Abstract
Previous investigations have shown that ≈35% of the 90 tumors analyzed to date contain mutations within the DNA polymeraseβ (pol β) gene. The existence of pol β mutations in a substantial fraction of human tumors studied suggests a link between DNA pol β and cancer. A DNA pol β variant, in which Lys-289 has been altered to Met, was identified previously in a colorectal carcinoma. The K289M protein was expressed in mouse L cells containing the λ cII mutational target. The λ DNA was packaged and used to infect bacterial cells to obtain the spontaneous mutation frequency. We found that expression of K289M in the mouse cells resulted in a 2.5-fold increase in the mutation frequency. What was most interesting was that expression of K289M in these cells resulted in a 16-fold increase in the frequency of C to G or G to C base substitutions at a specific site within the cII target. By using this cII target sequence, kinetic analysis of the purified K289M protein revealed that it was able to misincorporate dCTP opposite template C and dGTP opposite template G with significantly higher efficiency than the wild-type pol β protein. We provide evidence that misincorporation of nucleotides by K289M results from altered positioning of the DNA within the active site of the enzyme. Our data are consistent with the interpretation that misincorporation of nucleotides resulting from altered DNA positioning by the K289M protein has the potential to result in tumorigenesis or neoplastic progression.
Mutations in the gene encoding DNA polymerase β (pol β) have been identified in human colorectal, prostate, lung, and breast carcinomas and mouse lymphomas (1–5). Thus far, only 90 tumors have been analyzed for mutations within the pol β coding sequence, and mutations are present in 35% of these tumors. The pol β tumor-associated mutations are found only in the tumor, and not in normal tissue from the same patient, implying that they represent sporadic mutations underlying neoplastic disease. Furthermore, the mutations identified in these tumors are not present in the pol β gene of 124 normal individuals (6). Also of interest is that pol β is located within the proximal region of the short arm of chromosome 8 (p12-p11), a region that is frequently lost in a variety of human tumors, including colorectal and prostate carcinomas (7). These studies suggest a link between mutations within the pol β gene and carcinogenesis. Another piece of evidence that is consistent with a role for pol β in cancer is its interaction with the tumor suppressor protein p53 (8, 9). The p53 protein stabilizes pol β at an abasic site. An alteration of the p53-pol β interaction could result in less efficient DNA repair, which may contribute to the development of neoplastic disease. Most interestingly, a pol β mutant with an 87-bp deletion, which has been found in primary colorectal, lung, and breast adenocarcinomas (3, 5, 10), is dominant to the WT enzyme and disrupts its base excision repair (BER) activity if expressed in human cell lines (11, 12). It is quite possible that other pol β mutants found in tumor samples have a similar biological impact.
BER is an important eukaryotic DNA repair pathway that functions to remove DNA damage, including abasic sites, oxidative lesions, and methylated bases (13, 14). It is estimated that BER repairs as many as 10,000 DNA lesions per cell per day (15). During BER, base damage is excised by specific DNA glycosylases, followed by incision of the DNA backbone by an apurinic/apyrimidinic endonuclease. Pol β then excises the deoxyribose phosphate and fills in the resulting single nucleotide gap (16, 17). Pol β also functions in long-patch BER where it performs strand displacement synthesis (18). Pol β lacks 3′ to 5′ proofreading activity, which contributes to its low fidelity. Some pol β mutants can significantly increase the mutation frequency by committing errors during DNA synthesis, as documented by our laboratory (19–21), and those of others, for example (22, 23). Errors committed by pol β in cells may initiate or promote tumorigenesis.
We chose the colorectal cancer-associated pol β K289M variant for our initial study because Lys-289 is on helix N of pol β, which also contains Arg-283 and Met-282. We and others have shown that alteration of Arg-283 and Met-282 results in enzymes that synthesize DNA inaccurately (22, 24). The K289M pol β alteration was detected in the cDNA of one of six human colorectal carcinomas studied by Banerjee and colleagues (3). The WT pol β sequence was also detected in this tumor, suggesting that both WT and K289M are expressed in the colorectal carcinoma or that the tumor tissue was contaminated with normal mucosa. The K289M alteration was not detected in normal mucosa from the same patient and has not been detected as a polymorphism in normal individuals (6). This finding suggests that K289M could have a role in carcinogenesis. In this study, we provide evidence that the K289M variant of pol β induces mutations within interrupted runs of nucleotides and that K289M misincorporates nucleotides by altered positioning of the DNA within its active site. This study suggests that K289M could introduce mutations into the genome that underlie the development or progression of neoplastic disease.
Materials and Methods
Reagents. Deoxynucleoside triphosphates, adenine triphosphate, and [γ-32P]ATP (>5,000 Ci/mmol, 10 mCi/ml) (1 Ci = 37 GBq) were purchased from New England Biolabs, Sigma and Amersham Pharmacia, respectively. The oligonucleotides used in the preparation of the DNA substrates were synthesized at the Keck Molecular Biology Center at Yale University.
Strains, Cell Lines, and Cell Culture. For cloning of pol β, we used Escherichia coli DH5αMCR with the genotype mcrA (mrr-hsdRMS-mcrBC)φ80ΔlacZ(M15) (lacZYA-argF)U169 deoR recA1 endA1 phoA supE44 thi-1 gyrA96 relA1. For protein expression, we used BL21(DE3) with the genotype F-ompT hsdSb (rb- mb-) gal dcm.
Mouse LN12 cells were a gift from P. Glazer (25). There are ≈100 copies of the λ genome incorporated within LN12 cell genome. LN12 cells were maintained in DMEM (Invitrogen) with 10% calf serum donor (Invitrogen), 400 μg/ml G418 (Invitrogen), and Pen/Strep (Invitrogen) at 37°C in a humidified 5% CO2 incubator.
