Abstract
CD28 is one of the most important costimulatory receptors necessary for full T lymphocyte activation. The CD28 receptor can enhance T cell antigen receptor (TCR) signals, as well as deliver independent signals. Indeed, CD28 engagement by B7 can generate TCR-independent signals leading to IκB kinase and NF-κB activation. Here we demonstrate that the TCR-independent CD28 signal leads to the selective transcription of survival (Bcl-xL) and inflammatory (IL-8 and B cell activation factor, but not proliferative (IL-2), genes, in a NF-κB-dependent manner. CD28-stimulated T cells actively secrete IL-8, and Bcl-xL up-regulation protects T cells from radiation-induced apoptosis. The transcription of CD28-induced genes is mediated by the specific recruitment of RelA and p52 NF-κB subunits to target promoters. In contrast, p50 and c-Rel, which preferentially bind NF-κB sites on the IL-2 gene promoter after anti-CD3 stimulation, are not involved. Thus, we identify CD28 as a key regulator of genes important for both survival and inflammation.
CD28 is one of the most important costimulatory receptors necessary for full T lymphocyte activation. Early studies on CD28 demonstrated that it provides a potent signal for the up-regulation of several cytokines, acting at the level of both transcription and message stability. More recent studies have evidenced that CD28 plays a key role in enhancing T cell activation by the T cell antigen receptor (TCR) (1). As an integral component of the immunological synapse, CD28 plays a critical role in the recruitment of signaling molecules to the TCR (2). In particular, it has been recently demonstrated that CD28 enhances close contact between T cells and antigen-presenting cells and facilitates TCR signal transduction by amplifying phospholipase Cγ1 activation and Ca2+ response (3). The consequence of this action is an augmentation of early TCR-mediated signal (4). Recent evidence showing that CD28 can mediate adhesion signals beyond costimulatory ones (3) has led to new interest and induced a new wave of investigation of the intriguing problems concerning the molecular mechanisms as well as the targets of CD28 as an independent signaling unit. CD28 stimulation can induce cytoskeletal rearrangements in T cells (5) or up-regulate IL-2–4 only when Vav and the adapter SLP-76 are overexpressed (6). However, the mediators and the molecular mechanisms, which govern the process whereby CD28 may deliver an autonomous signal, remain unknown. We have recently demonstrated that CD28 engagement by B7 can generate TCR-independent signals leading to IκB kinase (IKK) and NF-κB activation (7) and that Vav-1 acts as the upstream regulator of this signaling pathway (8).
NF-κB/Rel transcription factors are critical regulators of the positioning of immune responses to integrate information from both innate and adaptive immune signaling pathways. In mammals, this family consists of five members that form homo- and heterodimeric complexes including NF-κB1 (p50 and its precursor p105), NF-κB2 (p52 and its precursor p100), RelA (p65), RelB, and c-Rel. RelA, c-Rel, and RelB contain transcriptional activation domains (TADs) and can activate transcription. p50 and p52 lack a TAD and therefore may form transcriptionally active heterodimers in association with RelA, c-Rel, and RelB (9). NF-κB activity is regulated by inhibitory proteins belonging to the IκB family, including IκBα,IκBβ, and IκBε. A protein kinase complex, known as the IKK signalsome, phosphorylates IκBα, IκBβ, and IκBε, thus leading to their proteolytic degradation and the release of NF-κB into the nucleus. The IKK complex contains two serine kinases, IKKα and IKKβ, and a third subunit, IKKγ/NEMO, with regulatory functions (10–15). In addition to the classical pathway of IκB degradation, NF-κB can be up-regulated by upstream signaling pathways involving the processing of the NF-κB2/p100 precursor.
Recent data show that IKKα can activate a second evolutionarily conserved pathway leading to NF-κB activation through the phosphorylation-dependent degradation of NF-κB2 molecules and the release of the RelB/p52 dimer (16). This noncanonical pathway is involved in the expression of several chemokine genes that regulate inflammation, lymphoid organ development, and homeostasis (17, 18). We demonstrated that in T cells CD28 activated NF-κB, cooperating with Vav-1 and IKKα (19). The effect of CD28 on the noncanonical NF-κB2 pathway has not been tested. This is an important issue for the potential role that CD28 target genes may have in normal and pathological conditions.
In this article we show that, in primary T cells, CD28 engagement by B7 and in the absence of any other signal induces the transcriptional activation of IL-8, Bcl-xL, and B cell activation factor (BAFF) genes in a NF-κB-dependent manner. CD28-mediated transcriptional activation of IL-8 and Bcl-xL genes was also accompanied by active secretion of IL-8 and up-regulation of Bcl-xL protein expression. The ability of CD28 to up-regulate Bcl-xL expression correlated with increased resistance to radiation-induced cell death. Furthermore, we find that CD28 stimulation leads to the preferential nuclear translocation of RelA and p52. Consistently, chromatin immunoprecipitation (ChIP) experiments revealed that p52 and RelA, but not RelB and p50, are recruited to CD28 target promoters. In contrast, RelA and p52 subunits are not recruited to the NF-κB site on the IL-2 promoter, which preferentially binds RelA and p50 after anti-CD3 stimulation. These data evidence that, when engaged separately from TCR, CD28 activates distinct NF-κB subunits regulating a definite subset of genes.
