Abstract
FLT3 is one of the most frequently mutated genes in acute leukemias. However, the role in leukemogenesis of wt Flt3, which is highly expressed in many hematological malignancies, is unclear. We show here that in mouse models established by retroviral transduction of leukemic fusion proteins deletion of Flt3 strongly inhibits MLL-ENL and to lesser extent p210BCR-ABL-induced leukemogenesis, but has no effect in MLL-AF9 or AML1-ETO9a models. Flt3 acts at the level of leukemic stem cells (LSCs), as a fraction of LSCs in MLL-ENL, but not in MLL-AF9-induced leukemia, expressed Flt3 in vivo, and Flt3 expression on LSCs was associated with leukemia development in this model. Furthermore, efficiency of MLL-ENL, but not of MLL-AF9-induced leukemia induction was significantly enhanced after transduction of Flt3+ compared to Flt3− wt myeloid progenitors. However, Flt3 is not required for immortalization of bone marrow cells in vitro by MLL-ENL and does not affect colony-formation by MLL-ENL LSCs in vitro, suggesting that in vitro models do not reflect the in vivo biology of MLL-ENL leukemia with respect to Flt3 requirement. We conclude that wt Flt3 plays a role in leukemia initiation in vivo, which is, however, not universal.
Keywords: Leukemia, Flt3, Leukemic Stem Cells, Mixed-Lineage Leukemia
INTRODUCTION
Leukemogenesis is a multistep process involving genomic rearrangements that generate aberrant transcription factors or signaling proteins, mutations in tumor suppressors or genes encoding signaling proteins, epigenetic changes, and dysregulated microRNA function and expression[1]. FLT3, a receptor tyrosine kinase expressed on early multipotential and lymphoid hematopoietic progenitors in the mouse and also on HSCs in humans[2–5], is one of the most frequently mutated genes in AML. Mutations cause constitutive activation, and are often associated with worse prognosis[2]. FLT3 inhibitors have thus far shown disappointing clinical results[6, 7]. The role of wild-type (wt) Flt3 in leukemogensis has not been examined in depth, however. Wt FLT3 is highly expressed in many hematological malignancies[2], in particular acute myeloid (AML) and lymphoid (ALL) with MLL (Mixed Lineage Leukemia) rearrangements[8–10]. Furthermore, Flt3 kinase blockers induce apoptosis in some MLL cell lines with high wt FLT3 expression or with mutated FLT3[11]. In contrast, another report showed minimal cytotoxicity of FLT3 inhibitors in infant ALL with MLL rearrangements in vitro[10]. These studies focused on leukemic blasts and on cell lines however, and not on in vivo models of leukemic stem cells[8]. In acute (AML) and chronic (CML) myeloid leukemias, leukemic stem cells (LSCs) are critical for initiation and propagation of leukemia[12]. Targeting the LSC is likely critical to achieve complete molecular remission and cure of leukemia[13, 14]. Data on the role of wt Flt3 signaling in leukemogenesis in vivo are conflicting. Overexpression of wt Flt3 collaborates with NUP98-HOX in the induction of AML in mice, suggesting a role in pathogenesis[15]. However, latency and mortality of leukemia after transplantation of Flt3−/− and wt BM transduced with MLL-ENL [16], MLL-CBP [16], and Meis/Hoxa9 [17], downstream targets of several MLL fusion proteins[18–20], was similar, arguing against a pathogenic role of wt Flt3 in vivo in this model. Given this controversy, we examined the role of wt Flt3 in leukemogenesis more closely.
MATERIALS AND METHODS
Mice
C57BL/6J mice (CD45.2+ B6) were purchased from The Jackson Laboratory and C57BL/6.SJL-PtprcaPep3b/BoyJ (CD45.1+ B6) mice from the National Cancer Institute. Flt3−/− mice were obtained from Dr. I. Lemischka (Mount Sinai School of Medicine). Animals were housed in a specific pathogen-free facility. Experiments and animal care were performed with approval from the Institutional Animal Care and Use Committee of Mount Sinai School of Medicine and Columbia University Medical Center.
Mouse genotyping
PCR genotyping of Flt3−/ − mice was performed with primers for wild type allele (Flt3F 5’-tccacgttgttccctctacc-3’, Flt3R 5’-tatgtgggcaatttggctct-3’) and for mutant allele (Neo514F 5’-tgccgcgctgttctcctct-3’, Neo1164R 5’-aagcggccattttccaccat-3’).
Antibodies and cytokines
FITC-conjugated anti-B220, PE-conjugated anti-FLT3, -Mac1, -IgG2a, and IgG1, PECy7-conjugated streptavidin, APC-eFluor 780-conjugated anti-c-kit, -CD45.1, and APC-conjugated anti-B220 were purchased from eBiosciences. PE-conjugated anti-CD16/32 and -B220, PerCP-Cy5.5–conjugated anti-CD45.2, -B220, -hNGFR, and -streptavidin, APC- conjugated anti-c-kit and -Gr1, APC-Cy7-conjugated anti-CD19, Pacific Blue-conjugated anti- B220, Alexa Fluor 647-conjugated anti-pSTAT5(pY694), and -CD34, and biotinylated anti-hNGFR were purchased from BD Biosciences, PECy7-conjugated anti-CD150, Pacific Blue-conjugated anti-Sca1 were purchased from BioLegend. Biotinylated Mouse Lineage Mixture (MLM15) was purchased from Invitrogen. Biotinylated anti-Thy1.1 was purchased from Southern Biotech. Recombinant mouse SCF, IL-11, IL-6, IL-3, Flt3L, and GM-CSF were purchased from PeproTech.