Mouse embryonic fibroblast cells 36.3 and 38Δ4 were gifts from S. Wilson (26). 36.3 is the WT cell line; 38Δ4 is a pol β knockout cell line from a matched littermate. They were maintained in DMEM with Glutamax I (Invitrogen), 10% FBS (Invitrogen), Pen/Strep (Invitrogen), and 80 μg/ml Hygromycin B (Sigma) at 34°C in a humidified 10% CO2 incubator.
Cloning and Transfection of K289M. WT pol β was cloned into the vector pcDNA3/TO/myc-His A (Invitrogen) by using standard methods. When tetracycline (Tet) is added to the medium, the pol β protein is expressed, and its expression is repressed in the absence of Tet. Transfection into the LN12 cells was carried out with FuGENE 6 reagent (Roche) according to the manufacturer's directions. Individual clones were isolated and expanded to generate stable cell lines.
Expression Analysis. Individual cell clones were seeded at a density of 6 × 105 into duplicate wells of six-well plates. When cells grew to ≈80% confluence, one of the duplicates was induced with 1 μg/ml Tet. Forty-eight hours later, both wells were washed two times with 1× PBS, and 200 μl of 1× SDS loading buffer that had been warmed to 80°C was added into the wells. After scraping the wells, cell lysates were pipetted into Eppendorf tubes and boiled for 10 min, and 16 μl of extract was resolved in a 10% SDS/PAGE gel. Protein transfer and Western blotting were performed as described (27).
λ cII Mutagenesis Assay. High molecular weight genomic DNA was purified as described (28). Packaging extracts were prepared as described (25), except that a new lysogen, NM759: E. coli K12 recA56Δ(mcrA) e14° Δ (mrr-hsd-mcr)(λimm434 cIts b2 red3 Dam15 Sam7)/λ was used instead of BHB2690 for the preparation of the sonication extract (29). Phage packaging was carried out as described (30). cII mutants were obtained from three independent packaging reactions. The number of plaques obtained from packaged DNA from noninduced cells and induced cells was 534,000 and 388,000, respectively. To identify the types of mutations produced by cells expressing K289M, each mutant was isolated, and the cII gene was amplified by PCR with the following primers: 5′-ACC ACA CCT ATG GTG TAT GCA-3′ and 5′-GTC ATA ATG ACT CCT GTT GA-3′. After DNA purification, the DNA sequences of these mutants were determined by the Keck Center for Biotechnology at Yale School of Medicine.
Protein Purification. WT and K289M pol β were purified as described (31).
DNA Substrates. The DNA substrates used in the biochemical assays described below are displayed in Table 1. The primer oligonucleotides were gel-purified as described (31) and were radiolabeled at the 5′ end by standard methods by using T4 polynucleotide kinase (New England BioLabs) and [γ-32P]ATP. The oligonucleotides 45A-22-22, CII45-CIIU-CIID, and CII45T-CIIU-CIIDA were annealed as described (20). The primer:template molar ratio for (CL-CP-CG) was 1.2:1:1.8 and 1.2:1:1.5 for (GL-GP-GG) in 50 mM Tris, pH 8.0, 250 mM NaCl. The mixture was incubated sequentially at 95°C (5 min), slow cool to 45°C (30 min), slow cool to 37°C (30min), and immediately transferred to ice. Annealing of primer was confirmed on an 18% polyacrylamide native gel followed by autoradiography as described (32).
Table 1. Primer-templates for burst, misincorporation, and gel mobility-shift assays.
| Oligonucleotides | Sequence |
|---|---|
| 45A-22—22 | 5′ GCCTCGCAGCCGTCCAACCAAC CAACCTCGATCCAATGCCGTCC |
| 3′ CGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGG | |
| CL-CP-CG | 5′ GAACTCCATCTGGATTT TTCAGAACGCTCGGTTGC |
| 3′ CTTGAGGTAGACCTAAACAAGTCTTGCGAGCCAACG | |
| GL-GP-GG | 5′ GACACCGAGCGTTCAGAA AAATCCAGATGGAGTTC |
| 3′ CGTTGGCTCGCAAGACTTGTTTAGGTCTACCTCAAG | |
| CII45-CIIU-CIID | 5′ TTGCGACTTATCAACGCCCACA CTTCCGCTGTCTTCTCAGTTCC |
| AACGCTGAATAGTTGCGGGTGTCGAAGGCGACAGAAGAGTCAAGG | |
| CII45T-CIIU-CIIDA | 5′ TTGCGACTTATCAACGCCCACA ATTCCGCTGTCTTCTCAGTTCC |
| AACGCTGAATAGTTGCGGGTGTCTAAGGCGACAGAAGAGTCAAGG | |
| BER substrate | 5′ TACCGCGGCCGGCCGAUCAAGCTTATTGGGTAC |
| ATGGCGCCGGCCGGCTAGTTCGAATAACCCATG |
The templating base is underlined.
Single-Turnover Misincorporation Assays. We determined the equilibrium dissociation constant for dNTP binding, Kd, and the maximum rate of polymerization, kpol, for correct and incorrect dNTPs for each enzyme as described (20). Single turnover conditions were determined empirically to be a ratio of enzyme to DNA of 15:1 for K289M. For correct incorporation reactions, substrate concentrations were typically 0–200 μM for WT or 0–500 μM for K289M, and reaction times were 0–5 s for WT and 0–20 s for K289M.
The kinetics of misincorporation were determined manually under the above single-turnover conditions as described (20). For incorrect incorporation, substrate concentrations were typically 0–2 mM, and reaction times were 0–3,600 s. The product quantification and data analysis were performed as described (20).
DNA Binding Assay. The apparent dissociation constant KD (DNA) was measured by using a gel mobility shift assay as described (19), and single nucleotide gapped (CL-CP-CG) DNA substrate. Fifteen protein concentrations ranging from 1 μM to 0.25 nM were incubated with 0.1 nM DNA that had been radiolabeled in buffer as described (20).