Materials and Methods
T Cells, Antibodies, and Reagents. Human primary CD4+ T cells were enriched from peripheral or cord blood mononuclear cells by MACS microbead sorting (Miltenyi Biotec, Milan) and cultured in RPMI medium 1640 supplemented with 5% human serum (Euroclone, Wetherby, England), l-glutamine, penicillin, and streptomycin (GIBCO/BRL). The purity of the sorted population was 95–99%. Murine L cells and L cells expressing human B7.1 (Dap3/B7) were previously described (20). CTLA4–Ig fusion protein was purchased from Chimerigen (Allston, MA). Anti-p65/RelA (C-20), anti-RelB (C19), anti-c-Rel (B6), anti-RNA polymerase II, anti-Bcl-xL (H-5), and anti-actin antibodies were purchased from Santa Cruz Biotechnology. Anti-NF-κB1/p50 (06-886) and anti-NF-κB2/p52 (05-361) were purchased from Upstate Biotechnology (Lake Placid, NY). Anti-human CD28.2, IKKα, and anti-CD3 (UCHT1) antibodies were from BD Pharmingen. NF-κB inhibitors used were Calpain inhibitor I (N-acetyl-leucyl-leucyl-norleucinal, ALLnL; Sigma), PGA1 (Cayman Chemical, Ann Arbor, MI), aspirin (Alexis, Lausen, Switzerland), and l-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK; Sigma).
Cell Stimulation, Immunoprecipitation, and Immunoblotting. T cells (30 × 106) were stimulated with adherent Dap3/B7 cells for the times indicated previously (8), and both cytoplasmic and nuclear extracts were prepared as previously described (8). Extracts were precleared for 1 h with protein A Sepharose (Amersham Pharmacia) and then immunoprecipitated for 2 h with specific antibodies preadsorbed on protein A/G Sepharose beads. Proteins were resolved by 8% SDS/PAGE and blotted onto nitrocellulose membranes. Blots were incubated with the indicated primary antibodies, extensively washed, and, after incubation with horseradish peroxidase (HRP)-labeled goat anti-rabbit or HRP-labeled goat anti-mouse (Amersham Pharmacia), developed with the enhanced chemiluminescence detection system (Amersham Pharmacia).
Semiquantitative RT-PCR. The PCR mixture contained 50 mM KCl, 10 mM Tris·HCl, 2.5 mM MgCl2, 0.2 mM dNTPs, and 0.2 μM 5′ and 3′ oligonucleotide primers; 2.5 units of Taq polymerase (Perkin–Elmer/Cetus) was amplified in 0.5-ml GeneAmp tubes in a final volume of 50 μl. PCRs were amplified by 35 cycles at 94°Cfor 1 min, 60°C for 30 s, and 72°C for 30 s. PCR was conducted in the automated DNA Thermal Cycler GeneAmp PCR System 2400 (Perkin–Elmer/Cetus). Primer sequences were as follows: Bcl-xL, 5′-ATTGGTGAGTCGGATCGCAGC-3′ and 5′-AGAGAAGGGGGTGGGAGGGTA-3′; BAFF, 5′-AGTCAGAACAGCAGAAATAAGCGT-3′ and 5′-TGAATTAGATGTCCCATGGCGTA-3′; and IL-2, 5′-ATGTACAGGATGCAACTCCTGTCTT-3′ and 5′-GCTAGTGTTGAGATGATGCTTTGAC-3′. GAPDH-, IL-8-, and CCR7-specific primers have been previously described (21, 22). PCR products were size-fractionated by agarose electrophoresis and normalized according to the amount of GAPDH detected in each sample.
ChIP Assay. ChIP assays were performed as previously described (23). Briefly, after fixing in 1% formaldehyde, T cells were lysed for 5 min in 50 mM Tris (pH 8.0), 2 mM EDTA, 0.1% Nonidet P-40, and 10% glycerol supplemented with protease inhibitors. Nuclei were resuspended in 50 mM Tris (pH 8.0), 1% SDS, and 5 mM EDTA. Chromatin was sheared by sonication, centrifuged, and diluted 10 times in 50 mM Tris (pH 8.0), 0.5% Nonidet P-40, 0.2 M NaCl, and 0.5 mM EDTA. After preclearing with a 50% suspension of salmon sperm-saturated protein A, lysates were incubated at 4°C overnight with the indicated antibodies. Immune complexes were collected with sperm-saturated protein A, washed three times with high-salt buffer [20 mM Tris (pH 8.0), 0.1% SDS, 1% Nonidet P-40, 2 mM EDTA, and 500 mM NaCl] and five times with TE buffer (10 mM Tris/1 mM EDTA). Immune complexes were extracted in TE buffer containing 1% SDS, and protein–DNA cross-links were reverted by heating at 65°C overnight. DNA was extracted by phenol/chloroform, and ≈1/20th of the immunoprecipitated DNA was used in each PCR. The primers used were as follows: IL-2 promoter, 5′-GAGGGATTTCACCTACATCCA-3′ and 5′-TGCAATGCAAGACAGGAGTT-3′; and Bcl-xL promoter, 5′-GCACCACCTACATTCAAATCC-3′ and 5′-CGATGGAGGAGGAAGCAAGC-3′. Sequences of IL-8 promoter-specific primers were kindly provided by G. Natoli (Institute for Research in Biomedicine, Bellinzona, Switzerland) and are available on request.
