Abstract
The unrestrained growth of tumor cells is generally attributed to mutations in essential growth control genes, but tumor cells are also influenced by signals from the environment. In multiple myeloma (MM), the factors and signals coming from the bone marrow microenvironment are possibly even essential for the growth of the tumor cells. As targets for intervention, these signals may be equally important as mutated oncogenes. Given their oncogenic potential, WNT signals form a class of paracrine growth factors that could act to influence MM cell growth. In this paper, we report that MM cells have hallmarks of active WNT signaling, whereas the cells have not undergone detectable mutations in WNT signaling genes such as adenomatous polyposis coli and β-catenin (CTNNB1). We show that the malignant MM plasma cells overexpress β-catenin, including its N-terminally unphosphorylated form, suggesting active β-catenin/T cell factor-mediated transcription. Further accumulation and nuclear localization of β-catenin, and/or increased cell proliferation, was achieved by stimulation of WNT signaling with either Wnt3a, LiCl, or the constitutively active S33Y mutant of β-catenin. In contrast, by blocking WNT signaling by dominant-negative T cell factor, we can interfere with the growth of MM cells. We therefore suggest that MM cells are dependent on an active WNT signal, which may have important implications for the management of this incurable form of cancer.
Keywords: β-catenin, T cell factor, lymphoma, plasma cell
Multiple myeloma (MM), one of the most common hematological malignancies in adults, is a neoplasm of terminally differentiated B cells; i.e., plasma cells. The tumor cells expand in the bone marrow (BM), ultimately leading to pancytopenia and osteolytic bone destruction. At present, the disease is still incurable, with a median survival of ≈3–4 years. The transition of a plasma cell to a fully transformed, aggressive myeloma is a multistep process, which requires the acquisition of mutations in several protooncogenes and tumor suppressor genes (1). Most of this evolution takes place in the BM, indicating that signals from the BM microenvironment, which may include paracrine growth factors, play a critical role in sustaining the growth and survival of MM cells during tumor progression (1, 2).
WNT signals form one class of paracrine growth factors that could act to influence MM cell growth. WNT signal transduction components, in particular adenomatous polyposis coli (APC), and β-catenin, are often mutated in cancers and sustained overexpression of WNT genes can cause cancer (3–6). In addition, WNT proteins themselves are able to promote the proliferation of progenitor or stem cells (7–10). WNT genes encode a family of 19 secreted glycoproteins, which promiscuously interact with several Frizzled receptors. This interaction leads to intracellular signals that control gene expression, cell behavior, cell adhesion, and cell polarity, during both embryonic development and postnatal life (11, 12). The key event in this signaling pathway is the stabilization of β-catenin. In the absence of WNT signals, a dedicated complex of proteins, including the tumor suppressor gene product APC, axin, and glycogen synthase kinase-3β (GSK3β) controls phosphorylation of specific serine and threonine residues in the N-terminal region of β-catenin. This GSK3β-mediated phosphorylation marks β-catenin for ubiquitination and degradation by the proteasome. Signaling by WNT factors blocks GSK3β activity, resulting in the accumulation of nonphosphorylated β-catenin, which will translocate to the nucleus. Here, it interacts with T cell factor (TCF) transcription factors (13, 14) to drive transcription of target genes (9, 15, 16). It is now well established that unrestrained β-catenin/TCF activity plays a major role in many human cancers (4, 5). Mutations of the APC tumor suppressor gene or of the sequences encoding the crucial GSK3β phosphorylation sites in the N-terminal domain of β-catenin have been found in the vast majority of colorectal cancers, as well as many other cancer types (4–6). The critical consequence of these mutations is the elevation of the levels of β-catenin, leading to the formation of constitutive nuclear β-catenin/TCF complexes and altered expression of TCF target genes (5, 9). Target genes which likely cooperate in neoplastic transformation include CCND1 (cyclin D1) (15), MYC (9, 15), CD44 (17), and MET (18).
Members of the TCF/LEF family of transcription factors were initially discovered in models of early lymphocyte development. In mice and humans, T lineage cells express both TCF1 and LEF1 (7, 19–22), and studies in knockout mice have demonstrated that these factors are essential for the maintenance of progenitor T cell compartments (7, 23, 24). Similarly, during early B cell development in the fetal liver and adult BM, LEF1 is involved in the regulation of pro-B cell proliferation and survival (8). By inference, these observations suggest a role for WNT signaling in the control of cell proliferation and survival during lymphocyte development. Indeed, recent in vitro and in vivo studies, demonstrating that WNT factors and β-catenin also affect lymphocyte progenitor fate as well as stem cell self-renewal, confirm this role (8, 10, 25, 26). The involvement of the WNT pathway in the regulation of the survival and expansion of progenitor and stem cells, in combination with its oncogenic potential in nonlymphoid cells, prompted us to test whether deregulation of the WNT pathway occurs in lymphoid neoplasia. Whereas the specificity of WNT signals with respect to target cells is relatively unknown, there are now powerful methods to examine whether cells are activated by a WNT signal. These tools include measuring the levels of the β-catenin protein, in particular, a nonphosphorylated form of β-catenin, that is generated by active WNT signaling (27, 28). In the nucleus, WNT signaling proceeds through the transcription factor TCF, and by interfering with TCF activity [using a dominant-negative form of TCF (ΔTCF4)], one can examine to what extend the behavior and growth of cells depends on an active WNT signal. Here, we show that WNT signaling is active in MMs and that WNT signals are involved in the control of MM growth.