BER Assay. Whole cell extract (WCE) was prepared as described (33). Purification, 5′ end labeling, and annealing of oligonucleotides to create the BER substrate were performed as described (21). The BER assay was performed as described (34). After being resolved in a 20% denaturing gel, the products were exposed and quantified by using a Molecular Dynamics Storm 840 PhosphorImager. In this assay, repaired substrates are 17 nt in length if pol β has extended the primer by a single nucleotide and are 33 nt in length if the substrate has been extended and ligated. Unrepaired substrates are 16 nt in length. The presence of bands that are between 17 and 33 nt in length, or smaller than 16 nt, are thought to result from the presence of nuclease activities in the WCE.
Results
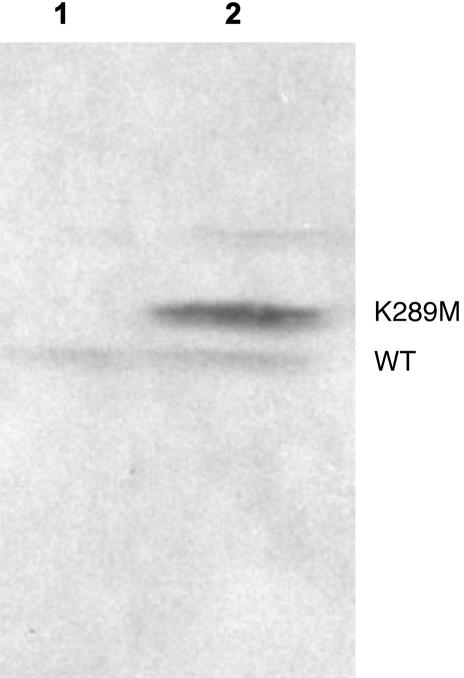
Expression of K289M Is Inducible in LN12 Cells. To express K289M in mouse cells, we cloned it into the pcDNA3/TO/myc-His A inducible vector and transfected it into mouse LN12 cells. After transfection, each clone was isolated, expanded, and screened by Western blotting to identify clones that were inducible for the expression of K289M, as shown in Fig. 1. In the noninduced control, only the WT protein (lane 1) could be detected. In cells induced to express K289M (lane 2), this protein was detected at a slightly increased molecular weight, due to the presence of a myc tag at the C terminus of K289M. Quantification of the bands indicates that K289M is expressed at ≈2-fold over endogenous WT pol β.
Fig. 1.
The K289M protein is induced by treatment of the cells with Tet. Primary antibody was pol β monoclonal antibody. Lane 1, cell extract without Tet (noninduced); lane 2, cell extract with Tet (induced) for 2 days.
Expression of K289M Increases in the Spontaneous Mutation Frequency. To determine whether expression of K289M results in mutagenesis in the LN12 cells, genomic DNA was extracted from the cells, packaged, and used to infect bacterial host cells. The spontaneous mutation frequency in cells induced to express K289M was 2.3 × 10-4 which is 2.5-fold higher than the frequency of 9.0 × 10-5, obtained from cells not expressing K289M.
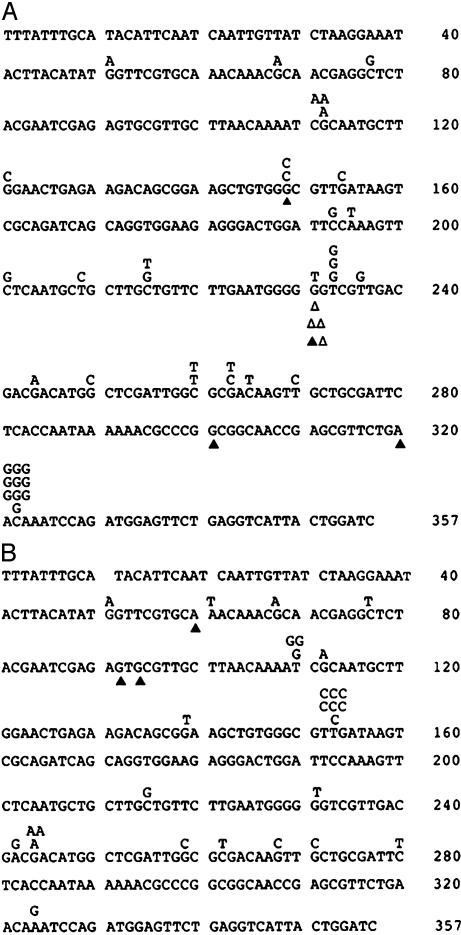
The Mutation Spectrum of K289M Is Different from the WT. To determine the types of mutations induced by K289M, we sequenced 47 of the cII mutants from cells expressing K289M and 42 of the mutants from cells expressing WT pol β. For WT, 11 of the 42 mutants sequenced did not have a mutation within the cII gene, suggesting that another λ gene had been altered in these mutants. As shown in Fig. 2, there are two putative hotspots in the mutation spectrum resulting from expression of the K289M protein, which are absent in the mutation spectrum obtained without the expression of K289M. The first is at position 232, which consists of four deletions, one insertion, and one G to T transversion. The second putative hotspot, at position 322, consists of 10 C:G to G:C transversions, which have rarely been observed in pol β mutation spectra (35, 36). The frequency of the C to G transversions at position 322 in the K289M-expressing cells is 4.8 × 10-5, which is 16 times higher than the C to G transversion frequency (2.9 × 10-6) that we observed in the WT (noninduced) spectrum. We used the Adams and Skopek algorithm (37) to determine whether the spectra were significantly different from each other and obtained P < 0.00059, strongly suggesting that these spectra were different. We then removed the mutations at position 322 in both spectra and obtained P < 0.01, which suggests that this putative hotspot contributes much to the significant differences between the spectra.