Results
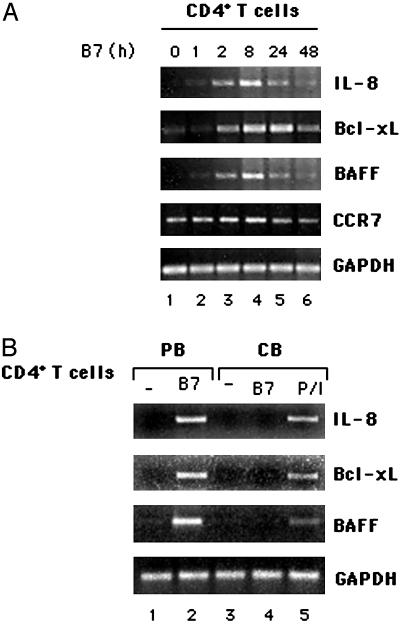
CD28 Engagement by B7 Induces the Transcription of NF-κB-Regulated Survival and Chemokine Genes in Primary CD4+ T Cells. NF-κB has been termed the central mediator of immune responses. Indeed, the NF-κB family promotes the expression of >100 target genes encoding antiapoptotic molecules, cytokines, and chemokines (24). In this context, we have recently demonstrated that CD28 can deliver a unique signal leading to the activation of NF-κB transcription factor in the absence of TCR (7, 8). Thus, we assessed the effect of CD28 stimulation on the expression of survival and chemokine genes likely regulated by NF-κB. We found that CD28 engagement by B7 induced a significant up-regulation of Bcl-xL, IL-8, and BAFF mRNAs with very similar kinetics. Indeed, the transcription of these genes was induced within 1–2 h of stimulation, reached a maximum between 8 and 18 h, and started to decrease after 24 h (Fig. 1A). Conversely, other chemokine genes, such as CCL21, CCL19, and SDF-1α were not induced (data not shown), and the basal expression of CCR7 was not increased after CD28 engagement (Fig. 1A). The expression of all these genes was not observed when B7-negative murine fibroblasts were used as stimulators, was induced after stimulation of T cells with anti-CD28 antibodies, and was inhibited by adding CTLA4–Ig (Fig. 7, which is published as supporting information on the PNAS web site). The majority of CD4+ T cells from peripheral blood of adult human individuals are memory cells (≈60–70%), and the strong responses observed to CD28 ligation may occur primarily in memory CD4+ T cells. To assess this issue we analyzed IL-8, Bcl-xL, and BAFF mRNAs in CD4+ T cells from cord blood, which are primarily naive T cells (>90%). CD28 ligation failed to activate transcription of target genes in naive CD4+ T cells (Fig. 1B), thus indicating that CD28 alone induces the expression of target genes only in memory cells. CD28-induced gene expression was accompanied by both IL-8 secretion and the increase of Bcl-xL protein levels (Fig. 8, which is published as supporting information on the PNAS web site). The up-regulation of Bcl-xL induced by CD28 stimulation in the absence of TCR engagement was related to protection from radiation-induced apoptosis (Fig. 9, which is published as supporting information on the PNAS web site). These data evidence a correlation between the capacity of CD28 itself to induce the expression of Bcl-xL and to protect T cells from radiation-induced apoptosis.
Fig. 1.
CD28 engagement by B7 induces IL-8, Bcl-xL, and BAFF mRNA expression in primary human T cells. (A) RT-PCR analysis of IL-8-, Bcl-xL-, BAFF-, and CCR7-specific mRNA expressions in human primary T cells stimulated for different times with adherent Dap3/B7 cells. (B) Peripheral and cord blood CD4+ T cells were stimulated for 8 h with adherent Dap3/B7 cells (lanes 1–4). Cord blood CD4+ T cells were also stimulated with phorbol 12-myristate 13-acetate (PMA, 10 ng/ml) plus ionomycin (0.5 μg/ml) as positive control (P/I, lane 5). IL-8, Bcl-xL, and BAFF mRNA expressions were analyzed by RT-PCR. Data represent three independent experiments.
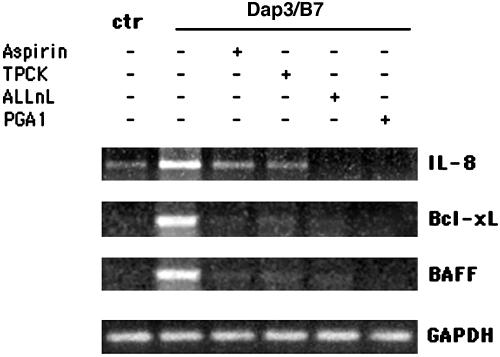
To verify whether CD28-induced transcriptional activation of target genes depended on NF-κB activity, we used different NF-κB inhibitors (25–28). Although they did so at different extents, all NF-κB inhibitors used were able to interfere with CD28-induced increase of mRNA expression of IL-8, Bcl-xL, and BAFF (Fig. 2). These findings represent evidence that CD28 can deliver a signal leading to the induction of NF-κB regulated genes and the expression of the relative proteins.
Fig. 2.
Inhibitors of the NF-κB pathway interfere with CD28-induced gene expression. Human T cells were pretreated for 2 h with 10 mM aspirin, 25 μM l-1-tosylamido-2-phenylethyl chloromethyl (TPCK), 10 μM N-acetyl-leucylleucyl-norleucinal (ALLnL), or 30 μM PGA1 and then stimulated for 6 h with adherent Dap3/B7 cells. IL-8, Bcl-xL, and BAFF mRNA expressions were analyzed by RT-PCR. Data represent three independent experiments.