Materials and Methods
Antibodies. Mouse mAbs used were: anti-CD138, BB4 (IgG1) (Instruchemie, Hilversum, The Netherlands); anti-β-catenin (clone 14, IgG1) (BD Biosciences, Erembodegem, Belgium); anti-active (nonphosphorylated) β-catenin (anti-ABC, IgG1) (27, 28); anti-active (nonphosphorylated) β-catenin (clone 8E4, IgG1) (Alexis Biochemicals, Lausanne, Switzerland); anti β-actin (clone AC15, IgG1) (Sigma); allophycocyanin-conjugated anti-CD19 (IgG1); FITC-conjugated anti-IgD (IgG1); FITC-conjugated anti-CD45RA (IgG2b) (all BD Biosciences); biotin-conjugated anti-CD38 (IgG1) (Caltag, Burlingame, CA). Polyclonal antibodies used were: rabbit anti-human ERK2 (C14, Santa Cruz Biotechnology); Dynabead-conjugated goat anti-mouse IgG (Dynal, Wirral, U.K.); horseradish peroxidase-conjugated rabbit anti-mouse; FITC-conjugated rabbit anti-mouse; and horseradish peroxidase-conjugated goat anti-rabbit (all DAKO).
Myelomas, Cell Cultures, and Transfections. Primary myeloma samples and normal BM samples were obtained during routine diagnostic procedures. Mononuclear cells were harvested by standard Ficoll/Paque density gradient centrifugation (Amersham Pharmacia Biosciences, Roozendaal, The Netherlands). Plasma cells were sorted by positive selection using anti-CD138 (clone BB4, Instruchemie) and Dynabead-conjugated goat anti-mouse IgG (Dynal). Positive sorting yielded populations plasma cells that were >97% pure (CD38+ and CD45RA-) as defined by fluorescence-activated cell sorter (FACS) analysis by using a FACScalibur flow cytometer (BD Biosciences). MM cell lines XG1 (29), LME1 (2), and UM6 (30) were cultured in Iscove's medium (Invitrogen Life Technologies, Breda, The Netherlands) as described (2). MM cell lines UM1 and UM3 (30), L363 (31), NCI H929 (32), and OPM1 (33) were cultured in RPMI medium 1640 (Invitrogen Life Technologies) containing 10% fetal clone I serum (HyClone), 100 units/ml penicillin, and 100 μg/ml streptomycin. Conditioned medium from parental and Wnt3a-transfected L cells was prepared as described (34). Wnt3a was purified as described by Willert et al. (35). Transient transfection was performed by electroporation of TOP/FOPFLASH (36), constructs expressing constitutively active β-catenin (β-catenin S33Y) (13), and ΔTCF4 (37), using the nucleofector technology in combination with solution v and program t-01 (Amaxa, Cologne, Germany). HeLa cell lysates were from BD Biosciences.
Isolation of B Cell Populations and Plasma Cells. B cells were isolated from human tonsils as described (38). Total B cell fractions were >97% pure, as determined by FACS analysis. Subpopulations were identified according to Pascual et al. (39), employing CD19, CD38, and IgD as discriminatory criteria. Analysis and sorting was performed by using a FACSVantage flow cytometer (BD Biosciences).
Luciferase Reporter Assay. Cells were transfected, and efficiency was determined and normalized by transfection by using pEGFP-N3 (Invitrogen Life Technologies) and subsequent FACS analysis. After 24 h, cells were stimulated with Wnt3a (100 ng/ml) for 18 h, were harvested, were lysed, and luciferase activity was determined as described (18).
Immunohistochemistry and Fluorescence Microscopy. After the indicated stimulus, cells were washed in PBS, diluted to 1 × 106 cells per ml, and 100,000 cells were spun onto slides before fixation. For detection of active, nonphosphorylated β-catenin and total β-catenin, mAb 8E4 (Alexis Biochemicals), and C14 (BD Biosciences) were used. In immunohistochemical studies, the slides were subsequently incubated with biotin-conjugated rabbit anti-mouse for 30 min, and horseradish peroxidase-conjugated avidin-biotin complex for 30 min. The substrate was visualized with 3,3-amino-9-ethylcarbazole (Sigma). In immunofluorescence studies, the primary antibodies were detected by using biotin-conjugated goat anti-mouse, followed by streptavidin-FITC (both from DAKO), and were analyzed by confocal laser scan microscopy. Nuclei were stained with propidium iodide (Molecular Probes).
Western Blot Analysis. A total of 5 ×105 cells were directly lysed in sample buffer, were separated by SDS/10% PAGE, and were blotted. Equal loading was confirmed by staining the part of the blot <50 kDa with either anti-ERK2 or anti-β-actin. The upper part (>50 kDa) was stained for anti-active β-catenin (with either anti-ABC or 8E4), or anti-β-catenin. Primary antibodies were detected by horseradish peroxidase-conjugated rabbit anti-mouse or goat anti-rabbit antibodies, followed by detection using an enhanced chemiluminescence kit (Amersham Pharmacia Biosciences).
Proliferation Assay. 24 h after transfection, viable, GFP-positive cells were sorted by using a MoFlow flow cytometer (Cytomation, Freiburg, Germany). Either GFP-positive or nonsorted cells were plated in 96-well flat bottom tissue culture plates (Costar, Cambridge, MA) at a density of 100,000 cells per ml. Stimuli were added, and cells were cultured in 2% serum for 2–3 days in a total volume of 200 μl. The cell culture was pulsed with 0.5 μCi (1 Ci = 37 GBq) methyl-[3H]thymidine (87 μCi/mmol, Amersham Pharmacia Biosciences) during the last 4 h of culture. Results are expressed as cpm.
Further information on RT-PCR analysis of WNT expression and mutation analysis of CTNNB1 and APC can be obtained in Supporting Text, which is published as supporting information on the PNAS web site.