Fig. 2.
Mutation spectrum of K289M. (A) Induced. (B) Noninduced. Single letter indicates base substitution. Shown are symbols for deletion (▵) and insertion (▴).
The K289M Mutant Is a Misincorporation Mutator at Position 322 of the cII Sequence. The C to G or G to C transversions at position 322 in the cII gene could result from misincorporation of dCTP opposite template C or dGTP opposite template G by K289M during BER in the LN12 cells. To test this hypothesis, we purified the K289M protein and determined that the K289M protein was active in a pre-steady-state burst experiment (data not shown). Next, we used a single turnover kinetic assay to measure the rates of misincorporation of dCTP opposite C and dGTP opposite G. For these experiments, we used the DNA primer-templates CL-CP-CG and GL-GP-GG, which correspond to the exact DNA sequence and complementary sequence, respectively, of the putative hotspot position 322 of cII gene.
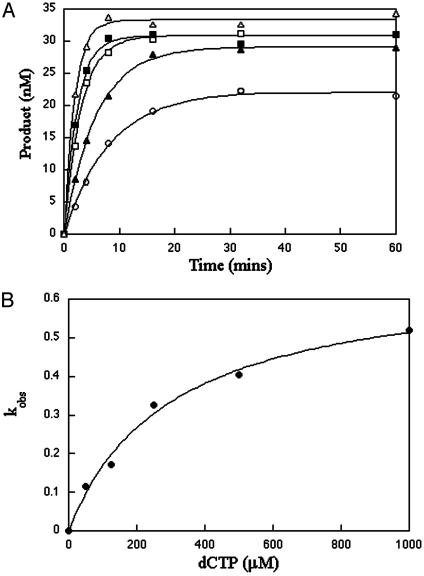
The Kd and kpol values were determined by measuring the rate of product formation at varying concentrations of dNTP. Fig. 3A illustrates an example of dCTP misincorporation opposite C for WT at five different concentrations of dCTP at 37°C. By fitting each set of data to the single exponential rate equation, the kobs was determined for each dCTP substrate concentration. These kobs values were plotted against the dCTP concentrations to yield the Kd and kpol parameters for WT (Fig. 3B) and K289M (not shown).
Fig. 3.
Single turnover experiments of misincorporation opposite cytosine. (A) Misincorporation of dCTP opposite C for WT at 37°C. A preincubated solution containing enzyme (750 nM) and gapped DNA (50 nM) was mixed with 10 mM MgCl2 and 50 mM (○), 125 mM (▴), 250 mM (□), 500 mM (▪), and 1,000 mM (▵) dCTP. The reactions were quenched and monitored as described in ref. 20. Data were fit to the single-exponential equation to obtain kobs.(B) Secondary kinetic plot of kobs against dCTP concentration for WT (•). The solid line represents the best fit of the data to the hyperbolic equation. Values Kd and kpol are listed in Table 3.
The fidelity values and rates obtained for WT and K289M are shown in Table 2. A 26-fold loss in fidelity was observed for K289M relative to WT for misincorporation of dCTP opposite C. This loss is largely due to a reduced ability to discriminate between dGTP vs. dCTP at the level of kpol, which is thought to reflect predominantly the rate of phosphodiester bond formation. For WT, the maximum rate of polymerization of the correct dNTP is 3,110 times faster than the incorrect one whereas K289M displays only a 34-fold difference, resulting in an ≈90-fold (3,110/34) loss in the discrimination at the level of kpol.
Table 2. The K289M mutant misincorporates dCTP opposite template C and cGTP opposite template G in the λcII target sequence.
| kpol, s—1 | Kd, μM | kpol(c)/kpol(i)* | Kd(i)†/Kd(c) | kpol/Kd, M—1s—1 | F (× 102)‡ | |
|---|---|---|---|---|---|---|
| β-WT | ||||||
| C:G§ | 31.1 ± 1.9 | 11.50 ± 3.0 | 270 × 104 | |||
| C:C¶ | 0.0100 ± 0.0007 | 299.2 ± 50.4 | 3110 | 26 | 33 | 820 |
| G:C∥ | 73.3 ± 0.2 | 80.3 ± 0.7 | 91 × 104 | |||
| G:G** | 0.0200 ± 0.0023 | 324.7 ± 39.6 | 3665 | 4 | 68 | 134 |
| K289M | ||||||
| C:G§ | 0.375 ± 0.025 | 3.3 ± 1.3 | 11 × 104 | |||
| C:C¶ | 0.0110 ± 0.0009 | 289.7 ± 48.5 | 34 | 88 | 35 | 31 |
| G:C∥ | 2.54 ± 0.19 | 353.6 ± 37.1 | 0.7 × 104 | |||
| G:G** | 0.0050 ± 0.0002 | 39.1 ± 7.2 | 508 | 0.1 | 128 | 0.55 |
The kpol for correct (c) divided by incorrect (i)
The Kd for incorrect (i) dNTP divided by correct (c)
Fidelity (F) was calculated as described in ref. 11
Kinetic constants for incorporation of dGTP opposite C
Kinetic parameters for misincorporation of dCTP opposite template C
Kinetic constants for incorporation of dCTP opposite G
Kinetic parameters for misincorporation of dGTP opposite template G. The DNA substrates were CL-CP-CG and GL-GP-GG, for incorporation opposite templates C and G, respectively
Similar findings were observed in the case of misincorporation of dGTP opposite template G as shown in Table 2. Here, the fidelity of K289M (55) was ≈240-fold lower than WT (13,400), with the primary defect being the inability to discriminate at the level of Kd. The Kd is the equilibrium dissociation constant and reflects the initial interaction of the enzyme with the dNTP substrate. Thus, the hotspot present at position 322 could result from the ability of K289M to misincorporate nucleotides within the specific sequence context present at that site, which would be consistent with our in vivo mutation spectrum. Alternatively, K289M could be a general misincorporation mutator polymerase.