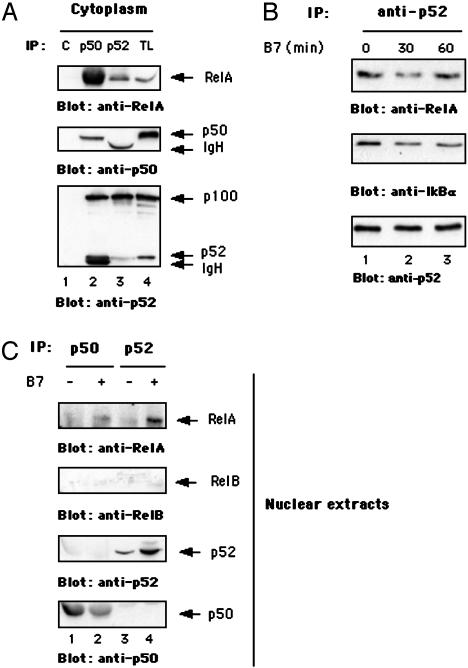
CD28 Engagement by B7 Induces the Nuclear Translocation of RelA and p52. To characterize the NF-κB proteins involved in the transcription of CD28 target genes, CD4+ human primary T lymphocytes were stimulated for different times with adherent Dap3/B7 cells. We first analyzed the cytoplasmic degradation of both IκBα and p100. As shown in Fig. 3, CD28 engagement by B7 induced a partial degradation of IκBα that began 15 min after stimulation; IκBα returned to a normal level 2 h later (Fig. 3A Middle). No changes in IκBα degradation were observed at later times (Fig. 3B Middle), although a second protein likely to be a cleaved form of IκBα was detected by the anti-IκBα antibody. In contrast, no detectable variation in p100 processing was observed at any time (Fig. 3 A and B Top), which may be a result of the high basal level of p100 process cytoplasm of unstimulated T cells (Fig. 3 A and B Top, leftmost lanes). We next examined the nuclear translocation of several NF-κB subunits. We found that CD28 stimulation induced a significant nuclear translocation of both RelA and p52 that increased within 30 min and remained high over 24 h (Fig. 4). A slight increase in the nuclear level of p50 was observed 30 min after stimulation. Conversely, no significant increase in the nuclear levels of RelB and c-Rel was detected. c-Rel transcription factor has been known to bind to and activate the CD28 response element (RE) within the IL-2 promoter, and CD28 costimulation accelerates the kinetics of c-Rel nuclear translocation (29). Thus, the total absence of c-Rel in the nucleus of CD28-stimulated T cells indicates that CD28, when engaged by B7 independently of TCR, can activate a distinct pool of NF-κB subunits, potentially involved in the regulation of a different subset of genes. To check the purity of our nuclear extracts, anti-IKKα immunoblotting was performed (Fig. 4 Bottom).
Fig. 3.
CD28 induces the early degradation of IκBα without affecting p100 processing. Cytoplasmic extracts from human primary T cells stimulated with adherent Dap3/B7 cells were analyzed by Western blotting with anti-IκBα- and anti-p52 antibodies. (A Middle) An early and partial degradation of IκBα was observed after CD28 engagement. (A and B Top) In contrast, no detectable p100 processing was found at any time. The blots were reprobed with anti-RelA antibodies to verify equal loading of proteins. Data represent three independent experiments.
Fig. 4.
CD28 ligation induces nuclear translocation of RelA and p52. Nuclear fractions from human T cells stimulated with adherent Dap3/B7 cells for different times were analyzed by Western blotting for the presence of RelA, p52, p50, RelB, and c-Rel. Expression of IKKα was analyzed to verify the purity of the nuclear extracts. Data represent three independent experiments.
RelA-containing NF-κB dimers are normally retained in the cytoplasm by the IκB subunits, whose processing is regulated by the canonical NF-κB pathway (30). After degradation of IκB subunits, RelA-containing dimers translocate into the nucleus. In contrast, RelB/p52 nuclear translocation has been known to depend on the processing of p100 (16, 18). Because we did not observe detectable cytoplasmic degradation of p100 after CD28 stimulation (Fig. 3), we verified the possibility that the release of cytoplasmic p52 could be associated with the proteolysis of IκBα. Thus, we analyzed the presence of RelA/p52 complexes in the cytoplasm of T cells. We found that RelA forms heterodimers with both p100/p52 and p50 (Fig. 5A Top, lanes 2 and 3). A fraction of cytoplasmic p50 was also associated with p100/p52 (Fig. 5A Bottom, lane 2), which may account for the slight and transient increase of nuclear p50 levels observed 30 min after stimulation (see Fig. 4 and below). RelA/p52 complexes contained IκBα that was partially degraded after CD28 stimulation (Fig. 5B Middle, lanes 2 and 3 vs. lane 1). Immunoprecipitation of p52 from the nuclear extracts of T cells demonstrated that CD28 stimulation triggered an increase in nuclear RelA/p52, but not RelB/p52, dimers (Fig. 5C, lane 4 vs. lane 3). Moreover, although cytoplasmic RelA/p50 complexes were more abundant than RelA/p52 dimers (Fig. 5A Top, lane 2 vs. lane 3), RelA was not detected in p50 immunoprecipitates from the nuclear extracts of CD28-stimulated T cells (Fig. 5C, lane 2 vs. lane 1). Altogether these data indicate that CD28 triggers the nuclear translocation of preexisting RelA/p52 heterodimers.
Fig. 5.
CD28 engagement by B7 induces the nuclear translocation of preexisting RelA/p52 heterodimers. (A) Cytoplasmic extracts from human primary T cells were subjected to immunoprecipitation (IP) with unrelated antibody (C) or anti-p50- or anti-p52-specific antibodies. Immunoprecipitations and total lysates (TL) were analyzed for the presence of RelA, p52, and p50 by Western blotting. (B) Cytoplasmic extracts from human T cells, stimulated for 30 and 60 min with adherent Dap3/B7 cells, were immunoprecipitated with anti-p52 antibody. Anti-IκBα, -RelA, and -p52 Western blotting was performed. (C) Human T cells were stimulated for 30 min with adherent Dap3/B7 cells, and both RelA and RelB Western blotting was performed on nuclear anti-p50 and anti-p52 immunoprecipitates. Each immunoprecipitate was analyzed for p50 and p52 content. Data represent three independent experiments.