Cell sorting and flow cytometry
Bone marrow (BM) cells were prepared by flushing or crushing the femora and tibia of mice with cold DMEM (Cellgro) containing 2% FBS (Atlanta Biologicals) and penicillin/streptomycin (Cellgro). BM cells were stained and isolated using FACSAriaII (Beckon Dickinson), MoFlo (CytoMation) sorters, or InFlux cell sorter (Beckon Dickinson). Flow cytometric analysis was performed on a 5 or 6-laser LSRII or on a 3-laser FACSCantoII with DiVa software (BD Biosciences) and analyzed using FlowJo software (Tree Star). For analysis of BrdU incorporation, cells were exposed to BrdU for 2 hours and then fixed, treated with DNase to expose BrdU epitopes, and stained with APC-conjugated anti-BrdU antibodies (BD Biosciences) as recommended by the manufacturer.
In vitro culture
Cells were plated in triplicate in 35-mm non-tissue culture plates in 1.5% methylcellulose (Sigma) with IMDM (Cellgro) containing 30% FBS (Atlanta Biologicals), IL-3 (10ng/ml), SCF (50ng/ml), IL-6 (10ng/ml), GM-CSF (10ng/ml), and / or Flt3L (50ng/ml). Colony formation was scored after 7 days. Immortalized cells isolated from tertiary round methylcellulose cultures were maintained in RPMI1640 (Cellgro) with IL-3 (6 ng/ml), IL6 (10 ng/ml), SCF (20 ng/ml), GM-CSF (10 ng/ml), with or without Flt3L (10 ng/ml).
Plasmid construct and retroviral transduction
pMSCV-MLL-ENL and pMSCV-AML-ETO9a-IRES-GFP were purchased from Addgene. pMSCV-MLL-AF9-IRES-YFP and pMSCV-p210BCR-ABL-IRES-hNGFR were obtained from Dr. Y. Aifantis (Howard Hughes Medical Institute, New York University School of Medicine). Murine Flt3 coding region (NM_010229.2) was synthesized by GenScript and cloned into pUC57 plasmid. cDNAs were cloned into pMSCV-IRES-GFP or pMSCV-IRES-hNGFR backbones. Plasmids were transfected into Plat-E cells (Cell Biolabs) with Lipofectamin LTX (Invitrogen). For retroviral transduction, 6 to 8-week old donor mice were injected with 150 mg/kg, IP, of 5-fluorouracil (Sigma). After 5 days BM cells were cultured in RPMI1640 with IL-3 (6 ng/ml), IL6 (10 ng/ml), SCF (20 ng/ml), IL11 (10 ng/ml) for 24 hours, followed by incubation with viral supernatant and 4 μg/ml of polybrene (Sigma). Plates were then centrifuged at 2500 rpm for 90 minutes at 30°C.
Quantitative RT-PCR
RNA was isolated using RNAeasy micro kit (Qiagen), and reverse-transcribed using SuperScript III (Invitrogen), according to the manufacturer's instructions. Gene expression were measured by TaqMan PCR using Gene Expression MasterMix and Gene Expression Assays (Applied Biosystems). All probes were purchased from inventoried stocks of Taqman Gene Expression Assays (Applied Biosystems). 18S RNA was used as internal control. Thermal cycling conditions included an initial step at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute on a ViiA 7 Real-Time PCR System (Applied Biosystems). Comparative Ct method (ΔΔCt) was used for analysis. All samples were analyzed in triplicate.
Transplantation assays
Transduced BM cells were injected into lethally irradiated (550 cGy followed by 700 cGy at 3 hours interval) recipient mice. In transplant analysis using immortalized cells or purified cells, 2.0 × 105 helper BM cells were co-injected. Moribund mice were euthanized and BM cells were analyzed with flow cytometer to confirm development of leukemia.
Statistics
Student 2-tailed t test for paired samples was used, and data represent mean ± SEM. For survival analysis, Log-rank (Mantel-Cox) test was used.
RESULTS
Differential requirement for Flt3 signaling in mouse models of leukemia
Flt3−/ − and wt littermate BM cells (CD45.2+) were transduced with bicistronic retroviral vectors expressing p210BCR-ABL, AML-ETO9a, MLL-AF9 or MLL-ENL (Fig. S1) and a reporter gene (YFP, GFP or hNGFR) separated by an internal ribosomal entry site (IRES) sequence, and transplanted into congenic CD45.1+ wt recipients, a well-established strategy to model human leukemia in the mouse[21–25]. Cell number was adjusted so that a similar number of transduced (reporter gene expressing) wt and Flt3−/ − cells were injected in each independent experiment (table S1).
MLL-ENL and MLL-AF9 induce a myelomonocytic AML [21, 22, 24, 26], consisting of Mac1+Gr1+ cells with intermediate forward and side scatter (Figure 1a). MLL-AF9 induced leukemia with similarly short latency and 100% mortality after transplantation of both Flt3−/ − and wt littermate transduced BM (Figure 1a). In the MLL-ENL model, however, only 1 out of 34 recipients of transduced Flt3−/ − BM developed leukemia while 30% of recipients of wt littermate BM died (Figure 1a, P=0.005). Transduction efficiency (0.45–2.13%) and therefore, the number of transduced cells injected (3,600–16,800) were relatively low in these particular cohorts. Similar data were obtained in cohort (n=10) with higher transduction efficiency (7%) (Table S1). As latency and mortality are cell dose-dependent, we performed another repeat experiment with hNGFR as a reporter instead of GFP, which yielded a much higher transduction efficiency (25.4%), and injected a higher dose of purified transduced cells (2.5x105) together with 2x105 helper BM cells. As expected, latency was much shorter and mortality was 100% when wt BM cells were transplanted. However, only 20% mortality was observed when Flt3−/ − BM cells were transplanted (Figure 1b, solid lines). The lower mortality in mice receiving Flt3−/ − MLL-ENL-transduced BM therefore did not depend on transduction efficiency or dose of transduced cells. Furthermore, we retrovirally restored Flt3 expression in Flt3−/ − BM cells, and injected 2.5x105 purified Flt3:GFP+ MLL-ENL:hNGFR+ cells (Figure 1b, upper panels). Mortality and latency in these recipients were similar to that of mice receiving MLL-ENL-transduced wt BM cells (Figure 1b, lower panel). All leukemic cells in these mice expressed Flt3 on the cell surface (Fig. 1c). Furthermore, Flt3−/ − MLL- ENL:hNGFR+Flt3:GFP+ leukemic BM cells expressed more Flt3 mRNA than primary BM cells, and than Flt3−/ − Flt3−/ − MLL-ENL:hNGFR+ leukemic BM cells that were not transduced with Flt3 (Fig. 1d). Overexpression of Flt3 in wt cells did not induce mortality or leukemia over the observation period (not shown). Together, these data definitively prove that Flt3 strongly enhances leukemogenesis in the MLL-ENL model, and exclude subtle variation in genetic background as the reason for the observed differences in MLL-ENL leukemogenesis in Flt3−/ − and wt littermate BM.