K289M Misincorporates dCTP Opposite Template C at Another Site Within the cII Gene. To determine whether K289M is a general misincorporation mutator in vitro, we ascertained whether it is able to misincorporate dCTP opposite template C at position 143 within the cII gene, a site that was not mutated in cells induced and not induced to express K289M. Careful perusal of the template sequence shows that the base that is 5′ to the templating C is a G. As shown in Table 3, for the sequence context 5′GC, the fidelity of K289M (3, 100) for misincorporation of dCTP opposite template C at position 143 in the cII gene is ≈10-fold less than that of WT (33,000) in the single turnover kinetic assays. The kinetic basis for this loss of fidelity is during ground state binding. Thus, it seems that K289M is able to misincorporate dCTP opposite C at sites within the cII gene other than position 322, the in vivo hotspot. In Table 3, we also show that WT has a 17-fold lower fidelity (1,900) for misincorporation of dATP opposite C than for misincorporation of dCTP opposite C (33,000), due to its reduced discrimination during both the binding and chemical steps. K289M also misincorporates dATP opposite C, but its fidelity is only ≈2-fold lower than WT, and ≈3-fold lower than its fidelity for misincorporation of dCTP opposite C at this site. This finding suggests that K289M retains the ability to misincorporate dATP opposite C, but that it has acquired the ability to insert dCTP opposite C, either by direct misincorporation or misalignment.
Table 3. K289M misincorporates nucleotides by repositioning of the DNA.
| Sequence context* | kpol, s—1 | Kd, μM | kpol(c)/kpol(i)† | Kd(i)/Kd(c)‡, M—1s—1 | kpol/Kd | F (×102)§ | |
|---|---|---|---|---|---|---|---|
| β-WT | |||||||
| 5′GC¶ | C:G∥ | 10.72 ± 0.45 | 23.5 ± 3.5 | 45.6 × 104 | |||
| 5′GC | C:C** | 0.0045 ± 0.0004 | 328.8 ± 78.0 | 2382 | 14 | 13.7 | 333 |
| 5′GC | C:A†† | 0.0340 ± 0.0016 | 140.6 ± 25.3 | 315 | 6 | 242 | 19 |
| 5′TC‡‡ | C:G∥ | 12.11 ± 0.89 | 10.1 ± 2.9 | 119.9 × 104 | |||
| 5′TC | C:C** | 0.00472 ± 0.00038 | 148.6 ± 51.5 | 2,565 | 15 | 31.76 | 380 |
| 5′TC | C:A†† | 0.023 ± 0.0003 | 384.6 ± 12.7 | 527 | 38 | 59.8 | 200 |
| K289M | |||||||
| 5′GC¶ | C:G∥ | 5.53 ± 0.17 | 71.0 ± 7.3 | 7.76 × 104 | |||
| 5′GC | C:C** | 0.00126 ± 0.00003 | 49.51 ± 10.2 | 4,389 | 0.69 | 25.5 | 31 |
| 5′GC | C:A†† | 0.009 ± 0.0004 | 106.9 ± 19.0 | 155 | 14 | 84.0 | 9 |
| 5′TC‡‡ | C:G∥ | 1.02 ± 0.03 | 12.6 ± 2.7 | 8.0 × 104 | |||
| 5′TC | C:C** | 0.00375 ± 0.00015 | 492.3 ± 46.4 | 272 | 39 | 7.6 | 110 |
| 5′TC | C:A†† | 0.006 ± 0.0006 | 254 ± 66 | 170 | 20 | 23.6 | 33 |
The sequence context at position 143. The templating base is underlined and the base that is 5′ to the templating base is shown
The kpol for correct (c) divided by incorrect (i)
The Kd for incorrect (i) dNTP divided by correct (c)
Fidelity (F) was calculated as described in ref. 11
The templating base is underlined. The DNA substrate was CII45-CIIU-CIID
Kinetic constants for incorporation of dGTP opposite template C
Kinetic parameters for misincorporation of dCTP opposite template C
Kinetic parameters for misincorporation of dATP opposite template C
The templating base is underlined. The DNA substrate was CII45T-CIIU-CIIDA
Because the base 5′ to the templating C is a G, we surmised that K289M could be repositioning the DNA within its active site such that original templating C could assume a somewhat extrahelical position, resulting in the 5′ G becoming the templating base. To test this misalignment hypothesis, we altered the 5′ base of the template from G to T, and, using the CII45T-CIIU-CIIDA DNA substrate, asked whether K289M could misincorporate dCTP opposite template C if the base 5′ to this C was a T. As shown in Table 3 for the 5′TC sequence context, K289M has a fidelity (11,000) that is only 3-fold less than that of WT (38,000) when T is 5′ to the templating C. This result demonstrates that the fidelity of K289M improves when the base 5′ to the template C is a T. Thus, our data are consistent with the interpretation that K289M repositions the DNA within its active site during catalysis.
As a final test of the repositioning hypothesis, we asked whether K289M could misincorporate dATP opposite C when the base 5′ to the C was a T, using the CII45T-CIIU-CIIDA DNA substrate. As shown in Table 3 for the 5′TC sequence context, K289M had an ≈6-fold lower fidelity (3,300) than WT (20,000) for misincorporation of dATP opposite C when the 5′base was a T, which is consistent with the repositioning hypothesis.