Results
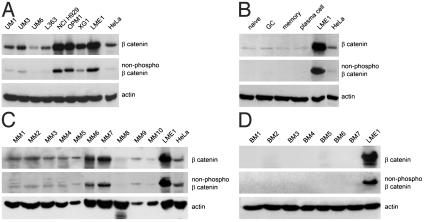
Myeloma Cells Overexpress β-Catenin. Regulation of β-catenin levels plays a central role in WNT signaling. Our initial evidence that the WNT pathway might play a role in the pathogenesis of MM came from a screen of B lineage malignancies for expression of β-catenin. We used both a pan anti-β-catenin antibody and an antibody that specifically recognizes the nonphosphorylated N-terminal Ser-37 and Thr-41 epitope of β-catenin. Only these nonphosphorylated β-catenin species are able to transduce WNT signals to the nucleus (27, 28). Interestingly, all of the myeloma cell lines tested expressed β-catenin, whereas the majority of the myeloma cell lines also contained detectable levels of the active, nonphosphorylated form of the molecule (Fig. 1A). We then compared the β-catenin expression levels in MM with the levels in normal B cell subpopulations and plasma cells, isolated from human tonsils by FACS analysis, using CD19, CD38, and IgD as markers (39). β-catenin levels were very low in all B cell subsets examined; i.e., in naïve (IgD+/CD38-), germinal center (CD38+), and memory (IgD-/CD38-) B cells, as well as in plasma cells (CD38high). In these normal B cell populations, the nonphosphorylated form of β-catenin was undetectable (Fig. 1B).
Fig. 1.
Myeloma cell lines and primary myeloma cells overexpress β-catenin. (A) β-catenin expression by myeloma cell lines. Cells were lysed and were immunoblotted by using a monoclonal anti-β-catenin antibody (Top), or an antibody against nonphosphorylated β-catenin (8E4) (Middle). (B) β-catenin expression in normal B cell subsets and plasma cells. B cell subsets and plasma cells were sorted from human tonsils as described in Materials and Methods. Cells were lysed and were immunoblotted as described above. (C) β-catenin expression in primary MMs. BM mononuclear cells from myeloma patients were lysed and were immunoblotted as described above. (D) β-catenin expression in normal BM. Total BM-derived mononuclear cells were isolated from control individuals, were lysed and were immunoblotted as described above. (A–D) Equal cell numbers of MM cell line LME1 were loaded as a reference for β-catenin expression. Stainings with anti-β-actin served as loading controls (Bottom).
To assess whether β-catenin accumulation also occurs in primary myelomas, we studied β-catenin expression in BM samples from MM patients. The samples studied were selected for extensive tumor infiltration and all contained >75% malignant plasma cells. β-catenin was expressed in 9 of 10 of these primary MM samples (MM1–10, Fig. 1C). Most cases also showed detectable expression of the active nonphosphorylated form of β-catenin (MM1–10, Fig. 1C). By contrast, β-catenin was not expressed in any of the nonneoplastic control BM samples examined (BM1–7, Fig. 1D). These results demonstrate that myeloma cell lines and most primary myelomas overexpress β-catenin, including the nonphosphorylated form of the molecule, an observation indicative for active WNT signaling.
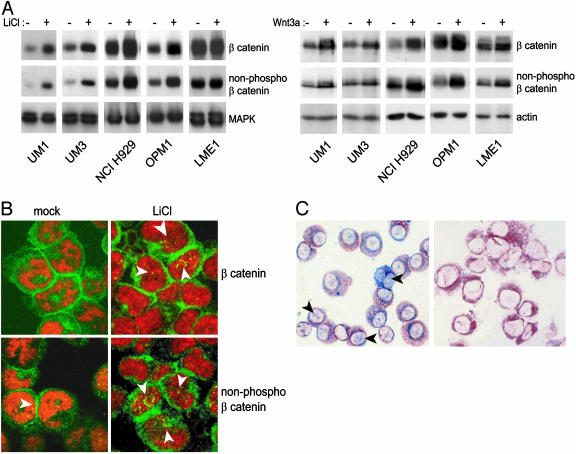
WNT Signaling Regulates β-Catenin Levels and Localization. We did not detect APC or CTNNB1 mutations in any of the MM cell lines used in this study (data not shown). We subsequently performed experiments to test whether MM cells could respond to WNT signaling. Initially, we tested the effects of LiCl, which inhibits GSK3β and mimics WNT signaling by stabilizing β-catenin (40). Stimulation with LiCl markedly increased the amounts of total and nonphosphorylated β-catenin in the myeloma cell lines tested, as measured by immunoblotting (Fig. 2A Left). The accumulation was most pronounced in cells with relatively low baseline β-catenin levels. To substantiate this finding, we determined whether specific WNT proteins could also regulate β-catenin levels. To this end, we stimulated MM cells with conditioned medium derived from L cells secreting Wnt3a (34). Indeed, this stimulation also resulted in β-catenin accumulation, including accumulation of nonphospho β-catenin (Fig. 2A Right; ref. 37). Similar results were also obtained by using purified Wnt3a (data not shown)
Fig. 2.
