Abstract
Cisplatin is one of the most widely used cancer chemotherapy agents, but its mechanism of action is not fully understood. Current models suggest that cell killing by cisplatin occurs in a cell-autonomous manner by means of formation of platinum-DNA adducts that, if not removed by DNA repair, block transcription and replication. Here, we show that there is a separate cell-interdependent pathway of cisplatin killing in which damaged cells can transmit a death signal to neighboring cells. This signal is produced within the damaged cell by the kinase function of the Ku70, Ku80, and DNA-dependent protein kinase complex and is conveyed to the recipient cell by direct cell-to-cell communication through gap junctions. These findings suggest that DNA-dependent protein kinase activity and gap junction expression in human cancers may influence the clinical response to cisplatin. In addition, strategies to manipulate these cellular components in conjunction with cisplatin treatment may provide new approaches to cancer therapy.
Keywords: DNA repair, signal transduction, connexin
Cisplatin has been used in cancer chemotherapy for over 30 years. It is highly effective in the treatment of testicular cancers and is active against carcinomas of the lung, head and neck, cervix, and ovary, among others. However, clinical responses to cisplatin are variable, and many tumors develop resistance over time (1). Consequently, there is intense interest in elucidating the mechanisms by which cisplatin kills cancer cells.
Cisplatin produces a variety of platinum-DNA adducts, including intra- and inter-strand crosslinks, and it is generally accepted that these lesions underlie most of the cytotoxic effects of the drug (1). Cisplatin adducts are recognized by a number of cellular proteins, including the DNA damage-recognition factors XPC/hHR23b and MSH2/MSH6 and the high-mobility group protein HMG1 (2). The adducts are subject to repair by several pathways, including nucleotide excision repair (NER) and homologous recombination (HR) (3, 4). Numerous reports have suggested that the efficiency of NER-mediated repair and the extent of HMG1 binding can influence the effects of cisplatin in cancer cells (4–6). Cisplatin resistance can also result from diminished cellular uptake associated with altered expression of a copper transporter (7, 8) and from inactivation by intracellular glutathione (1).
Intracellular signaling pathways have also been implicated in the cisplatin response. p53 is phosphorylated and p73 is induced in cells after cisplatin treatment, and c-abl kinase is activated (9, 10). In some cases, these signaling events are dependent on an intact DNA mismatch repair (MMR) pathway (9, 10). Cells deficient in DNA MMR show moderate cisplatin resistance (11, 12), consistent with a model in which the DNA MMR pathway recognizes platinum-DNA adducts and participates in proapoptotic signaling.
In an effort to identify other repair factors that might respond to cisplatin damage, we examined cells deficient in elements of the Ku/DNA-dependent protein kinase (DNA-PK) complex. This complex includes the Ku70/Ku80 heterodimer and the catalytic subunit, DNA-PKcs. The complex participates in nonhomologous end joining (NHEJ) and VDJ recombination (13). DNA-PK is a member of the phosphatidylinositol 3-kinase (PI3-kinase)-related family and is proposed to have a role in cell signaling after DNA damage. In vitro, the Ku70/Ku80 heterodimer can bind to DNA ends at double-strand breaks and to DNA fragments with cisplatin-DNA adducts (14).
Here, we report that cells deficient in Ku80 or in DNA-PKcs are markedly resistant to cisplatin compared with matched wild-type counterparts, but that this difference is manifest only when the cells are at high density. In low-density cultures, no survival differences were seen. In contrast, MMR-deficient cells show moderate resistance regardless of density, suggesting that the MMR-mediated and DNA-PK-mediated responses are distinct. The density dependence of the DNA-PK-mediated response prompted further experiments examining a role for cell-to-cell communication, and we present evidence demonstrating that intercellular communication by means of gap junctions is required for the DNA-PK-mediated cytotoxic response to cisplatin.
Materials and Methods
Cell Lines. Ku80+/+ and Ku80-/- immortalized mouse embryonic fibroblasts (MEFs) were from G. Li (Memorial Sloan–Kettering Cancer Center, New York) (15). Xrs6 and xrs6-hamKu80 were purchased from the European Collection of Cell Cultures (16). WB-F344 and WB-aB1 were from E. Azzam (New Jersey Medical School, Newark) by permission of J. Trosko (Michigan State University, East Lansing) (17). Severe combined immunodeficient (SCID) (50D) and SCID plus human DNA-PK (100E) mouse fibroblasts were obtained from C. Kirchgessner (Stanford University, Stanford, CA) (18). MCF-7 cells were from the American Type Culture Collection.
Plasmid Constructs. Connexin43 cDNA was generated by RT-PCR of total RNA (TRIzol) from human AG1522 fibroblasts (Coriell Cell Repositories, Camden, NJ) and cloned into pcDNA3.1 (Invitrogen). The connexin43 cDNA was stably transfected (FuGENE6, Roche) into MCF-7 cells by using 0.1 mg/ml zeocin selection. Positive clones were identified by Western blot (Connexin43 Ab clone CXN-6, Sigma) and by lucifer yellow (Molecular Probes) dye transfer using the scrape/loading technique (17). Ku80-/- MEFs were made resistant to zeocin by stable transfection of pcDNA4 (Invitrogen) and selection in 0.2 mg/ml zeocin.