The Affinity of K289M for Gapped DNA Is Decreased Compared with pol β-WT. One mechanism to explain the apparent altered positioning of the DNA within the active site of K289M is that K289M could have a lower affinity for DNA. Therefore, a gel mobility-shift assay was conducted to estimate the affinity of pol β for single base gapped DNA (CL-CP-CG). The apparent Kd for WT and K289M proteins, respectively, are 0.077 ± 0.012 nM and 1.2 ± 0.24 nM. These results demonstrate that the K289M mutant had a 16-fold lower apparent affinity for 1 base gapped DNA (CL-CP-CG) than WT.
K289M Functions in BER. To determine whether K289M can participate in BER, we constructed a uracil-containing DNA substrate, treated it with uracil DNA glycosylase and apurinic/apyrimidinic endonuclease to generate an abasic site, and incubated it with WCE prepared from pol β+/+ or pol βΔ/Δ mouse embryonic fibroblast cell lines, in the presence or absence of pol β-WT or K289M as indicated in Fig. 4, which is published as supporting information on the PNAS web site. The BER activities of these reactions were calculated as the relative ratio of the intensity of the 17-mer plus the intensity of bands from 18 to 33 nucleotides in length divided by the intensity of the total bands. As shown in Fig. 4, lane 3, the WT WCE was able to repair 81% of the substrate within 20 min whereas the WCE prepared from cells deleted of pol β (lane 4) could repair only 14% of the substrate, probably through DNA repair pathways other than short-patch BER. When added into the WCE prepared from cells deleted of pol β, both the WT and K289M repaired at least 30% of the substrate in as little as 2 s, and at least 85% in 2 min.
Discussion
The frequent detection of somatic pol β mutations in human cancers suggests that alteration of the pol β gene may be linked to cancer. In the current study, our goal was to characterize the biological and biochemical phenotype of the K289M pol β variant. Here, we demonstrate that expression of the K289M variant in mouse LN12 cells resulted in the induction of mutations within an interrupted run of like nucleotides. We also provide evidence that the K289M protein itself has lower fidelity than WT pol β. Our data suggest that K289M induces mutations during BER in the cells, even in the presence of WT pol β, and are consistent with the interpretation that the K289M Pol β protein plays a role in the onset or progression of neoplasia.
K289M Misincorporates Nucleotides. To determine the specificity of mutations induced by K289M, we compared the types of cII mutants we obtained from cells induced to express K289M to cells that expressed only WT pol β. One site that was mutated at a high frequency in cells expressing K289M was position 322 of the cII gene. At this position, we observed C to G transversion mutations at a 16-fold higher frequency for K289M than WT pol β. This finding suggested to us that the K289M pol β variant was directly responsible for misincorporation at this site, perhaps during BER.
By using purified proteins, we showed (Table 2) that K289M had 26-fold and 243-fold less fidelity for incorporation of dCTP opposite C and dGTP opposite G, respectively, than the WT protein. Therefore, we suggest that the pol β cancer-associated K289M mutant is directly responsible for the mutations observed within the AACAAA (TTTGTT) sequence context in the cII gene.
K289M Misincorporation Is Due to Altered Positioning of the Template. Our results (Table 3) showed that the fidelity of K289M is also significantly decreased for insertion of dCTP opposite C at position 143 of the cII gene when compared with WT. This finding suggested that K289M was generally able to misincorporate dCTP opposite C. However, we noticed that the base 5′ to the templating C was a G. This finding indicated that C was assuming an altered, perhaps extrahelical, position within the active site of K289M, permitting G to become the templating base and leading to the correct incorporation of dCTP opposite G, and our results suggest this to be the case. Previous studies have suggested that WT pol β is prone to misalignment-mediated misincorporation (38, 39), and it seems that, in the sequence contexts examined in this study, K289M exhibits a much more robust ability to misalign the template. These data are consistent with the interpretation that the DNA within the active site of K289M is more prone to assuming an altered position than it is in the active site of WT, ultimately resulting in misincorporation by K289M. Consistent with the misalignment-mediated mechanism, we found that K289M has a lower affinity for single nucleotide gapped DNA than WT, permitting us to conclude that Lys-289 is important for the interaction of pol β with the DNA.
Altered positioning of the DNA may also explain the mechanism of misincorporation at position 322. However, the only way the mechanism operating at position 143 of the cII gene could operate at position 322 would be if two of the bases within the template assumed an extrahelical-type position, resulting in incorporation of C opposite the G at position 319, which is not likely. We did not observe any evidence for this slippage in our kinetic assays (data not shown). A likely mechanism for the mutations at position 322 is that K289M does not hold tightly to the DNA, permitting it to assume an altered structure within the active site that is permissive for misincorporation of dCTP.
K289M Misincorporates Nucleotides During BER. Our in vivo results show that, when we express K289M in LN12 cells, we observe a mutational hotspot at position 322. However, our in vitro results demonstrate that K289M is apparently able to produce misalignment-mediated errors at positions within the cII gene that were not sites of mutation in the cells. A likely scenario to explain these results is that, during BER in the cells, K289M misincorporates at sites that are prone to misalignment. The resulting nick may not be ligated (40) or may not be corrected by APE-1 (41), mismatch repair, or another activity that has not yet been described. Thus, the mutation spectrum we observe results from misalignment-mediated misincorporation by K289M and repair of these mistakes. We suggest that the errors inserted by K289M at position 322 are not corrected or repaired. The lack of correction at position 322 may be due to the nature of the sequence context surrounding position 322 of the cII gene.
K289M and Colon Carcinoma. Limited analysis of genes that are altered in colon cancer shows that the AACAAA sequence found at position 322 is also present at four sites within the adenomatous polyposis coli (APC) gene, which is known to be mutated in a majority of sporadic colon carcinomas (42). Inspection of the p53 gene, mutation of which is also associated with colon cancer, revealed no sequences with the AACAAA motif. In addition, the most common hotspot for mutation of the APC gene in human colon carcinomas is at the ATAAAAGAAAAGATT sequence, which may have similarities to position 322 of the cII gene in that it consists of an interrupted run of A residues (43). Muniappan and Thilly (43) found that copying this sequence with WT pol β resulted in deletion of AAAAG or AAAGA, which is a mutation that is found in tumors. Unfortunately, the DNA from the tumor containing the K289M variant is not available for analysis of the APC gene.