Figure 1. Requirement for Flt3 in mouse models of leukemia.
(a) Phenotypes of leukemic cells and survival in mice transplanted with non-sorted wt or Flt3−/ − BM transduced with MLL-ENL, MLL-AF9, AML1-ETO9a or p210BCR-ABL. Representative plots gated on reporter-positive cells in PB (when >30%). As Flt3−/ − cells transduced with MLL-ENL rarely induced leukemia, Flt3 expression in wt MLL-ENL blasts was compared with GFP+Gr1+Mac1+ PB cells from recipients of Flt3−/ − BM. (b) Survival of mice transplanted 2x105 helper BM cells and sorted 2.5x105 wt or Flt3−/ − Flt3:GFP+MLL-ENL:hNGFR+ or MLL-ENL:hNGFR+-transduced BM cells (isolated according to the gates in the upper panels). (c) Expression of Flt3:GFP and MLL-ENL:hNGFR in the PB of leukemic mice transplanted with Flt3−/ − Flt3:GFP+MLL-ENL:hNGFR+ transduced BM cells. (d) Flt3 mRNA expression in primary BM cells, Flt3−/ − BM cells transduced with MLL-ENL:hNGFR, and leukemic cells from mice transplanted with Flt3−/ − BM cells transduced with MLL-ENL:hNGFR and Flt3:GFP (n=3).
Similar to the MLL-AF9 model, deletion of Flt3 did not affect latency and mortality in leukemia induced by AML-ETO9a, a splice variant of AML1-ETO that induces AML with a very immature lin−kitloFlt3−phenotype in mice[27] (Figure 1a). However, in mice transplanted with BM transduced with p210BCR-ABL, which induces a chronic myeloid leukemia (CML)-like disease with short latency and rapid accumulation of mature granulocytes, Flt3 deletion prolonged survival (P=0.03, Fig. 1a, pooled data from two independent cohorts).
We conclude that wt Flt3 plays a role in leukemogenesis, which is, however, not universal.
Multilineage engraftment of wt and Flt3−/ − MLL-ENL BM cells
Although deletion of Flt3 does not affect engraftment of murine HSCs and myelopoiesis [5], we assessed whether MLL-ENL-transduced Flt3−/ − BM cells engrafted. MLL-ENL:GFP+ cells initially established multilineage (T, B and myeloid) hematopoiesis both in recipients of wt and of Flt3−/ − BM (Figure 2a,b). In a fraction of recipients of wt cells, the myeloid MLL-ENL:GFP+ population increased steeply after 15 weeks as leukemia developed. Although some of these mice displayed relatively high lymphoid reconstitution, lymphoid leukemia was never observed.
Figure 2. Flt3−/ − MLL-ENL:GFP cells establish long-term, multilineage hematopoiesis.
(a) Representative flow cytometric analysis of the reconstitution of the PB of recipients of Flt3−/ − BM transduced with MLL-ENL:GFP 6 weeks after transplantation. (b) Fraction of MLL-ENL:GFP+ , myeloid (Mac1+Gr1+), T (Thy1.1+), and B (B220+) cells in the PB of mice transplanted with wt or Flt3−/ − MLL-ENL:GFP-transduced BM cells.
These data rule out the possibility that decreased leukemogenesis by MLL-ENL in Flt3−/ − BM cells is caused by impaired engraftment of transduced cells.
Differential expression of Flt3 in MLL-ENL and MLL-AF9 LSCs in vivo
To explain the differential role of Flt3 in leukemogenesis, we first examined Flt3 expression on PB leukemic cells. Both in the MLL-ENL and in the MLL-AF9 model PB leukemic cells expressed Flt3 (Figure 1a), while in the AML-ETO9a model and the p210BCR-ABL model, Flt3 was absent. Therefore, the differential Flt3 requirement among these models cannot be explained by variation in the expression of Flt3 on peripheral leukemic cells.