Cisplatin Treatment and Survival Assays. Cisplatin was obtained from Sigma (P4394), and stock solutions were prepared fresh at 0.5 mg/ml in PBS. All exposures to cisplatin were performed for 1 hr in the dark. Lindane (Sigma, H4500) and oleamide (Sigma, O-122) were dissolved in DMSO at 50 mg/ml and incubated either simultaneously with or 1 hr before the 1-hr incubation with cisplatin. The final concentrations of lindane and oleamide in media were each 50 μM. Wortmannin (Sigma, W1628) was added to cells at 20 μM 1 hr before cisplatin exposure and remained during the 1-hr treatment with cisplatin.
Cell survival was assayed either by visualization of monolayer growth or by colony formation. To quantify survival by monolayer growth, cells were seeded at defined densities in 60- or 100-mm dishes and treated with cisplatin for 1 hr, with or without other agents as indicated. After treatment, the cells were washed with PBS, replenished with fresh medium, and left undisturbed until staining with crystal violet 6 days posttreatment for monolayer visualization.
To assay for survival by colony formation, cells at high density were seeded such that confluency would be 70–100% at time of cisplatin exposure, ≈30,000 cells per cm2. After the 1-hr exposure to cisplatin with or without other agents, cells were washed twice with PBS, harvested by trypsinization, counted, serially diluted, and seeded into six-well dishes, and colony formation was determined by staining with crystal violet 10–14 days after treatment. Colonies containing 50 or more cells were scored. Cells at low density were seeded at ≈500 cells per cm2, typically in six-well dishes. After cells had attached (4 hr to overnight) at the desired seeding density, they were treated with cisplatin for 1 hr (with or without other agents), washed twice with PBS, replenished with fresh media, and left undisturbed until staining for colony counting 10–14 days later as described above. Alternatively, cells initially seeded at varying densities were in all cases harvested by trypsinization after cisplatin treatment, reseeded, and counted for colony formation after 10–14 days (see Figs. 2B and 10). Colony formation was normalized to plating efficiency of the non-cisplatin-treated cells in all cases. Error bars in the survival analyses indicate SD based on three independent experiments in all cases. There was no significant difference in plating efficiency between the low- and high-density cultures in the untreated samples (data not shown). Also, lindane did not alter the plating efficiency of the cells (see Fig. 9).
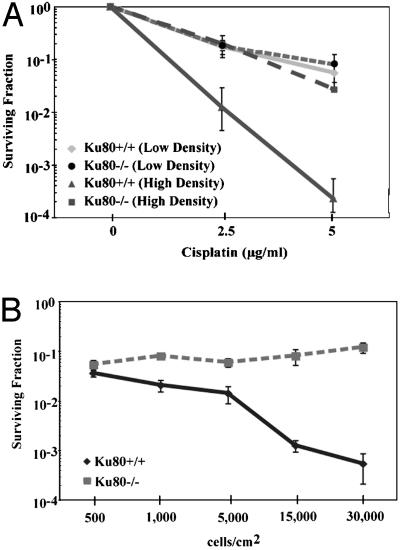
Fig. 2.
Cisplatin survival of Ku80+/+ MEFs but not of Ku80-/- MEFs is density dependent. (A) Clonogenic survival of cells treated with cisplatin at low density (500 cells per cm2) or at high density (30,000 cells per cm2). Note that Ku80+/+ cells (solid lines) are more sensitive at high density; the survival of the Ku80-/- cells (dashed lines) does not change with density. (B) Clonogenic survival of Ku80+/+ (solid line) and Ku80-/- (dashed line) MEFs over a range of cell densities after treatment with 5 μg/ml cisplatin. Cells were treated at the indicated densities, washed, harvested by trypsinization, reseeded, and stained and counted 10–14 days later.
To measure monolayer growth or colony formation exclusively by Ku80-/- cells in mixed cell populations also containing Ku80+/+ cells, the Ku80-/- cells were stably transfected with pcDNA4 vector to confer resistance to zeocin. Wild-type cells were mixed with Ku80-/-(pcDNA4) cells at various ratios and treated with 5 μg/ml cisplatin, and either monolayer growth after 6 days or clonogenic survival after 10–14 days was assayed in the presence of 0.2 mg/ml zeocin such that only Ku80-/-(pcDNA4) cells would be able to survive.