In summary, we provide evidence that the presence of the K289M pol β variant protein in cells is linked to cancer. How might the K289M protein function to induce or promote a carcinogenic phenotype in cells in the presence of WT pol β, given its somewhat lower catalytic efficiency and DNA affinity? One possibility is that the presence of K289M does not have a direct role in enhancing mutations in cells and in promoting the onset or progression of neoplasia. We do not favor this possibility, given the fact that the K289M protein is less accurate than WT Pol β and induces a mutational spectrum that is different from that of WT pol β. We feel that our data are consistent with the possibility that K289M is able to enhance mutations in cells and play a role in carcinogenesis for the following reasons. First, tumor cells are known to undergo loss of heterozygosity (LOH) at rates that are significantly higher than normal cells. Thus, once LOH occurs in the tumor, the WT pol β would no longer be present to compete with K289M during BER, and we suggest that gap filling by K289M would lead to mutations. Second, pol β is known to form multimeric complexes on DNA (44), and it has been proposed that this form of binding may stabilize pol β. Thus, in cells expressing equal amounts of WT and K289M, one would expect that a complex of K289M and WT would be bound to the gapped DNA, assuming that loading of these polymerases by apurinic/apyrimidinic endonuclease is equally efficient, as suggested by the fact that they are both functional in the in vitro BER assay (Fig. 4). Being a member of this complex could stabilize the K289M protein on the DNA and ultimately result in misinsertion by K289M. Third, if we assume that 10,000 lesions occur in each cell per day, as suggested by Lindahl (15) and others, K289M would be expected to remain associated with 625 of those gaps, given the fact that K289M has a DNA binding affinity that is 16-fold less than WT. K289M would then misinsert nucleotides within a subset of those gaps, which could ultimately result in mutations. If the mutations induced by K289M are within a key growth control gene, tumorigenesis or tumor promotion could be initiated. Finally, cells expressing K289M at 2.5-fold higher levels than WT also have a 2.5-fold increased overall mutation frequency. However, the mutation frequency at position 322 for K289M is 16-fold over that of WT and seems to be significant in our statistical evaluation. This frequency is at least 6-fold over the frequency that would be expected if the levels of expression of K289M were the only reason for the observed mutations at position 322. Thus, we suggest that K289M might have a higher catalytic efficiency or DNA binding affinity in vivo, due to its association with other proteins. Unpublished results from our laboratory, showing that expression of the K289M protein in mouse cells results in a transformed phenotype as measured by focus formation and growth in low serum (J.B.S., unpublished data), support the idea that the presence of the K289M variant is linked to cancer.
Supplementary Material
Acknowledgments
We thank Dr. Peter Glazer for the LN12 cells and Latha Narayanan for advice on the packaging reactions. This research was supported by National Institutes of Health Grant CA16038. J.B.S. is a Donahue Investigator.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: pol β, DNA polymerase β; BER, base excision repair; Tet, tetracycline; WCE, whole cell extract; UDG, uracil DNA glycosylase.
References
- 1.Dobashi, Y., Shuin, T., Tsuruga, H., Uemura, H., Torigoe, S. & Kubota, Y. (1994) Cancer Res. 54, 2827-2829. [PubMed] [Google Scholar]
- 2.Iwanaga, A., Ouchida, M., Miyazaki, K., Hori, K. & Mukai, T. (1999) Mutat. Res. 435, 121-128. [DOI] [PubMed] [Google Scholar]
- 3.Wang, L., Patel, U., Ghosh, L. & Banerjee, S. (1992) Cancer Res. 52, 4824-4827. [PubMed] [Google Scholar]
- 4.Kubota, Y., Murakami-Murofushi, K., Shimada, Y., Ogiu, T. & Oikawa, T. (1995) Cancer Res. 55, 3777-3780. [PubMed] [Google Scholar]
- 5.Bhattacharyya, N., Chen, H. C., Grundfest-Broniatowski, S. & Banerjee, S. (1999) Biochem. Biophys. Res. Commun. 259, 429-435. [DOI] [PubMed] [Google Scholar]
- 6.Mohrenweiser, H., Xi, T., Vazquez-Matias, J. & Jones, I. M. (2002) Cancer Epidemiol. Biomarkers Prev. 11, 1054-1064. [PubMed] [Google Scholar]
- 7.Macoska, J. A., Trybus, T. M., Sakr, W. A., Wolf, M. C., Benson, P. D., Powell, I. J. & Pontes, J. E. (1994) Cancer Res. 54, 3824-3830. [PubMed] [Google Scholar]
- 8.Seo, Y. R., Fishel, M. L., Amundson, S., Kelley, M. R. & Smith, M. L. (2002) Oncogene 21, 731-737. [DOI] [PubMed] [Google Scholar]