We next concentrated our studies on LSCs. Human and mouse LSCs have been identified in populations that are not normally self-renewing, such as granulocyte-macrophage (GMP) and common myeloid progenitors (CMP) [21–24]. In CML, however, the LSC is derived from very early progenitors, while the LSC underlying blast crisis bears a GMP phenotype[14]. We focused on the MLL-ENL and MLL-AF9 leukemia models because of the nearly absolute difference in Flt3 requirement between them. We defined LSCs here broadly as lin−Sca1−c-kit+ cells. Both in MLL-ENL and MLL-AF9-induced leukemia, virtually all reporter positive lin−Sca1−c-kit+ cells expressed CD16/32, consistent with a GMP-like phenotype (Figure 3a). Expression of CD34 was low however, but appeared somewhat higher in MLL-ENL than in MLL-AF9 cells (Figure 3a). In contrast, the remaining non-leukemic lin−GFP− compartment contained LSK cells, while the lin−Sca1−kit+ fraction showed a normal distribution of megakaryocyte/eyrthroid progenitors (CD34−CD16/32−), CMPs (CD34+CD16/32−) and GMPs (CD34+CD16/32+) (not shown). In recipients of wt BM, leukemic MLL-AF9:GFP+ lin−Sca1−c-kit+ cells were predominantly Flt3−, while 8 to 45% of leukemic MLL-ENL:GFP+ lin−Sca1−c-kit+ cells expressed Flt3 (Figure 3a,b), a finding consistent with a role for Flt3 in the function of LSCs in the MLL-ENL, but not in the MLL-AF9 model. We next more carefully analyzed the lin−Sca1−kit+ compartment in leukemic and non-leukemic mice. Among non-leukemic mice (< 30% GFP+ myeloid cells in the PB and clinically healthy) transplanted with MLL-ENL:GFP-transduced BM and analyzed between days 90 and 110 (see Fig. 1a), the fraction of lin−Sca1−c-kit+ cells as a percentage of the total GFP+ population was significantly lower in recipients of Flt3−/ − than of wt BM (Figure 3c), supporting a role for Flt3 in the generation of MLL-ENL-expressing lin−Sca1−c-kit+ cells. Furthermore, among recipients of MLL-ENL-transduced wt BM analyzed 90 to 110 days after transplantation, significantly fewer lin−Sca1−c-kit+ cells expressed detectable Flt3 in non-leukemic than in leukemic mice (Figure 3d), again supporting a role for Flt3 in the generation of putative LSC in vivo.
Figure 3. Flt3 expression on leukemic stem cells.
(a) Expression of CD34, c-kit, Sca1, CD16/CD32 and Flt3 on leukemic BM lin−Sca1−kit+ cells in mice transplanted with wt MLL-ENL:GFP and MLL-AF9:GFP-transduced BM. (b) Fraction of leukemic lin−Sca1−kit+ cells expressing Flt3 in mice transplanted with wt MLL-ENL:GFP and MLL-AF9:GFP-transduced BM. (c) Fraction of lin−kit+ cells among MLL-ENL:GFP+ cells in mice transplanted with wt or Flt3−/ − MLL-ENL:GFP-transduced BM prior to the onset of leukemia in recipients of wt BM (90–110 days after transplantation). (d) Fraction of Flt3+ cells in non-leukemic and leukemic (>30% GFP+ myeloid cells in PB) mice transplanted with wt MLL-ENL:GFP-transduced BM. (e) Survival of mice transplanted with 2x105 helper BM and 2,000 Flt3+ or Flt3− MLL-ENL: GFP+ wt CMP/GMPs (left) or MLL-AF9: GFP+ wt CMP/GMPs (right). (f) Survival of mice transplanted with 2x105 helper BM and 1,000 Flt3+ or Flt3− MLL-ENL: GFP+ lin−kit+ cells from leukemic primary recipients.
To further confirm the role of Flt3 in leukemogenesis, we isolated Flt3+ or Flt3− wt CMP/GMPs (lin−Sca1−kit+CD34+) cells, from wt BM., and transduced these with MLL-ENL:GFP or MLL-AF9:GFP. We transplanted 2,000 transduced Flt3+ or Flt3− CMP/GMPs with 2x105 helper BM cells into lethally irradiated syngeneic recipients. Mortality was significantly higher in mice transplanted with MLL-ENL:GFP+Flt3+ CMP/GMPs than in mice transplanted with MLL-ENL:GFP+Flt3− CMP/GMPs (Figure 3e, left). In contrast the mortality of mice transplanted with Flt3+ or Flt3− MLL-AF9:GFP+ CMP/GMPs was similar (Figure 3e, right). These data demonstrate that Flt3 acts at the level of leukemia-initiating cells in the MLL-ENL leukemia.
We next examined whether Flt3 is required for leukemia maintenance in the MLL-ENL model. We isolated Flt3+ or Flt3− MLL-hNGFR+ lin−Sca1−kit+ cells from leukemic mice and examined their in vivo secondary leukemia-initiating capacity. In contrast to the data obtained in primary transplantation, however, 1,000 Flt3+ and Flt3− cells were equally efficient at inducing leukemia and mortality after secondary transplantation (Figure 3f). These data suggest that Flt3 is not required for maintenance of LSCs, possibly because of acquisition of secondary mutations.
Taken together, our observations show that Flt3 strongly enhances leukemia initiation in the MLL-ENL, but not in the MLL-AF9 model, but that Flt3 does not appear required for the maintenance of LSCs.
MLL-ENL, but not MLL-AF9 increases Flt3 responsiveness of putative LSCs in vitro
BM cells immortalized by MLL-ENL and MLL-AF9 form replatable colonies (AML-CFUs) in semisolid media[21, 22, 24, 26]. It is believed that AML-CFUs may reflect LSCs in vivo. We transduced wt and Flt3−/ − BM cells with MLL-ENL and MLL-AF9 constructs. We plated MLL-ENL or MLL-AF9 immortalized cells in methylcellulose cultures supported by IL3, IL6, KL and GM-CSF with or without Flt3L. Cells were subsequently replated in secondary and tertiary cultures. MLL-ENL:GFP+ and MLL-AF9:GFP+ cells formed large colonies, while reporter-negative cells only formed small colonies that showed no replating capacity (not shown). Serial replating capacity was similar in wt and Flt3−/ − BM cells both in the MLL-ENL and MLL-AF9 models, even when Flt3L was present in the cytokine cocktail. Furthermore, the frequency of colony-initiating cells was similar in wt and Flt3−/ − BM cells transduced with MLL-ENL or ML-AF9 (Figure 4a). BM cells immortalized in vitro and propagated in the presence of IL3, IL6, KL, GM-SCF and Flt3L contained a population of lin−kit+ cells that uniformly expressed CD16/32, while CD34 was low to absent (Figure 4b). Although it has been shown that AML-CFCs express Mac and Gr1 in the MLL-AF9 model[26], AML-CFUs were highly enriched in the lin−kit+ fraction in both MLL-ENL and MLL-AF9-transduced cells (Figure 4c). In contrast to their phenotype in vivo, however, in vitro propagated lin−kit+ cells generated after transduction with either MLL-AF9 or MLL-ENL did not express detectable Flt3 (Figure 4d), explaining why addition of Flt3L or genetic deletion of Flt3 did not affect AML-CFU formation in MLL-ENL-immortalized cells. Finally, similar to normal lin−kit+ cells (Fig. 4e, left), among reporter-positive lin−kit+ cells isolated from mice with MLL-ENL leukemia, the Flt3+ fraction was enriched in CFUs even though Flt3L was not present in the growth factor culture (Figure 4e), right. This is again surprising, as leukemic Flt3+ and Flt3− lin−kit+ cells show similar leukemogenic capacity in secondary recipients (see Figure 3f). Thus, in vitro culture of MLL-ENL-immortalized BM cells does not reflect the role of Flt3 in MLL-ENL leukemogenesis in vivo.