IL-8 and Bcl-xL Promoters Selectively Recruit RelA and p52 After CD28 Stimulation. We next analyzed the recruitment of NF-κB subunits to IL-8 and Bcl-xL genes induced by CD28 in primary T cells. To this aim we used ChIP assays. Interestingly, both IL-8 (Fig. 6A) and Bcl-xL promoters (Fig. 6B) recruited p52 and RelA in the early phase of CD28-induced transcriptional activation, which was monitored by analyzing the promoter occupancy by RNA polymerase II. p52 recruitment to both promoters was transient and completely absent 2 h after stimulation. In contrast, RelA was further recruited to IL-8 and Bcl-xL promoters after p52 removal and persisted for a longer time(>8 h). Because RNA polymerase II was still bound on both promoters after p52 replacement, it is conceivable that RelA is involved in supporting their transcription. Indeed, the IL-8 promoter has been described as highly responsive to RelA homodimers in vitro (31) and has been known to selectively recruit RelA in vivo in lipopolysaccharide-stimulated dendritic cells (23). However, the IL-8 promoter has also been shown to bind p52 (31). Our data evidence that RelA/p52 heterodimers are recruited to the IL-8 promoter in the early phase of CD28-induced transcriptional activation and then are rapidly replaced by RelA homodimers. Clearly we cannot exclude the possibility that RelA/p52 heterodimers and RelA homodimers bind to distinct NF-κB sites on the same promoter. A similar kinetic was observed for the Bcl-xL promoter (Fig. 6B). No RelB or p50 recruitment to the IL-8 or Bcl-xL promoter was observed in CD28-stimulated T cells. Moreover, according to the inability of CD28 stimulation alone to activate IL-2 transcription (4, 20), neither RelA nor p52 was found on the IL-2 promoter (Fig. 6C), which contains a canonical NF-κB binding site but is not activated after CD28 stimulation alone (Fig. 6C Right). In contrast, when T cells were stimulated with anti-CD3 antibody, RelA, c-Rel, and p50 were preferentially recruited to the IL-2 promoter (Fig. 6D). Moreover, in contrast to CD28 stimulation alone, CD3 stimulation alone was not able to induce the transcription of IL-8 and Bcl-xL genes (Fig. 6D Right) or the recruitment of NF-κB subunits to their promoters (data not shown).
Fig. 6.
CD28 stimulation of human T cells induces the selective recruitment of RelA and p52 to the IL-8 and Bcl-xL promoters. Human primary T cells were stimulated with adherent Dap3/B7 cells for different times. ChIP assays were performed by using anti-RelA, -p52, -RelB, -p50 and -RNA polymerase II (pol II) antibodies or no antibody (NA) as control. (A and B) Immunoprecipitated DNA was analyzed by PCR with IL-8 (A) and Bcl-xL (B) promoter-specific primers. (C) RelA and p52 recruitment to IL-2 promoter in human T cells stimulated with Dap3/B7 for different times. (Right) The kinetics of the induction of IL-2 mRNA (evaluated by RT-PCR) is shown. (D) RelA, p50, c-Rel, or p52 recruitment to the IL-2 promoter in human T cells stimulated with anti-CD3 (UCHT1) antibody. (Right) The kinetic of induction of IL-2, IL-8, and Bcl-xL mRNAs (evaluated by RT-PCR) is shown. Data represent three independent experiments.
These findings indicate that CD28 leads to the nuclear translocation of both RelA/p52 heterodimers and RelA homodimers with functional transcriptional activity on the IL-8 and Bcl-xL promoters.
Discussion
The existence of autonomous CD28 signaling was proposed by Schwartz (32) in 1992, when he observed that CD28 was able to act as a TCR-independent signaling unit and to deliver in trans costimulation. In an attempt to identify and characterize the signaling pathways activated by CD28 in a TCR-independent manner, we found that NF-κB is the most relevant CD28 biochemical target (7, 8, 19). The importance of NF-κB for T cell costimulation has been emphasized for several years (33). However, the significance of CD28-mediated NF-κB activation, as well as the identification of NF-κB-regulated genes, remained undefined. In this article we present evidence that CD28 can deliver a unique signal leading to the up-regulation of NF-κB-regulated genes. Indeed, we demonstrate that after the engagement of CD28 by its natural ligand, B7, Bcl-xL, IL-8, and BAFF genes are transcribed in primary T cells. In contrast, other known NF-κB-regulated chemokine genes are not induced. The functional consequence of CD28-mediated IL-8 and Bcl-xL gene expression is the secretion of IL-8 and up-regulation of Bcl-xL protein levels. Moreover, T cells that have received CD28 signal are more resistant to radiation-induced cell death. We also show that the induction of these genes occurs through the sequential recruitment of RelA/p52 heterodimers and RelA homodimers. The former are likely to be involved in the initiation of transcription, and the latter in sustaining the activation of both genes. Transcriptional active RelA/p52 heterodimers have been observed in B cells after CD40 and lipopolysaccharide stimulation (17, 18). Here we demonstrate that this complex is active also in human T cells after CD28 stimulation.
The human T cell leukemia virus Tax protein (34, 35) that specifically activates IKKα (36) induces both IL-8 and Bcl-xL genes. Lipopolysaccharide-induced BAFF expression is lost in IKKαAA knockin mice (18). In this context, we have recently demonstrated that a complex containing Vav-1 and IKKα is involved in CD28-mediated NF-κB activation (19). CD28-induced RelA and p52 nuclear translocations are associated with partial IκBα degradation, whereas p100 processing is not detected (Fig. 2). A common concept is that the canonical (RelA) and noncanonical (p52) pathways are independent and are regulated by different components of the IKK complex. Indeed, the release of RelA-containing dimers depends on the β and γ subunits of the IKK complex (10–15). In contrast, p52/RelB nuclear translocation requires the processing of its precursor, p100, which is mediated by IKKα (16). Furthermore, IKKα may induce IκB degradation in response to RANK ligand (37). One possibility is that CD28-mediated activation of IKKα regulates the preferential degradation of RelA/p52-associated IκBα by directly phosphorylating IκBα (37). This event would lead to the nuclear translocation of RelA/p52-containing, but not RelA/p50-containing, dimers, which are commonly released after IKKβ-mediated degradation of IκBα (10–15). Indeed, cytoplasmic RelA/p50 dimers are more abundant than RelA/p52 complexes (Fig. 5A). In contrast, in the nucleus of CD28-stimulated T cells, RelA/p52 heterodimers were preferentially found (Fig. 5C). Alternatively, CD28 may mediate a slow and persistent degradation of p100, undetectable by our anti-p52 antibody, especially at the high basal level of p100 processing observed in unstimulated T cells, (Fig. 3). Clearly, only when efficient techniques to specifically silence IKKα and IKKβ genes in primary human T cells are developed will the specific role of these kinases in receptor-mediated NF-κB activation be elucidated.