WNT signaling induces β-catenin accumulation and nuclear translocation in myeloma cells. (A) Accumulation of β-catenin in MM cells in response to LiCl, which mimics a WNT signal, or Wnt3a. Cells were incubated for 2 h in the absence or presence of 20 mM LiCl (Left) or in the absence or presence of Wnt3a conditioned medium (Right). To assess β-catenin accumulation, cells were lysed and were immunoblotted by using a monoclonal anti-β-catenin antibody (Top), or an antibody against nonphosphorylated β-catenin (8E4; Middle). The bottom part of the blot was stained with anti-extracellular signal-regulated kinase (MAPK), or anti-β-actin to verify equal loading. (B) LiCl induces accumulation of β-catenin and increased nuclear localization of nonphosphorylated β-catenin. NCI H929 cells were incubated for 2 h in the absence or presence of 20 mM LiCl. Cytospins were prepared and were stained with an antibody against total β-catenin (Top), or nonphosphorylated β-catenin (8E4; Bottom), followed by a FITC-conjugated secondary antibody and analyzed by confocal laser scan microscopy. (C) Expression and nuclear localization of β-catenin in primary MMs. Immunohistochemical double staining of BM-derived mononuclear cells by using antibodies against Ig light chains (brown) and nonphosphorylated β-catenin (blue; Left). Note nuclear staining of β-catenin (arrows). As a control, an isotype-matched antibody in combination with the anti-Ig light-chain antibodies is shown (Right).
Apart from causing β-catenin accumulation, WNT signaling also affected the localization of β-catenin. Before LiCl stimulation, low amounts of β-catenin were detected in the cytoplasm and nucleus of MM cells by confocal laser scan microscopy. Stimulation with LiCl led to an increase in the total amount of β-catenin, located to the plasma membrane at cell–cell contact sites, as well as in the nucleus (Fig. 2B). Stimulation with Wnt3a had similar effects (data not shown). Interestingly, most of the nonphosphorylated β-catenin was localized in the nucleus (Fig. 2B). Nonphosphorylated β-catenin was also present in the nucleus of primary myelomas (Fig. 2C). Nuclear localization of nonphosphorylated β-catenin was also detected in primary myelomas, but the levels varied among individual cells within a given tumor (Fig. 2C). This variation possibly reflects heterogeneity in the differentiation of individual tumor cells with higher WNT signaling in tumor stem cells (41).
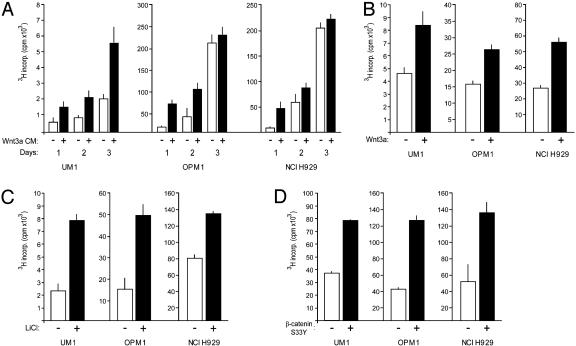
WNT Signaling Controls Proliferation of Myeloma Cells. We subsequently explored the effects of WNT signaling on MM proliferation. Myeloma cells responded to stimulation with Wnt3a-conditioned medium with a 2- to 4-fold increase in proliferation. This enhanced proliferation was already observed within the first 24 h (Fig. 3A). During culture, cell numbers increased, whereas viability was not significantly affected by the presence of Wnt3a (data not shown). Importantly, similar results were also obtained with purified Wnt3a (Fig. 3B), which has recently become available (35), demonstrating the specificity of the effect of Wnt3a and ruling out indirect effects of other growth factors released as a result of autocrine stimulation of the Wnt3a-transfected L cells. Treatment with LiCl, which inhibits GSK3β resulting in β-catenin accumulation (Fig. 2A), gave rise to a similar increase in proliferation (Fig. 3C). Furthermore, enhanced proliferation was also observed after expression of the β-catenin S33Y mutant (Fig. 3D). Taken together, these data clearly demonstrate that activation of various components of the canonical WNT signaling pathway in myeloma cells induces proliferation.
Fig. 3.
Stimulation of WNT signaling promotes proliferation of MM cells. (A) Exogenous Wnt3a promotes proliferation of MM cells. The MM cell lines UM1, OPM1, and NCI H929 were cultured in the presence of L cell-conditioned medium (open bars) or conditioned medium derived from Wnt3a-transfected L cells (Wnt3a CM, filled bars) and [3H]thymidine incorporation was measured after 1, 2, and 3 days of culture. Error bars represent SD of triplicate measurements. (B) Purified Wnt3a promotes proliferation of MM cells. Cells were cultured in the absence (open bars) or presence (filled bars) of purified Wnt3a (100 ng/ml) in serum-free medium. [3H]thymidine incorporation was measured after 2 days of culture. (C) LiCl stimulation promotes proliferation of MM cells. Cell were cultured in the absence (open bars) or presence (filled bars) of 2 mM LiCl in serum-free medium. [3H]Thymidine incorporation was measured after 3 days of culture. (D) β-catenin S33Y promotes proliferation of MM cells. Cells were transfected with empty vector (open bars) or with β-catenin S33Y (filled bars) and [3H]thymidine incorporation was measured after 2 days of culture.
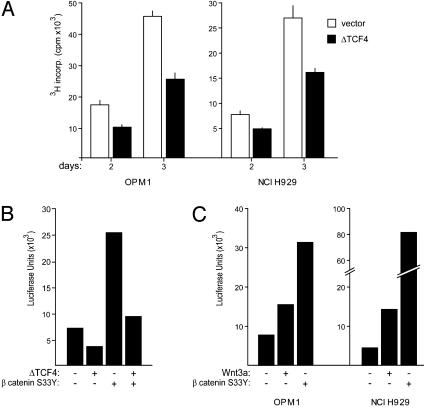
Finally, we investigated the effect of disruption of β-catenin/TCF activity on MM proliferation. As is shown in Fig. 4A, transfection with ΔTCF4 strongly inhibited proliferation of the MM cell lines OPM1 and NCI H929, which both contain large amounts of (active) β-catenin. These data demonstrate that WNT signaling regulates proliferation in MM cells.