Figure 4. Characteristics of AML-CFUs in vitro.

(a) Secondary and tertiary replating efficiency of AML-CFU grown from wt and Flt3−/ − BM cells immortalized MLL-ENL or MLL-AF9 in the presence of IL3, IL6, KL and GM-CSF, with and without Flt3L (n=3). (b) Expression of CD34, Sca1, c-kit, and CD16/CD32 on wt BM cells transduced and immortalized with MLL-ENL:GFP and MLL-AF9:GFP and cultured for 3 weeks in the presence of GM-CSF, KL, IL3, and IL6. (c) AML-CFUs in lin−kit+ and lin+ cells after immortalization of wt BM cells with MLL-ENL and cultured for three weeks in the presence of GM-SCF, IL3, IL6 and KL (n=3). (d) Expression of Flt3 on lin−kit+ cells immortalized by MLL-ENL or MLL-AF9 and grown in the presence of IL3, IL6, KL, GM-CSF, and Flt3L. Red: isotype control. (e) CFU formation in normal (left) and leukemic reporter-positive (right) Flt3+ and Flt3− lin−kit+ cells isolated from MLL-ENL leukemic mice (n = 4).
As we have established that Flt3 strongly enhances leukemia-initiation by MLL-ENL, but not MLL-AF9, we next explored whether these MLL fusions would in turn differentially affect Flt3 responsiveness. As Flt3 is not expressed on MLL-ENL lin−kit+ cells in vitro, we examined the effect of MLL-AF9 and MLL-ENL on Flt3 responsiveness by transducing Flt3−/ − BM cells with Flt3 and either MLL-ENL or MLL-AF9, and culturing these in liquid cultures supported by IL3, IL6, GM-SCF, KL with or without Flt3L. In MLL-ENL, but not in MLL-AF9-immortalized Flt3-expressing cells, addition of Flt3L increased the lin−kit+ fraction (Figure 5a,b). These findings were further corroborated using BrdU pulse (Figure 5c). Flt3L only enhanced BrdU incorporation in lin−kit+ cells expressing Flt3:GFP and MLL-ENL:hNGFR, and not in lin−kit+ cells expressing Flt3:GFP and MLL-AF9:hNGFR. Furthermore, Flt3L did not affect BrdU incorporation in lin+ cells in either model. To confirm that overexpression of Flt3 is sufficient to induce activation, we measured expression of Flt3 and activation of Stat5 by Flt3L. Flt3−/ − MLL-ENL:hNGFR+Flt3:GFP+ and MLL-AF9:hNGFR+Flt3:GFP+ cells have increased expression of Flt3 (Figure 5d). When these cells were stimulated with Flt3L, phosphorylation activation of Stat5 was observed by phosphoflow (Figure 5e), indicating that these expression levels are sufficient to induce activation of signaling downstream of Flt3.
Figure 5. MLL-ENL enhances Flt3 responsiveness.
(a) Representative example of the effect of Flt3L on the frequency of lin−kit+ cells grown in the presence of IL3, IL6, GM-CSF and KL from Flt3−/ − BM cells transduced with MLL-ENL and Flt3. (b,c ) Effect of Flt3L on the frequency of lin−kit+ cells (b) and BrdU incorporation in lin+ and lin−kit+ cells (c) grown in the presence of IL3, IL6, GM-CSF and KL from Flt3−/ − BM cells transduced with MLL-ENL or MLL-AF9 and Flt3 (n=4). (d) Flt3 mRNA expression in primary bone marrow cells, Flt3−/ − BM cells transduced with MLL-ENL and / or Flt3, and Flt3−/ − BM cells transduced with MLL-AF9 and / or Flt3. (e) pStat5 expression in primary bone marrow cells, Flt3−/ − BM cells transduced with MLL-ENL and / or Flt3, and Flt3−/ − BM cells transduced with MLL-AF9 and / or Flt3 with or without Flt3L (50ng/ml) stimulation.
These data indicate that MLL-ENL, but not ML-AF9 specifically renders putative LSCs more responsive to the proliferative effect of Flt3, and suggest reciprocal synergy between Flt3 and MLL-ENL at the level of putative LSCs.