The ability of CD28 to deliver survival signals through Bcl-xL in primary T cells has been known since 1995 (38). The NF-κB signaling pathway regulates CD28-induced Bcl-xL expression (39), but the NF-κB proteins involved in binding to the Bcl-xL promoter after CD28 stimulation remain undefined. Recent data from Zheng et al. (40) reveal that in double p50-/-/c-Rel-/- mice, TCR-induced T cell survival is impaired. We have found that p50 and c-Rel are not involved in CD28-induced Bcl-xL expression, which, conversely, is regulated by RelA/p52 heterodimers and RelA homodimers (Fig. 6B). This apparent discrepancy reinforces the notion that, when engaged separately from the TCR, CD28 leads to the activation of a distinct signaling cascade. Indeed, TCR-mediated regulation of NF-κB has been described as independent of serine/threonine kinase protein kinase B (PKB/AKT) (40). AKT is activated by CD28 in a phosphatidylinositol 3-kinase (PI3-kinase)-dependent manner (41, 42) and has been recently implicated in CD28-induced Bcl-xL expression (43). AKT-dependent activation of NF-κB relies on its ability to stimulate the transactivation potential of RelA (44). Although the molecular mechanisms by which AKT activates NF-κB in T cells are not known, evidence suggests that it may provide the unique CD28 signal leading to the regulation of survival genes (45).
IL-8 is a CXC chemokine that acts as a potent chemotactic factor for neutrophils, granulocytes, and T lymphocytes and is produced by different cell types, including T lymphocytes. IL-8 is produced by primary human T cells and T cell lines in a CD28-dependent fashion, and the NF-κB-like sequence in the promoter of the IL-8 gene functions as a CD28 RE (46). However, these data were obtained after combined stimulation of T cells with anti-CD3 plus anti-CD28 antibodies, and no detailed analysis of the NF-κB subunits involved in the in vivo transcription of the IL-8 promoter was made. The sequence of the CD28 RE in the IL-8 promoter has a striking similarity to that of the CD28 RE in the IL-2 promoter (46). However, in vitro experiments evidenced that IL-8 CD28 RE sequence differed from IL-2 CD28 RE sequence in the binding of NF-κB/Rel family proteins (47). The IL-2 CD28 RE, which has been known to bind in vitro c-Rel, RelA, and p50 (48), does not recruit RelA/p52 and RelA dimers after CD28 stimulation alone. In contrast, when T cells are stimulated with anti-CD3 antibody, the IL-2 promoter specifically recruits RelA, c-Rel, and p50 subunits in vivo as detected by ChIP assays (Fig. 6). Moreover, CD3 stimulation alone does not lead to the transcriptional activation of IL-8 and Bcl-xL genes. These data evidence that CD3 and CD28 differentially use the NF-κB machinery to achieve the transcription of specific genes. Saccani et al. (23) have recently shown that the IL-8 promoter selectively recruits RelA in lipopolysaccharide-stimulated dendritic cells. Similarly, we found that RelA is implicated in both the initiation and propagation of IL-8 transcription.
The exchange of NF-κB dimers at target promoters has been recently identified in dendritic cells as a mechanism to modulate NF-κB-dependent transcription (23). Depending on the stimulus used, the sequential recruitment of dimers to target promoters is a strategy that the system may use to reach maximal and sustained NF-κB-dependent gene expression, as well as to down-regulate transcription. Our findings that RelA/p52 dimers occupy IL-8 and Bcl-xL promoters in the early phase of transcription (whereas RelA homodimers are recruited later) suggest that both complexes may be sequentially recruited at the same NF-κB site. Alternatively, dimer exchange may occur at different NF-κB sites. In this case, RelA/p52 heterodimers are recruited early to a NF-κB site, and after removal RelA homodimers bind to an adjacent but distinct NF-κB site (23). The early recruitment of RelA/p52 may likely be essential for the correct assembly of a higher-order, three-dimensional transcription factor/enhancer DNA complex, termed enhanceosome, that leads to chromatin remodeling and stimulates the start of transcription, a role recently suggested for c-Rel at the IL-2 promoter (49). It is known that RelA homodimers, compared with heterodimers, poorly bind DNA (50). RelA/p52 heterodimers are then rapidly replaced by RelA homodimers, which have higher transcriptional activity on the IL-8 promoter through their enhancement of the recruitment of histone-modifying enzymes (23, 31).
In the last few years, plentiful data have accumulated characterizing chemokines and the receptors necessary to turn an innate immune response into an adaptive one. Less is known about the production by T lymphocytes of chemokines recruiting both innate and adaptive immune cells to the tissue. Particularly rare is information on the signals that activate chemokine transcription in T lymphocytes. This is an important issue, because it is a common opinion that chemokines are involved in the pathology of various autoimmune diseases (i.e., rheumatoid arthritis and psoriasis) and in cancer, thus representing a reasonable therapeutic target (51). Therefore, the management of these disorders is particularly linked to the knowledge of the mechanism that allows T lymphocytes to produce chemokines, which favor leukocyte activation/accumulation in the tissue of the target organ and maintain the inflammatory response associated with immune disorders. We provide evidence that CD28 stimulation selectively regulates proinflammatory and antiapoptotic genes without affecting proliferative gene transcription. These data may further understanding of the mechanisms regulating the accumulation of memory-infiltrating T cells under normal and pathological conditions and may have heuristic implications for therapeutic applications. Indeed, IL-8 and BAFF have been involved in both autoimmunity and cancer (52, 53). In this context, Shapiro et al. (54) have shown that the expression of endogenous CD28 in multiple myeloma was able to up-regulate the transcription of the IL-8 gene, thus suggesting a potential role in the progression of multiple myeloma. Finally, the blockade of CD28/B7 interaction with CTLA4–Ig is underway in clinical trials aimed to modulate T cell functions in rheumatoid arthritis patients (55).