Fig. 4.
ΔTCF4 inhibits MM proliferation and suppresses TCF reporter activity, whereas both β-catenin S33Yand Wnt3a stimulate TCF reporter activity. (A) ΔTCF4 inhibits proliferation of MM cells. MM cell lines OPM1 and NCI H929 were transfected with either ΔTCF4 (filled bars) or empty vector (open bars), both in combination with pEGFP constructs. After overnight culture, viable, GFP-positive cells were sorted and [3H]thymidine incorporation was measured after 2 and 3 days of culture. Error bars represent SD of triplicate measurements. (B) TCF reporter activity in MM cells. OPM1 cells were either transfected with luciferase reporter constructs (pTOPFLASH), alone or in combination with β-catenin S33Y and ΔTCF4, and were assayed for luciferase activity. (C) Wnt3a stimulates TCF reporter activity in MM cells. OPM1 and NCI H929 cells were transfected with pTOPFLASH, alone or in combination with β-catenin S33Y. The cells were either stimulated or not stimulated with purified Wnt3a, and were assayed for luciferase activity. As a control, a TCF reporter containing scrambled TCF-binding sites (pFOPFLASH) was used. The activity of this control reporter remained unchanged (data not shown).
Regulation of WNT Signaling in MMs. Our observation that stimulation of WNT signaling by Wnt3a, LiCl, or mutant S33Y β-catenin promotes MM proliferation, whereas disruption of the pathway by ΔTCF4 inhibits proliferation, indicates that WNT signaling is constitutively active, but not maximally activated and sensitive to regulation. To corroborate this conclusion, we directly monitored TCF transcriptional activity in the MM cell line OPM1 by transfecting a TCF reporter (pTOPFLASH). As a control, we used a reporter containing scrambled TCF-binding sites (pFOPFLASH). Consistent with our functional studies, OPM1 cells showed a moderate constitutive β-catenin/TCF activity. This activity was inhibited by cotransfection of ΔTCF4 (Fig. 4B). A strong reporter activity was obtained after cotransfection of the active β-catenin mutant S33Y. Moreover, TCF reporter activity was increased by stimulating MM cells with purified Wnt3a (Fig. 4C). pFOPFLASH activity was not affected by any of the applied stimuli (data not shown).
The above data imply that the WNT pathway in MM cells is intact and suggest changes in a regulatory component; e.g., the presence of an autocrine activation loop. To explore this possibility, we assessed the expression of WNT genes previously demonstrated to be expressed within the hematopoietic environment; i.e., WNT3a, 5a, 10b, and 16 (42–44). Neither normal B cells nor plasma cells expressed these WNTs (Table 1). By contrast, in all myeloma cell lines tested, we found expression of WNT5a and WNT10b, whereas WNT16 transcripts were found in one MM cell line (Table 1). Also, in highly purified primary MM cells, obtained by positive selection with anti-CD138, we detected expression of WNT5a and/or WNT10b and, in one case, WNT16 (Table 1). Interestingly, WNT5A and WNT10b expression was also found in BM stromal cells (Table 1), suggesting that these cells may function as a paracrine source of WNTs within the BM microenvironment.
Table 1. WNT expression in myelomas and normal B lineage cells.
|
WNT
|
||||
|---|---|---|---|---|
| Cell type | 3a | 5a | 10b | 16 |
| Naïve B cells | — | — | — | — |
| GC B cells | — | — | — | — |
| Memory B cells | — | — | — | — |
| Plasma cells | — | — | — | — |
| Total B cells | — | — | — | — |
| Fibroblast | — | + | + | + |
| BM stromal cells | — | + | + | — |
| MM cell lines | 0/8 | 4/8 | 8/8 | 1/8 |
| Purified primary MMs | 0/4 | 4/4 | 3/4 | 1/4 |
WNT expression was determined by RT-PCR as described in Materials and Methods. —, negative; +, positive. Numbers indicate no. positive/no. tested.
Discussion
In the present study, we identified the WNT pathway as a signaling route involved in MM growth control. This unexpected finding implies that the WNT pathway, which is essential for early T and B cell development and has recently been shown to play a role in the self-renewal of hematopoietic stem cells (10), is activated in this tumor of terminally differentiated B cells.
Regulation of β-catenin levels plays a central role in WNT signaling. Our initial evidence for a role of the WNT pathway in the pathogenesis of MM came from the observation that all of the MM cell lines, as well as most primary MM samples studied, expressed β-catenin (Fig. 1). Compared with the very low or undetectable β-catenin expression in normal B cell subsets, plasma cells, and control BM, β-catenin was vastly overexpressed in most of these MMs (Fig. 1). In addition to overexpressing β-catenin per se, the majority of MM cell lines and primary MMs also expressed detectable levels of β-catenin harboring nonphosphorylated N-terminal Ser/Thr residues. Recent studies (27, 28) have shown that only these nonphosphorylated β-catenin species are signaling competent and can transduce WNT signals to the nucleus. Our observation that stimulation of MM cells with LiCl, which mimics WNT signaling by inhibiting GSK3β, or with Wnt3a, led to further accumulation and nuclear translocation of nonphosphorylated β-catenin (Fig. 2), implies that these tumors have an intact WNT pathway and suggests a functional role of the WNT pathway in the biology of MM. Indeed, stimulation of WNT signaling, either by LiCl, β-catenin S33Y, or by Wnt3a, promoted the proliferation of myeloma cells (Fig. 3), whereas disruption of β-catenin/TCF activity by ΔTCF4 inhibited MM proliferation (Fig. 4).