Gene expression in Flt3+/+ and Flt3−/ − MLL-ENL lin−Sca1−kit+ cells
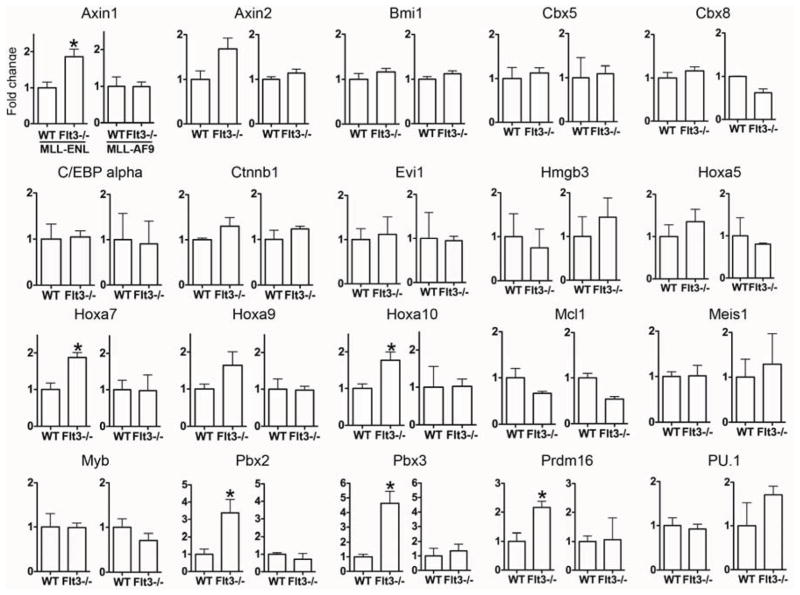
We measured expression of genes involved in the pathogenesis of AML in MLL- ENL:GFP+lin−Sca1−kit+ cells and MLL-AF9:GFP+lin−Sca1−kit+ cells before the onset of leukemia (MLL-ENL: between days 60 and 75, MLL-AF9: day 20). The expression of genes that are induced (MCL1)[28] or suppressed (PU.1, C/EBPα)[29] by Flt3-ITD and not by wt Flt3, and play a role in FLT3-ITD expressing leukemias was similar in Flt3−/ − and in Flt3+/+ cells (Figure 6). Surprisingly, however, genes involved in leukemia induced by MLL fusions were expressed more highly in Flt3−/ − than in Flt3+/+ MLL-ENL:GFP+lin−Sca1−kit+ cells. These genes included Pbx2 and Pbx3[20], HoxA genes (HoxA7, HoxA10)[30, 31], and Axin1[32]. For some, the increase was near significant (Axin2, Hoxa9) [19, 31, 33] (Figure 6). Axins are of interest, as Wnt signaling is required for LSC renewal in leukemias induced by overexpression of Meis1 and HoxA9 [32], two critical targets of MLL-fusions, including MLL-ENL[18]. In contrast to MLL-ENL, no differences in gene expression were observed between Flt3+/+ and Flt3−/ − MLL-AF9 LSCs.
Figure 6. Gene expression in Wt and Flt3−/− LSCs.
qPCR for genes indicated on top of each panel in MLL-ENL:GFP+lin−Sca1−kit+ BM cells from mice transplanted with WT and Flt3−/ − BM cells (60–75 days after transplantation) (left for each transcript; n=3 triplicate experiments) and MLL-AF9:GFP+lin−Sca1−kit+ BM cells from mice transplanted with WT and Flt3−/ − (right for each transcript; n=2 triplicate experiments) BM cells (20 days after transplantation). * P<0.05.
DISCUSSION
We have shown here that Flt3 signaling strongly enhances leukemogenesis in some mouse models of human leukemia, in particular of MLL-ENL leukemia. In the absence of Flt3, only rare recipients developed MLL-ENL leukemia, irrespective of the number of transduced cells injected, and therefore irrespective of latency and mortality, but not in the MLL-AF9 or AML-ETO9a models. In addition, there was a statistically significant increase in survival in the p210BCR-ABL model, where disease occurs with very short latency. The effect of Flt3 on leukemia-induction by MLL-ENL was not due to differences in the genetic background of Flt3−/ − and wt mice as Flt3−/ − mice were fully backcrossed onto the C57BL/6 background and as wt littermates were used as controls. Furthermore, restoration of Flt3 expression in MLL-ENL-transduced Flt3−/ − BM restored its leukemogenic capacity, while overexpression of Flt3 in wt BM had no effect. Overexpression of Flt3 has been shown to induce a slowly progressing myeloproliferative disease. The observation period in our experiments was too short to observe this disease however, as it only becomes detectable six months after transplantation[34]. Together, these data support the conclusion that Flt3 signaling strongly enhances MLL-ENL, and to a lesser extent p210BCR-ABL leukemogenesis.
This effect of Flt3 signaling on leukemia initiation in vivo is also not caused by enhanced engraftment of transduced cells. We found, somewhat surprisingly, that in the period preceding leukemia development, transplanted MLL-ENL-transduced wt and Flt3−/ − BM cells established multilineage hematopoiesis with equal efficiency. This was never observed before, although in at least one publication[22], there is some evidence of B and T cell reconstitution, even when mice are leukemic. It is likely that because we used a relatively low number of transduced BM cells to induce leukemia in this particular experimental cohort, latency was long, thus allowing us to observe multilineage reconstitution by transduced HSCs.
A fraction of MLL-ENL, but not of MLL-AF9, LSCs expressed Flt3, while deletion of Flt3 was associated with a significantly lower frequency of MLL-ENL:GFP+ lin−Sca1−kit+ LSCs. Furthermore, within the cohort of recipients of Wt MLL-ENL-transduced BM where, due to the low number of injected transduced cells, not all mice developed leukemia, expression of Flt3 on lin−Sca1−kit+ cells was significantly higher in leukemic than in non-leukemic mice. These observations suggest that the effect of Flt3 is situated at the level of LSCs in the MLL-ENL model. This idea was further corroborated by the much higher efficiency of leukemia induction by MLL-ENL-transduced Flt3+ CMP/GMPs than by their Flt3− counterparts, while no such difference was observed after transduction with MLL-AF9. Remarkably however, expression of Flt3 became irrelevant in secondary transplantations. These data suggest that Flt3 is predominantly required for the establishment of LSCs in vivo, and not, or less so, for their subsequent maintenance. It is not excluded that Flt3 becomes dispensable in secondary transplantations because of additional accumulated mutations that allow maintenance of LSCs in a Flt3-independent fashion.