Supplementary Material
Acknowledgments
We thank E. Dejardin for suggestions; G. Natoli and D. R. Green for critical reading and helpful discussion; and M. Framarino dei Malatesta (University of Rome La Sapienza) for providing cord blood. This work was supported in part by Istituto Pasteur Fondazione Cenci Bolognetti, University of Rome La Sapienza, and the Ministry of Universities and Scientific and Technological Research (MIUR-COFIN).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TCR, T cell antigen receptor; IKK, IκB kinase; BAFF, B cell activator factor; ChIP, chromatin immunoprecipitation; RE, response element.
References
- 1.Lanzavecchia, A., Iezzi, G. & Viola, A. (1999) Cell 96, 1-4. [DOI] [PubMed] [Google Scholar]
- 2.Wulfing, C. & Davis, M. M. (1998) Science 282, 2266-2269. [DOI] [PubMed] [Google Scholar]
- 3.Michel, F., Attal-Bonnefoy, G., Mangino, G., Mise-Omata, S. & Acuto, O. (2001) Immunity 15, 935-945. [DOI] [PubMed] [Google Scholar]
- 4.Tuosto, L. & Acuto, O. (1998) Eur. J. Immunol. 28, 2131-2142. [DOI] [PubMed] [Google Scholar]
- 5.Kaga, S., Ragg, S., Rogers, K. A. & Ochi, A. (1998) J. Immunol. 160, 24-27. [PubMed] [Google Scholar]
- 6.Raab, M., Pfister, S. & Rudd, C. E. (2001) Immunity 15, 921-933. [DOI] [PubMed] [Google Scholar]
- 7.Tuosto, L., Costanzo, A., Guido, F., Marinari, B., Vossio, S., Moretti, S., Levrero, M. & Piccolella, E. (2000) Eur. J. Immunol. 30, 2445-2454. [DOI] [PubMed] [Google Scholar]
- 8.Marinari, B., Costanzo, A., Viola, A., Michel, F., Mangino, G., Acuto, O., Levrero, M., Piccolella, E. & Tuosto, L. (2001) Eur. J. Immunol. 32, 447-456. [DOI] [PubMed] [Google Scholar]
- 9.Verma, I. M., Stevenson, J. K., Schwarz, E. M., Van Antwerp, D. & Miyamoto, S. (1995) Genes Dev. 9, 2723-2735. [DOI] [PubMed] [Google Scholar]
- 10.DiDonato, J. A., Rothwarf, D. M., Zandi, E. & Karin, M. (1997) Nature 388, 548-554. [DOI] [PubMed] [Google Scholar]
- 11.Reigner, C. H., Song, H. Y., Gao, X., Goedell, D. V., Cao, Z. & Rothe, M. (1997) Cell 90, 373-383. [DOI] [PubMed] [Google Scholar]
- 12.Rothwarf, D. M., Zandi, E., Natoli, G. & Karin, M. (1998) Nature 395, 297-300. [DOI] [PubMed] [Google Scholar]
- 13.Woronicz, J. D., Gao, X., Cao, Z., Rothe, M. & Goeddel, D. V. (1997) Science 278, 866-869. [DOI] [PubMed] [Google Scholar]
- 14.Yamaoka, S., Courtois, G., Bessia, C., Whiteside, S. T., Weil, R., Agou, F., Kirk, H. E., Kay, R. J. & Israel, A. (1998) Cell 93, 1231-1240. [DOI] [PubMed] [Google Scholar]
- 15.Zandi, E., Rothwarf, D. M., Delhase, M., Hyakawa, M. & Karin, M. (1997) Cell 91, 243-252. [DOI] [PubMed] [Google Scholar]
- 16.Senftleben, U., Cao, Y., Xiao, G., Greten, F. R., Krahn, G., Bonizzi, G., Chen, Y., Hu, Y., Fong, A., Sun, S.-C. & Karin, M. (2001) Science 293, 1495-1499. [DOI] [PubMed] [Google Scholar]
- 17.Coope, H. J., Atkinson, P. G. P., Huhse, B., Belich, M., Janzen, J., Holman, M. J., Klaus, G. G. B., Johnston, L. H. & Ley, S. C. (2002) EMBO J. 21, 5375-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dejardin, E., Droin, N. M., Delhase, M., Haas, E., Cao, Y., Makris, C., Zhi-Wei, L., Karin, M., Ware, C. F. & Green, D. R. (2002) Immunity 17, 525-535. [DOI] [PubMed] [Google Scholar]
- 19.Piccolella, E., Spadaro, F., Ramoni, C., Marinari, B., Costanzo, A., Levrero, M., Thompson, L., Abraham, R. T. & Tuosto, L. (2003) J. Immunol. 170, 2895-2903. [DOI] [PubMed] [Google Scholar]
- 20.Michel, F., Mangino, G., Attal-Bonnefoy, G., Tuosto, L., Olive, D. & Acuto, O. (2000) J. Immunol. 165, 3820-3829. [DOI] [PubMed] [Google Scholar]
- 21.Dieu, M.-C., Vanbervliet, B., Vicari, A., Bridon, J.-M., Oldham, E., Ait-Yahia, S., Briere, F., Zlotnik, A., Lebecque, S. & Caux, C. (1998) J. Exp. Med. 188, 373-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatano, Y., Katagiri, K. & Takayasu, S. (1999) Clin. Exp. Immunol. 117, 237-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saccani, S., Pantano, S. & Natoli, G. (2003) Mol. Cell 11, 1563-1574. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh, S., May, M. J. & Kopp, E. B. (1998) Ann. Rev. Immunol. 16, 225-260. [DOI] [PubMed] [Google Scholar]