During normal lymphocyte development, WNT signaling plays an essential role in early lymphopoiesis by contributing to precursor cell survival and expansion independent of, or in parallel to, preantigen receptor signaling (8, 22–26). Our present observation that the WNT pathway is active and controls proliferation in MM is unexpected, because this illegitimate activation involves the “wrong,” most terminal, end of the B cell differentiation spectrum. Our data present, to our knowledge, the first direct evidence for a role of the WNT pathway in the pathogenesis of lymphoid cancer. In the canonical cancers involving WNT signaling, such as colorectal cancer, pilomatricomas, and hepatoblastoma, deregulation and accumulation of β-catenin is typically due to truncating mutations in APC, or to mutations in the GSK-3β target residues in CTNNB1 (5), but the mechanism of nuclear β-catenin accumulation in several other tumor types is, at present, unclear. Although we have not yet identified the cause of β-catenin accumulation in MM, our current results make direct mutational activation of the WNT pathway unlikely. First, we did not detect APC or CTNNB1 mutations in any of the MM cell lines and primary MM samples studied. This finding does not exclude the presence of mutation in axin/conductin, or other WNT pathway components; however, such mutations appear to represent a rare cause of WNT activation in cancer. Second, whereas (i) the overexpression of (active) β-catenin, (ii) the spontaneous TCF reporter activity, and (iii) the growth inhibitory effects of ΔTCF4 all testify constitutive WNT activity in MM, the level of activation of the pathway in the MM cell lines clearly was suboptimal. This conclusion can be arrived at from the fact that exogenous WNT stimuli like Wnt3a and LiCl caused further β-catenin accumulation and translocation, and enhanced cell proliferation. The latter observations, specifically the growth-stimulatory effects of Wnt3a, point to an intact WNT pathway and therefore are difficult to reconcile with a direct mutational activation. Rather, the data suggest that the β-catenin accumulation and β-catenin/TCF activity results from changes involving a regulatory or upstream component; e.g., a member of the WNT family. Consistent with this idea, we observed that myeloma cell lines as well as primary myelomas, unlike normal B cell subsets and plasma cells, express WNT5a, WNT10b, and in some cases, WNT16, mRNAs (Table 1), indicating an autocrine activation loop. Although the functionality of this loop needs to be explored, it might explain the observed constitutive, but still inducible, activation of the WNT pathway. In this context, it is of interest that WNT16-mediated autocrine growth has recently been proposed by Murre and coworkers (44) to contribute to the development of t(1, 19)-positive pre-B-precursor acute lymphoblastic leukemia.
Although the above findings suggest an autocrine activation loop, paracrine stimulation of MM cells presumably also takes place within the BM microenvironment, because we observed that BM stromal cells express both WNT5a and WNT10b (Table 1). This observation corroborates studies by Austin et al. (42) and Van den Berg et al. (43), who reported expression of WNT5a and 10b in mouse and human BM, respectively, and demonstrated that these factors function as hematopoietic growth factors, promoting expansion of mixed colony-forming units and burst-forming units erythroid. It is conceivable that during progression of MM, a gain of WNT expression takes place, establishing an autocrine activation loop, thus leading to autonomous growth, and finally, allowing dissemination to extramedullary sites. It will therefore be of great interest to assess the expression of β-catenin and WNTs at early stages of MM.
A key finding of our current study is that WNT signaling can control MM proliferation. Because normal plasma cells are terminally differentiated, nondividing cells, activation of signaling route(s) promoting cell proliferation is a crucial step in their transformation to MM. Further studies are needed to establish whether deregulation of the WNT pathway is a general event in the initiation or progression of MM. The effects of WNT signaling on proliferation in our current study clearly involve the canonical WNT pathway, which regulates β-catenin/TCF-mediated transcription, because stimulation or inhibition by either Wnt3a, LiCl, S33Y β-catenin, or ΔTCF4, which affect the canonical WNT pathway at a number of distinct levels, had profound effects on proliferation (Figs. 3 and 4). During the preparation of this manuscript, a study by Qiang et al. (45) also reported WNT signaling in MM cell lines. Unlike our study, however, this study examined neither primary patient samples nor normal B cells and plasma cells. Moreover, the functional effects of Wnt3a stimulation reported by Qiang et al. (45); i.e., morphological changes and rearrangement of the actin cytoskeleton, were associated with a noncanonical WNT pathway dependent on RHO activation.
Our current study indicates that aberrant WNT signaling drives MM proliferation and could represent an important step in the pathogenesis of MM. In intestinal epithelium, which presents the paradigm for the role of WNT signaling in tumor-igenesis, the β-catenin/TCF complex constitutes the “master switch” that controls proliferation versus differentiation (9). Interestingly, a recently study by Reya et al. (10) indicates that WNT signaling also controls the self-renewal of hematopoietic stem cells by the induction of proliferation and the prevention of hematopoietic stem cell differentiation. In intestinal epithelium, the proliferative effects of WNT signaling are mediated through its control over MYC. In the presence of active β-catenin/TCF complexes, MYC is expressed and blocks the expression of the cell-cycle inhibitor p21CIP1/WAF1, leading to cell-cycle progression (9). MYC expression is frequently deregulated in MMs (46, 47), but whether MYC also plays a central role in WNT-induced proliferation in MM remains to be determined. Alternative WNT target genes that may also contribute to the growth-promoting effects of WNT signaling in MMs include the cell-cycle regulator CYCLIN D1 (15) and MET (18), the receptor tyrosine kinase for HGF, a potent myeloma growth and survival factor (2, 48).