It is remarkable that a widely used in vitro assay of LSCs, the AML-CFU assay, is not reflective of in vivo behavior in the MLL-ENL model. Even though ex vivo isolated Flt3+ MLL-ENL+ lin−kit+ cells were enriched in AML-CFUs, they were as effective as their Flt3− counterparts at inducing leukemia in secondary recipients. Furthermore, in contrast to LSCs in vivo, Flt3 is not expressed on lin−Sca1−kit+ cells in cultures of MLL-ENL transduced BM cells and does not affect AML-CFU potential. The cells are nevertheless capable of responding to Flt3, as retroviral expression of Flt3 in MLL-ENL-immortalized cells enhanced their proliferation in response to Flt3L. This was not the case in MLL-AF9-immortalized cells, again demonstrating the irrelevance of Flt3 signaling for MLL-AF9 leukemia.
The mechanism underlying the differential effect of Flt3 signaling in MLL-ENL and MLL-AF9 LSCs is not clear. Among MLL rearrangements, MLL-ENL and MLL-AF9 fusion proteins share overlapping pathogenetic mechanisms, including fusion with a nuclear protein, interaction with Menin, recruitment of the H3K9 methyltransferase hDOT1L and activation of MEIS and HOXA9[31, 35]. Although Flt3 has been shown to be dispensable for leukemogenesis induced by MEIS/HOXA9 overexpression[17], we find that Flt3 enhances MLL-ENL, but not MLL-AF9 leukemogenesis. Our data therefore suggest a partially distinct mechanism of action of MLL-ENL compared to MLL-AF9. A partially distinct mechanism of action of MLL-ENL and MLL-AF9 is also suggested by the different distribution of types of leukemia caused by these MLL fusions in humans. MLL-AF9 is more strongly associated with AML, while MLL-ENL is found in both AML and ALL[35]. As many genes believed to be essential for MLL-induced leukemogenesis are in fact expressed more highly in Flt3−/ − than in wt MLL-ENL lin−Sca1−kit+ cells, it is likely that Flt3 affects a mechanism essential for MLL-ENL, but not for MLL-AF9.
In a recent paper, Albouhair et al. [16] argue that Flt3 does not play a role in MLL-ENL leukemia. However, the MLL-ENL retroviral plasmid did not contain a reporter gene, and there is no mention of transduction efficiency. It is therefore not clear that the number of MLL-ENL positive cells transplanted was similar in Flt3+/+ and Flt3−/− cohorts. As we showed in Fig.1 and table S1, there is dose dependence in the initiation of MLL-ENL leukemia. Furthermore, the controls were not littermates. Thus, any differences in leukemogenic capacity may have been obfuscated. Similar to our data however, Albouhair et al[16] did not find differences in AML-CFU and in secondary engraftment of leukemias.
Another recent paper, however, did indicate a role for Flt3 signaling in MLL-AF9 leukemia. Jiang et al.[36] showed that miR-150 is downregulated in this model, and targets, among others, Flt3. shRNA inhibition of Flt3 caused a slight delay in mortality, while FLT3-ITD accelerated leukemia development. However, given its fundamentally different signaling characteristics[37–39], FLT3-ITD cannot simply be considered a hyperactive form of Flt3. Furthermore, shRNAs may have off-target effects, and no data on appropriate controls were presented. We believe that our data, using BM cells where Flt3 is deleted, convincingly demonstrate that Flt3 signaling is not required in this model, but strongly enhances leukemogenesis in the MLL-ENL model.
It is possible that in leukemias where LSCs express Flt3, treatment with Flt3 inhibitors might be beneficial by targeting LSCs. The clinical efficacy of Flt3 kinase inhibitors has been disappointing however, although most studies focused on leukemias with mutated Flt3, which might not be critical for LSC maintenance[6]. Given our evidence that Flt3 plays a role in leukemia initiation in vivo, rather than maintenance, it is possible that the level of Flt3 signaling might also predispose to the development of some leukemias. We have shown previously that Flt3 signaling in HSPCs shows mouse strain-dependent variation[40], and that a coding polymorphism in the gene, Prdm16, underlies this variation. Prdm16 allelic variation regulates Flt3 signaling through modulation of the transcriptional activity of peroxysome proliferator-activated receptor-γ (PPARγ), although it is unclear how PPARγ affects Flt3 signaling[41]. It is interesting to note however that the human populations show extensive germline coding variation in the same region of Prdm16 (Fig. S2, data extracted from the HapMap project). It will therefore be of interest to determine whether this coding variation is associated with leukemia predisposition.
In conclusion, we have shown that Flt3 plays a role in leukemogenesis in the MLL-ENL model, which is not accurately reflected in AML-CFU assays in vitro.
Supplementary Material
Acknowledgments
Support
This work was supported by NIH/NHLBI grant RO1 HL073760.