- 9.Zhou, J., Ahn, J., Wilson, S. H. & Prives, C. (2001) EMBO J. 20, 914-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharyya, N., Chen, H. C., Comhair, S., Erzurum, S. C. & Banerjee, S. (1999) DNA Cell Biol. 18, 549-554. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharyya, N. & Banerjee, S. (1997) Proc. Natl. Acad. Sci. USA 94, 10324-10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharyya, N. & Banerjee, S. (2001) Biochemistry 40, 9005-9013. [DOI] [PubMed] [Google Scholar]
- 13.Lindahl, T. (2001) Prog Nucleic Acid Res. Mol. Biol. 68, xvii-xxx. [DOI] [PubMed] [Google Scholar]
- 14.Wilson, S., Abbotts, J. & Widen, S. (1988) Biochim. Biophys. Acta 949, 149-157. [DOI] [PubMed] [Google Scholar]
- 15.Lindahl, T. (1993) Nature 362, 709-715. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto, Y. & Kim, K. (1995) Science 269, 699-702. [DOI] [PubMed] [Google Scholar]
- 17.Sobol, R. W., Horton, J. K., Kuhn, R., Gu, H., Singhal, R. K., Prasad, R., Rajewsky, K. & Wilson, S. H. (1996) Nature 379, 183-186. [DOI] [PubMed] [Google Scholar]
- 18.Podlutsky, A. J., Dianova, I. I., Podust, V. N., Bohr, V. A. & Dianov, G. L. (2001) EMBO J. 20, 1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosa, J. L. & Sweasy, J. B. (1999) J. Biol. Chem. 274, 35866-35872. [DOI] [PubMed] [Google Scholar]
- 20.Maitra, M., Gudzelak, A., Jr., Li, S. X., Matsumoto, Y., Eckert, K. A., Jager, J. & Sweasy, J. B. (2002) J. Biol. Chem. 277, 35550-35560. [DOI] [PubMed] [Google Scholar]
- 21.Shah, A. M., Li, S. X., Anderson, K. S. & Sweasy, J. B. (2001) J. Biol. Chem. 276, 10824-10831. [DOI] [PubMed] [Google Scholar]
- 22.Beard, W. A., Osheroff, W. P., Prasad, R., Sawaya, M. R., Jaju, M., Wood, T. G., Kraut, J., Kunkel, T. A. & Wilson, S. H. (1996) J. Biol. Chem. 271, 12141-12144. [DOI] [PubMed] [Google Scholar]
- 23.Osheroff, W. P., Beard, W. A., Yin, S., Wilson, S. H. & Kunkel, T. A. (2000) J. Biol. Chem. 275, 28033-28038. [DOI] [PubMed] [Google Scholar]
- 24.Shah, A. M., Conn, D. A., Li, S. X., Capaldi, A., Jager, J. & Sweasy, J. B. (2001) Biochemistry 40, 11372-11381. [DOI] [PubMed] [Google Scholar]
- 25.Glazer, P. M., Sarkar, S. N. & Summers, W. C. (1986) Proc. Natl. Acad. Sci. USA 83, 1041-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobol, R. W., Prasad, R., Evenski, A., Baker, A., Yang, X. P., Horton, J. K. & Wilson, S. H. (2000) Nature 405, 807-810. [DOI] [PubMed] [Google Scholar]
- 27.Clairmont, C. A., Narayanan, L., Sun, K. W., Glazer, P. M. & Sweasy, J. B. (1999) Proc. Natl. Acad. Sci. USA 96, 9580-9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leach, E., Gunther, E., Yeasky, T., Gibson, L., Yang-Feng, T. & Glazer, P. (1996) Mutagenesis 11, 49-56. [DOI] [PubMed] [Google Scholar]
- 29.Gunther, E. J., Murray, N. E. & Glazer, P. M. (1993) Nucleic Acids Res. 21, 3903-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu, X. S., Narayanan, L., Dunklee, B., Liskay, R. M. & Glazer, P. M. (2001) Cancer Res. 61, 3775-3780. [PubMed] [Google Scholar]
- 31.Kosa, J. L. & Sweasy, J. B. (1999) J. Biol. Chem. 274, 3851-3858. [DOI] [PubMed] [Google Scholar]
- 32.Li, S. X., Vaccaro, J. A. & Sweasy, J. B. (1999) Biochemistry 38, 4800-4808. [DOI] [PubMed] [Google Scholar]
- 33.Biade, S., Sobol, R. W., Wilson, S. H. & Matsumoto, Y. (1998) J. Biol. Chem. 273, 898-902. [DOI] [PubMed] [Google Scholar]
- 34.Podlutsky, A. J., Dianova, I. I., Wilson, S. H., Bohr, V. A. & Dianov, G. L. (2001) Biochemistry 40, 809-813. [DOI] [PubMed] [Google Scholar]
- 35.Kunkel, T. A. (1985) J. Biol. Chem. 260, 5787-5796. [PubMed] [Google Scholar]
- 36.Opresko, P. L., Sweasy, J. B. & Eckert, K. A. (1998) Biochemistry 37, 2111-2119. [DOI] [PubMed] [Google Scholar]
- 37.Adams, W. & Skopek, T. (1987) J. Mol. Biol. 194, 391-396. [DOI] [PubMed] [Google Scholar]
- 38.Boosalis, M. S., Mosbaugh, D. W., Hamatake, R., Sugino, A., Kunkel, T. A. & Goodman, M. F. (1989) J. Biol. Chem. 264, 11360-11366. [PubMed] [Google Scholar]
- 39.Efrati, E., Tocco, G., Eritja, R., Wilson, S. H. & Goodman, M. F. (1997) J. Biol. Chem. 272, 2559-2569. [DOI] [PubMed] [Google Scholar]
- 40.Bhagwat, A. S., Sanderson, R. J. & Lindahl, T. (1999) Nucleic Acids Res. 27, 4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou, K. M. & Cheng, Y. C. (2002) Nature 415, 655-659. [DOI] [PubMed] [Google Scholar]
- 42.Fodde, R., Smits, R. & Clevers, H. (2001) Nat. Rev. Cancer 1, 55-67. [DOI] [PubMed] [Google Scholar]
- 43.Muniappan, B. P. & Thilly, W. G. (2002) Cancer Res. 62, 3271-3275. [PubMed] [Google Scholar]
- 44.Jezewska, M. J., Rajendran, S. & Bujalowski, W. (2001) J. Biol. Chem. 276, 16123-16136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.