In summary, the data presented here implicate the canonical WNT signaling pathway in the pathogenesis of MM, by showing accumulation and nuclear localization of the active β-catenin, and by demonstrating that these deregulated levels of active β-catenin contribute to MM proliferation. These findings indicate that a pathway that normally drives proliferation of hematopoietic stem cells may become illegitimately activated in MM cells and identify the WNT pathway as a potential novel target for therapy in MM.
Supplementary Material
Acknowledgments
We thank Niels van de Donk, Berris van Kessel, and Richard Bende for primary myelomas, cDNAs, and helpful discussions; and Frank van Diepen and Anita Pfauth for assistance with cell sorting. This work was supported by grants from the Dutch Cancer Society and the Association for International Cancer Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MM, multiple myeloma; BM, bone marrow; APC, adenomatous polyposis coli; GSK, glycogen synthase kinase; FACS, fluorescence-activated cell sorter; TCF, T cell factor; ΔTCF4, dominant-negative TCF; β-catenin S33Y, constitutively active β-catenin.
References
- 1.Kuehl, W. M. & Bergsagel, P. L. (2002) Nat. Rev. 2, 175-187. [DOI] [PubMed] [Google Scholar]
- 2.Derksen, P. W., Keehnen, R. M., Evers, L. M., van Oers, M. H., Spaargaren, M. & Pals, S. T. (2002) Blood 99, 1405-1410. [DOI] [PubMed] [Google Scholar]
- 3.Nusse, R. & Varmus, H. E. (1982) Cell 31, 99-109. [DOI] [PubMed] [Google Scholar]
- 4.Bienz, M. & Clevers, H. (2000) Cell 103, 311-320. [DOI] [PubMed] [Google Scholar]
- 5.Polakis, P. (2000) Genes Dev. 14, 1837-1851. [PubMed] [Google Scholar]
- 6.Kinzler, K. W. & Vogelstein, B. (1996) Cell 87, 159-170. [DOI] [PubMed] [Google Scholar]
- 7.Verbeek, S., Izon, D., Hofhuis, F., Robanus-Maandag, E., te Riele, H., van de Wetering, M., Oosterwegel, M., Wilson, A., MacDonald, H. R. & Clevers, H. (1995) Nature 374, 70-74. [DOI] [PubMed] [Google Scholar]
- 8.Reya, T., O'Riordan, M., Okamura, R., Devaney, E., Willert, K., Nusse, R. & Grosschedl, R. (2000) Immunity 13, 15-24. [DOI] [PubMed] [Google Scholar]
- 9.van de Wetering, M., Sancho, E., Verweij, C., de Lau, W., Oving, I., Hurlstone, A., van der Horn, K., Batlle, E., Coudreuse, D., Haramis, A. P., et al. (2002) Cell 111, 241-250. [DOI] [PubMed] [Google Scholar]
- 10.Reya, T., Duncan, A. W., Ailles, L., Domen, J., Scherer, D. C., Willert, K., Hintz, L., Nusse, R. & Weissman, I. L. (2003) Nature 423, 409-414. [DOI] [PubMed] [Google Scholar]
- 11.Wodarz, A. & Nusse, R. (1998) Annu. Rev. Cell Dev. Biol. 14, 59-88. [DOI] [PubMed] [Google Scholar]
- 12.Moon, R. T., Bowerman, B., Boutros, M. & Perrimon, N. (2002) Science 296, 1644-1646. [DOI] [PubMed] [Google Scholar]
- 13.Molenaar, M., van de Wetering, M., Oosterwegel, M., Peterson-Maduro, J., Godsave, S., Korinek, V., Roose, J., Destree, O. & Clevers, H. (1996) Cell 86, 391-399. [DOI] [PubMed] [Google Scholar]
- 14.Behrens, J., von Kries, J. P., Kuhl, M., Bruhn, L., Wedlich, D., Grosschedl, R. & Birchmeier, W. (1996) Nature 382, 638-642. [DOI] [PubMed] [Google Scholar]
- 15.Tetsu, O. & McCormick, F. (1999) Nature 398, 422-426. [DOI] [PubMed] [Google Scholar]
- 16.He, T. C., Sparks, A. B., Rago, C., Hermeking, H., Zawel, L., da Costa, L. T., Morin, P. J., Vogelstein, B. & Kinzler, K. W. (1998) Science 281, 1509-1512. [DOI] [PubMed] [Google Scholar]
- 17.Wielenga, V. J., Smits, R., Korinek, V., Smit, L., Kielman, M., Fodde, R., Clevers, H. & Pals, S. T. (1999) Am. J. Pathol. 154, 515-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boon, E. M., Van der Neut, R., van de Wetering, M., Clevers, H. & Pals, S. T. (2002) Cancer Res. 62, 5126-5128. [PubMed] [Google Scholar]
- 19.Travis, A., Amsterdam, A., Belanger, C. & Grosschedl, R. (1991) Genes Dev. 5, 880-894. [DOI] [PubMed] [Google Scholar]
- 20.van de Wetering, M., Oosterwegel, M., Dooijes, D. & Clevers, H. (1991) EMBO J. 10, 123-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waterman, M. L., Fischer, W. H. & Jones, K. A. (1991) Genes Dev. 5, 656-669. [DOI] [PubMed] [Google Scholar]
- 22.van de Wetering, M., de Lau, W. & Clevers, H. (2002) Cell 109, 13-19. [DOI] [PubMed] [Google Scholar]
