. (2001) Cancer Res. 61, 7388-7393. [PubMed] [Google Scholar]
- 4.Aza-Blanc, P., Cooper, C. L., Wagner, K., Batalov, S., Deveraux, Q. L. & Cooke, M. P. (2003) Mol. Cell 12, 627-637. [DOI] [PubMed] [Google Scholar]
- 5.Chanda, S. K., White, S., Orth, A. P., Reisdorph, R., Miraglia, L., Thomas, R. S., DeJesus, P., Mason, D. E., Huang, Q., Vega, R., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 12153-12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su, A. I., Cooke, M. P., Ching, K. A., Hakak, Y., Walker, J. R., Wiltshire, T., Orth, A. P., Vega, R. G., Sapinoso, L. M., Moqrich, A., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pruitt, K. D. & Maglott, D. R. (2001) Nucleic Acids Res. 29, 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerlavage, A., Bonazzi, V., di Tommaso, M., Lawrence, C., Li, P., Mayberry, F., Mural, R., Nodell, M., Yandell, M., Zhang, J., et al. (2002) Nucleic Acids Res. 30, 129-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. (2002) Nature 420, 563-573.12466851 [Google Scholar]
- 10.Kent, W. J. (2002) Genome Res. 12, 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Florea, L., Hartzell, G., Zhang, Z., Rubin, G. M. & Miller, W. (1998) Genome Res. 8, 967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanapin, A., Batalov, S., Davis, M. J., Gough, J., Grimmond, S., Kawaji, H., Magrane, M., Matsuda, H., Schonbach, C., Teasdale, R. D., et al. (2003) Genome Res. 13, 1335-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lockhart, D. J., Dong, H., Byrne, M. C., Follettie, M. T., Gallo, M. V., Chee, M. S., Mittmann, M., Wang, C., Kobayashi, M., Horton, H., et al. (1996) Nat. Biotechnol. 14, 1675-1680. [DOI] [PubMed] [Google Scholar]
- 15.Hubbell, E., Liu, W. M. & Mei, R. (2002) Bioinformatics 18, 1585-1592. [DOI] [PubMed] [Google Scholar]
- 16.Edgar, R., Domrachev, M. & Lash, A. E. (2002) Nucleic Acids Res. 30, 207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent, W. J., Baertsch, R., Hinrichs, A., Miller, W. & Haussler, D. (2003) Proc. Natl. Acad. Sci. USA 100, 11484-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishop, J. O., Morton, J. G., Rosbash, M. & Richardson, M. (1974) Nature 250, 199-204. [DOI] [PubMed] [Google Scholar]
- 19.Hastie, N. D. & Bishop, J. O. (1976) Cell 9, 761-774. [DOI] [PubMed] [Google Scholar]
- 20.Hogenesch, J. B., Ching, K. A., Batalov, S., Su, A. I., Walker, J. R., Zhou, Y., Kay, S. A., Schultz, P. G. & Cooke, M. P. (2001) Cell 106, 413-415. [DOI] [PubMed] [Google Scholar]
- 21.Harmer, S. L., Hogenesch, J. B., Straume, M., Chang, H. S., Han, B., Zhu, T., Wang, X., Kreps, J. A. & Kay, S. A. (2000) Science 290, 2110-2113. [DOI] [PubMed] [Google Scholar]
- 22.DeRisi, J. L., Iyer, V. R. & Brown, P. O. (1997) Science 278, 680-686. [DOI] [PubMed] [Google Scholar]
- 23.Li, Q., Peterson, K. R., Fang, X. & Stamatoyannopoulos, G. (2002) Blood 100, 3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spellman, P. T. & Rubin, G. M. (2002) J. Biol. 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caron, H., van Schaik, B., van der Mee, M., Baas, F., Riggins, G., van Sluis, P., Hermus, M. C., van Asperen, R., Boon, K., Voute, P. A., et al. (2001) Science 291, 1289-1292. [DOI] [PubMed] [Google Scholar]
- 26.Roy, P. J., Stuart, J. M., Lund, J. & Kim, S. K. (2002) Nature 418, 975-979. [DOI] [PubMed] [Google Scholar]
- 27.Cohen, B. A., Mitra, R. D., Hughes, J. D. & Church, G. M. (2000) Nat. Genet. 26, 183-186. [DOI] [PubMed] [Google Scholar]
- 28.Bell, A. C. & Felsenfeld, G. (2000) Nature 405, 482-485. [DOI] [PubMed] [Google Scholar]
- 29.Hark, A. T., Schoenherr, C. J., Katz, D. J., Ingram, R. S., Levorse, J. M. & Tilghman, S. M. (2000) Nature 405, 486-489. [DOI] [PubMed] [Google Scholar]
- 30.Thorvaldsen, J. L., Duran, K. L. & Bartolomei, M. S. (1998) Genes Dev. 12, 3693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, J., Lu, X. & Stubbs, L. (1999) Hum. Mol. Genet. 8, 847-854. [DOI] [PubMed] [Google Scholar]
- 32.Georges, M., Charlier, C. & Cockett, N. (2003) Trends Genet. 19, 248-252. [DOI] [PubMed] [Google Scholar]
- 33.Hatada, I., Morita, S., Obata, Y., Sotomaru, Y., Shimoda, M. & Kono, T. (2001) J. Biochem. 130, 187-190. [DOI] [PubMed] [Google Scholar]
- 34.Cavaille, J., Seitz, H., Paulsen, M., Ferguson-Smith, A. C. & Bachellerie, J. P. (2002) Hum. Mol. Genet. 11, 1527-1538. [DOI] [PubMed] [Google Scholar]
- 35.Yevtodiyenko, A., Carr, M. S., Patel, N. & Schmidt, J. V. (2002) Mamm. Genome 13, 633-638. [DOI] [PubMed] [Google Scholar]
- 36.Seitz, H., Youngson, N., Lin, S. P., Dalbert, S., Paulsen, M., Bachellerie, J. P., Ferguson-Smith, A. C. & Cavaille, J. (2003) Nat. Genet. 34, 261-262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.