Footnotes
Conflict of interest disclosure: The authors declare no financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nature reviews Cancer. 2010;10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meshinchi S, Appelbaum FR. Structural and functional alterations of FLT3 in acute myeloid leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:4263–4269. doi: 10.1158/1078-0432.CCR-08-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kikushige Y, Yoshimoto G, Miyamoto T, et al. Human Flt3 is expressed at the hematopoietic stem cell and the granulocyte/macrophage progenitor stages to maintain cell survival. J Immunol. 2008;180:7358–7367. doi: 10.4049/jimmunol.180.11.7358. [DOI] [PubMed] [Google Scholar]
- 4.Mackarehtschian K, Hardin JD, Moore KA, Boast S, Goff SP, Lemischka IR. Targeted disruption of the flk2/flt3 gene leads to deficiencies in primitive hematopoietic progenitors. Immunity. 1995;3:147–161. doi: 10.1016/1074-7613(95)90167-1. [DOI] [PubMed] [Google Scholar]
- 5.Buza-Vidas N, Cheng M, Duarte S, Charoudeh HN, Jacobsen SE, Sitnicka E. FLT3 receptor and ligand are dispensable for maintenance and posttransplantation expansion of mouse hematopoietic stem cells. Blood. 2009;113:3453–3460. doi: 10.1182/blood-2008-08-174060. [DOI] [PubMed] [Google Scholar]
- 6.Kindler T, Lipka DB, Fischer T. FLT3 as a therapeutic target in AML: still challenging after all these years. Blood. 2010;116:5089–5102. doi: 10.1182/blood-2010-04-261867. [DOI] [PubMed] [Google Scholar]
- 7.Fathi A, Levis M. FLT3 inhibitors: a story of the old and the new. Current opinion in hematology. 2011;18:71–76. doi: 10.1097/MOH.0b013e3283439a03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armstrong SA, Kung AL, Mabon ME, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer cell. 2003;3:173–183. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 9.Armstrong SA, Staunton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 10.Stam RW, den Boer ML, Schneider P, et al. Targeting FLT3 in primary MLL-gene-rearranged infant acute lymphoblastic leukemia. Blood. 2005;106:2484–2490. doi: 10.1182/blood-2004-09-3667. [DOI] [PubMed] [Google Scholar]
- 11.Brown P, Levis M, Shurtleff S, Campana D, Downing J, Small D. FLT3 inhibition selectively kills childhood acute lymphoblastic leukemia cells with high levels of FLT3 expression. Blood. 2005;105:812–820. doi: 10.1182/blood-2004-06-2498. [DOI] [PubMed] [Google Scholar]
- 12.Becker MW, Jordan CT. Leukemia stem cells in 2010: current understanding and future directions. Blood reviews. 2011;25:75–81. doi: 10.1016/j.blre.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Ito K, Bernardi R, Morotti A, et al. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453:1072–1078. doi: 10.1038/nature07016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sloma I, Jiang X, Eaves AC, Eaves CJ. Insights into the stem cells of chronic myeloid leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2010;24:1823–1833. doi: 10.1038/leu.2010.159. [DOI] [PubMed] [Google Scholar]
- 15.Palmqvist L, Argiropoulos B, Pineault N, et al. The Flt3 receptor tyrosine kinase collaborates with NUP98-HOX fusions in acute myeloid leukemia. Blood. 2006;108:1030–1036. doi: 10.1182/blood-2005-12-007005. [DOI] [PubMed] [Google Scholar]
- 16.Albouhair S, Morgado E, Lavau C. Flt3 Does Not Play a Critical Role in Murine Myeloid Leukemias Induced by MLL Fusion Genes. PloS one. 2013;8:e72261. doi: 10.1371/journal.pone.0072261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgado E, Albouhair S, Lavau C. Flt3 is dispensable to the Hoxa9/Meis1 leukemogenic cooperation. Blood. 2007;109:4020–4022. doi: 10.1182/blood-2006-01-039586. [DOI] [PubMed] [Google Scholar]
- 18.Zeisig BB, Milne T, Garcia-Cuellar MP, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Molecular and cellular biology. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. The EMBO journal. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 22.Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guibal FC, Alberich-Jorda M, Hirai H, et al. Identification of a myeloid committed progenitor as the cancer-initiating cell in acute promyelocytic leukemia. Blood. 2009;114:5415–5425. doi: 10.1182/blood-2008-10-182071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 25.Pear WS, Miller JP, Xu L, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 26.Somervaille TC, Matheny CJ, Spencer GJ, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan M, Kanbe E, Peterson LF, et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 28.Yoshimoto G, Miyamoto T, Jabbarzadeh-Tabrizi S, et al. FLT3-ITD up-regulates MCL-1 to promote survival of stem cells in acute myeloid leukemia via FLT3-ITD-specific STAT5 activation. Blood. 2009;114:5034–5043. doi: 10.1182/blood-2008-12-196055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuki M, Schwable J, Steur C, et al. Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 2003;101:3164–3173. doi: 10.1182/blood-2002-06-1677. [DOI] [PubMed] [Google Scholar]
- 30.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 31.Thiel AT, Huang J, Lei M, Hua X. Menin as a hub controlling mixed lineage leukemia. BioEssays : news and reviews in molecular, cellular and developmental biology. 2012;34:771–780. doi: 10.1002/bies.201200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Krivtsov AV, Sinha AU, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faber J, Krivtsov AV, Stubbs MC, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hawley TS, Fong AZ, Griesser H, Lyman SD, Hawley RG. Leukemic predisposition of mice transplanted with gene-modified hematopoietic precursors expressing flt3 ligand. Blood. 1998;92:2003–2011. [PubMed] [Google Scholar]
- 35.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nature reviews Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 36.Jiang X, Huang H, Li Z, et al. Blockade of miR-150 Maturation by MLL-Fusion/MYC/LIN-28 Is Required for MLL-Associated Leukemia. Cancer cell. 2012;22:524–535. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choudhary C, Brandts C, Schwable J, et al. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood. 2007;110:370–374. doi: 10.1182/blood-2006-05-024018. [DOI] [PubMed] [Google Scholar]
- 38.Choudhary C, Olsen JV, Brandts C, et al. Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Molecular cell. 2009;36:326–339. doi: 10.1016/j.molcel.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt-Arras D, Bohmer SA, Koch S, et al. Anchoring of FLT3 in the endoplasmic reticulum alters signaling quality. Blood. 2009;113:3568–3576. doi: 10.1182/blood-2007-10-121426. [DOI] [PubMed] [Google Scholar]
- 40.Avagyan S, Glouchkova L, Choi J, Snoeck HW. A quantitative trait locus on chromosome 4 affects cycling of hematopoietic stem and progenitor cells through regulation of TGF-beta 2 responsiveness. J Immunol. 2008;181:5904–5911. doi: 10.4049/jimmunol.181.9.5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avagyan S, Aguilo F, Kamezaki K, Snoeck HW. Quantitative trait mapping reveals a regulatory axis involving peroxisome proliferator-activated receptors, PRDM16, transforming growth factor-beta2 and FLT3 in hematopoiesis. Blood. 2011;118:6078–6086. doi: 10.1182/blood-2011-07-365080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.