- 23.Schilham, M. W., Wilson, A., Moerer, P., Benaissa-Trouw, B. J., Cumano, A. & Clevers, H. C. (1998) J. Immunol. 161, 3984-3991. [PubMed] [Google Scholar]
- 24.Okamura, R. M., Sigvardsson, M., Galceran, J., Verbeek, S., Clevers, H. & Grosschedl, R. (1998) Immunity 8, 11-20. [DOI] [PubMed] [Google Scholar]
- 25.Gounari, F., Aifantis, I., Khazaie, K., Hoeflinger, S., Harada, N., Taketo, M. M. & von Boehmer, H. (2001) Nat. Immunol. 2, 863-869. [DOI] [PubMed] [Google Scholar]
- 26.Ioannidis, V., Beermann, F., Clevers, H. & Held, W. (2001) Nat. Immunol. 2, 691-697. [DOI] [PubMed] [Google Scholar]
- 27.Staal, F. J., Noort Mv, M., Strous, G. J. & Clevers, H. C. (2002) EMBO Rep. 3, 63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Noort, M., Meeldijk, J., van der Zee, R., Destree, O. & Clevers, H. (2002) J. Biol. Chem. 277, 17901-17905. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, X. G., Gaillard, J. P., Robillard, N., Lu, Z. Y., Gu, Z. J., Jourdan, M., Boiron, J. M., Bataille, R. & Klein, B. (1994) Blood 83, 3654-3663. [PubMed] [Google Scholar]
- 30.Kuipers, J., Vaandrager, J. W., Weghuis, D. O., Pearson, P. L., Scheres, J., Lokhorst, H. M., Clevers, H. & Bast, B. J. (1999) Cancer Genet. Cytogenet. 109, 99-107. [DOI] [PubMed] [Google Scholar]
- 31.Diehl, V., Schaadt, M., Kirchner, H., Hellriegel, K. P., Gudat, F., Fonatsch, C., Laskewitz, E. & Guggenheim, R. (1978) Blut 36, 331-338. [DOI] [PubMed] [Google Scholar]
- 32.Gazdar, A. F., Oie, H. K., Kirsch, I. R. & Hollis, G. F. (1986) Blood 67, 1542-1549. [PubMed] [Google Scholar]
- 33.Katagiri, S., Yonezawa, T., Kuyama, J., Kanayama, Y., Nishida, K., Abe, T., Tamaki, T., Ohnishi, M. & Tarui, S. (1985) In. J. Cancer 36, 241-246. [DOI] [PubMed] [Google Scholar]
- 34.Shibamoto, S., Higano, K., Takada, R., Ito, F., Takeichi, M. & Takada, S. (1998) Genes Cells 3, 659-670. [DOI] [PubMed] [Google Scholar]
- 35.Willert, K., Brown, J. D., Danenberg, E., Duncan, A. W., Weissman, I. L., Reya, T., Yates, J. R. & Nusse, R. (2003) Nature 423, 448-452. [DOI] [PubMed] [Google Scholar]
- 36.van de Wetering, M., Cavallo, R., Dooijes, D., van Beest, M., van Es, J., Loureiro, J., Ypma, A., Hursh, D., Jones, T., Bejsovec, A., et al. (1997) Cell 88, 789-799. [DOI] [PubMed] [Google Scholar]
- 37.Korinek, V., Barker, N., Morin, P. J., van Wichen, D., de Weger, R., Kinzler, K. W., Vogelstein, B. & Clevers, H. (1997) Science 275, 1784-1787. [DOI] [PubMed] [Google Scholar]
- 38.Koopman, G., Keehnen, R. M., Lindhout, E., Newman, W., Shimizu, Y., van Seventer, G. A., de Groot, C. & Pals, S. T. (1994) J. Immunol. 152, 3760-3767. [PubMed] [Google Scholar]
- 39.Pascual, V., Liu, Y. J., Magalski, A., de Bouteiller, O., Banchereau, J. & Capra, J. D. (1994) J. Exp. Med. 180, 329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stambolic, V., Ruel, L. & Woodgett, J. R. (1996) Curr. Biol. 6, 1664-1668. [DOI] [PubMed] [Google Scholar]
- 41.Rohrer, J. W., Vasa, K. & Lynch, R. G. (1977) J. Immunol. 119, 861-866. [PubMed] [Google Scholar]
- 42.Austin, T. W., Solar, G. P., Ziegler, F. C., Liem, L. & Matthews, W. (1997) Blood 89, 3624-3635. [PubMed] [Google Scholar]
- 43.Van Den Berg, D. J., Sharma, A. K., Bruno, E. & Hoffman, R. (1998) Blood 92, 3189-3202. [PubMed] [Google Scholar]
- 44.McWhirter, J. R., Neuteboom, S. T., Wancewicz, E. V., Monia, B. P., Downing, J. R. & Murre, C. (1999) Proc. Natl. Acad. Sci. USA 96, 11464-11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiang, Y. W., Endo, Y., Rubin, J. S. & Rudikoff, S. (2003) Oncogene 22, 1536-1545. [DOI] [PubMed] [Google Scholar]
- 46.Hallek, M., Leif Bergsagel, P. & Anderson, K. C. (1998) Blood 91, 3-21. [PMC free article] [PubMed] [Google Scholar]
- 47.Zhan, F., Hardin, J., Kordsmeier, B., Bumm, K., Zheng, M., Tian, E., Sanderson, R., Yang, Y., Wilson, C., Zangari, M., et al. (2002) Blood 99, 1745-1757. [DOI] [PubMed] [Google Scholar]
- 48.Derksen, P. W., De Gorter, D. J., Meijer, H. P., Bende, R. J., van Dijk, M., Lokhorst, H. M., Bloem, A. C., Spaargaren, M. & Pals, S. T. (2003) Leukemia 17, 764-774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.