- 25.Henkel, T., Machleidt, T., Alkalay, I., Kronke, M., Ben-Neriah, Y. & Bauerle, P. A. (1993) Nature 365, 182-185. [DOI] [PubMed] [Google Scholar]
- 26.Lin, Y. C., Brown, K. & Siebenlist, U. (1995) Proc. Natl. Acad. Sci. USA 92, 552-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossi, A., Kapahi, P., Natoli, G., Takahashi, T., Chen, Y., Karin, M. & Santoro, M. G. (2000) Nature 403, 103-108. [DOI] [PubMed] [Google Scholar]
- 28.Yin, M.-J., Yamamoto, Y. & Gaynor, R. B. (1998) Nature 396, 77-80. [DOI] [PubMed] [Google Scholar]
- 29.Bryan, R. G., Li, Y., Lai, J. H., Van, M., Rice, N. R., Rich, R. R. & Tan, T. H. (1994) Mol. Cell. Biol. 14, 7933-7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karin, M. & Ben-Neriah, Y. (2000) Annu. Rev. Immunol. 18, 621-663. [DOI] [PubMed] [Google Scholar]
- 31.Kunsch, C. & Rosen, C. A. (1993) Mol. Cell. Biol. 13, 6137-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz, R. H. (1992) Cell 71, 1065-1068. [DOI] [PubMed] [Google Scholar]
- 33.Kane, L. P., Lin, Y. & Weiss, A. (2002) Trends Immunol. 23, 413-420. [DOI] [PubMed] [Google Scholar]
- 34.Mori, N., Mukaida, N., Ballard, D. W., Matsushima, K. & Yamamoto, N. (1998) Cancer Res. 58, 3993-4000. [PubMed] [Google Scholar]
- 35.Tsukara, T., Kannagi, M., Ohashi, T., Kato, H., Arai, M., Nunez, G., Iwanaga, Y., Yamamoto, N., Ohtani, K., Nakamura, M. & Fujii, M. (1999) J. Virol. 73, 7981-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao, G., Cvijic, M. E., Fong, A., Harhaj, E. W., Uhlik, M. T., Waterfield, M. & Sun, S.-C. (2001) EMBO J. 20, 6805-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao, Y., Bonizzi, G., Seagroves, T. N., Greten, F. R., Johnson, R., Schmidt, E. V. & Karin, M. (2001) Cell 107, 763-775. [DOI] [PubMed] [Google Scholar]
- 38.Boise, L. H., Minn, A. J., Noel, P. J., June, C. H., Accavitti, M. A., Lindsten, T. & Thompson, C. B. (1995) Immunity 3, 87-98. [DOI] [PubMed] [Google Scholar]
- 39.Khoshnan, A., Tindell, C., Laux, I., Bae, D., Bennett, B. & Nel, A. E. (2000) J. Immunol. 165, 1743-1754. [DOI] [PubMed] [Google Scholar]
- 40.Zheng, Y., Vig, M., Lyons, J., Parijis, L. V. & Beg, A. A. (2003) J. Exp. Med. 197, 861-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kane, L. P., Abdres, P. G., Howland, K. C., Abbas, A. K. & Weiss, A. (2001) Nat. Immunol. 2, 37-44. [DOI] [PubMed] [Google Scholar]
- 42.Parry, R. V., Reif, G., Smith, D. M., Sanson, B. A., Hemmings, B. A. & Ward, S. G. (1997) Eur. J. Immunol. 27, 2495-2501. [DOI] [PubMed] [Google Scholar]
- 43.Burr, J. S., Savage, N. D., Messah, G. E., Kimzey, S. L., Shaw, A. S., Arch, R. H. & Green, J. M. (2001) J. Immunol. 166, 5331-5335. [DOI] [PubMed] [Google Scholar]
- 44.Madrid, L. V., Mayo, M. W., Reuther, J. Y. & Baldwin, A. S. (2001) J. Biol. Chem. 276, 18934-18940. [DOI] [PubMed] [Google Scholar]
- 45.Jones, R. G., Elford, A. R., Parsons, M. J., Wu, L., Krawczky, C. M., Yeh, W.-C., Hakem, R., Rottapel, R., Woodgett, J. R. & Ohashi, P. S. (2002) J. Exp. Med. 196, 335-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wechsler, A. S., Gordon, M. C., Dendorfer, U. & Le Claire, K. P. (1994) J. Immunol. 153, 2515-2523. [PubMed] [Google Scholar]
- 47.Civil, A., Rensink, I., Aarden, L. A. & Verwei, C. L. (1999) J. Biol. Chem. 274, 34369-34374. [DOI] [PubMed] [Google Scholar]
- 48.Ghosh, P., Tan, T.-H., Rice, N. R., Sica, A. & Young, H. A. (1993) Proc. Natl. Acad. Sci. USA 90, 1696-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao, S., Gerondakis, S., Woltring, D. & Shannon, M. F. (2003) J. Immunol. 170, 3724-3731. [DOI] [PubMed] [Google Scholar]
- 50.Nolan, G. P., Ghosh, S., Liou, H.-C., Tempst, P. & Baltimore, D. (1991) Cell 64, 961-969. [DOI] [PubMed] [Google Scholar]
- 51.Baggiolini, M. (2001) J. Int. Med. 250, 91-104. [DOI] [PubMed] [Google Scholar]
- 52.Taylor, P. C., Williams, R. O. & Maini, R. N. (2001) Curr. Opin. Immunol. 13, 611-616. [DOI] [PubMed] [Google Scholar]
- 53.Ware, C. F. (2000) J. Exp. Med. 192, F35-F38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shapiro, V. S., Mollenauer, M. N. & Weiss, A. (2001) Blood 98, 187-193. [DOI] [PubMed] [Google Scholar]
- 55.Taylor, P. C. (2003) Curr. Opin. Pharmacol. 3, 323-328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.