RNA Interference. To inhibit connexin43 expression by RNA interference, a target sequence consisting of 19 nucleotides was chosen in the ORF of mouse connexin43 (5′-TGGCTGCTCCTCACCAACG-3′). Oligonucleotides (64-mer) (Keck Facility, Yale University) were synthesized and ligated into the pSUPER RNA interference (RNAi) vector for stable transfection into wild-type immortalized MEFs. A zeocin-resistance plasmid, pcDNA4 (Invitrogen) was cotransfected along with pSUPER containing the connexin43 RNAi target sequence at a 1:10 ratio to allow isolation of stable clones. Clones showing decreased connexin43 protein expression were identified by Western blot. Control cells were transfected with the pSUPER vector lacking the connexin43-specific insert.
Results
In testing the role of selected DNA damage recognition factors in response to cisplatin, we measured the cisplatin survival of cells deficient in Ku80, a component, along with Ku70 and DNA-PKcs, of the DNA-PK complex that participates in DNA double-strand break repair and nonhomologous end joining (13). Surprisingly, we found in clonogenic survival assays that mouse and hamster cell lines deficient in Ku80 are markedly resistant to cisplatin compared with matched cells that are wild-type for Ku80 (Fig. 1A). The cell lines included: (i) MEFs derived from Ku80 knockout and wild-type littermate mice (Ku80-/- and Ku80+/+ MEFs, respectively) and (ii) Chinese hamster ovary (CHO)-derived cell lines mutant for Ku80 (xrs6) and a subclone of xrs6 complemented with a construct expressing wild-type Ku80 cDNA (xrs6 plus Ku80). Ku80 expression was tested by Western blot in all of these cell lines, and the expected patterns of expression in line with the genotypes were confirmed (Fig. 6, which is published as supporting information on the PNAS web site). In addition, the level of Ku80 expression in the xrs6 plus Ku80 cells was shown to be similar to that in wild-type CHO cells (19). In all cases, the survival analyses included data from at least three independent experiments, with each treatment performed in triplicate, with errors calculated as SD.
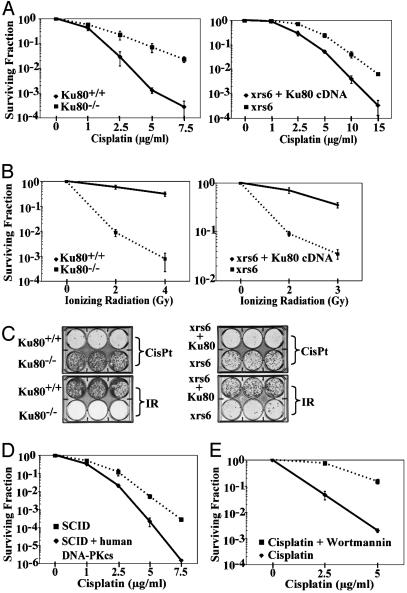
Fig. 1.
Cells deficient in Ku80 or in DNA-PKcs are resistant to cisplatin. (A) Clonogenic survival of matched pairs of MEFs and CHO cells either wild-type or mutant for Ku80 after exposure to cisplatin. (Left) Shown is survival of Ku80+/+ (solid line) vs. Ku80-/- (dashed line) MEFs. (Right) Shown is survival of xrs6 plus Ku80 cDNA (solid line) vs. xrs6 (dashed line) CHO cells to increasing doses of cisplatin. (B) Survival of Ku80+/+ vs. Ku80-/- MEFs and xrs6 plus Ku80 cDNA vs. xrs6 CHO cells to IR (IR; dose given in Gy). (C) Cell monolayers fixed and stained 6 days after either cisplatin (5 μg/ml) or IR exposure (4 Gy), as indicated, to directly visualize growth. (D) Clonogenic survival of SCID (DNA-PKcs mutant) mouse cells (dashed line) vs. SCID cells complemented by chromosome transfer with the human DNA-PKcs gene (solid line) after cisplatin exposure. (E) Clonogenic survival of wild-type MEFs treated either with cisplatin alone (solid line) or with cisplatin plus 20 μM wortmannin, a DNA-PK kinase inhibitor (dashed line), showing that inhibition of DNA-PK kinase activity results in increased resistance of cells to cisplatin. Error bars indicate SD based on three independent replicates in all cases.
To further correlate the difference in cisplatin response with Ku80 expression in otherwise isogenic cells, we transfected Ku80-/- MEFs with a vector expressing Ku80 cDNA. Expression of Ku80 in the transfectants was confirmed by Western blot and was shown to confer sensitivity to cisplatin (Fig. 7, which is published as supporting information on the PNAS web site). Therefore, three sets of comparisons in matched pairs of cell lines derived in different ways all demonstrate a role for Ku80 in cisplatin response. The resistance of the Ku80-deficient cells was unexpected because they are deficient in DNA repair and are otherwise sensitive to a number of DNA-damaging agents, particularly ionizing radiation (IR). The expected radiation sensitivity of the Ku80-deficient cells was confirmed (Figs. 1 B and C).
To determine whether other members of the DNA-PK complex are required for the cisplatin sensitivity seen in the wild-type cells, we examined cells deficient in the catalytic subunit of DNA-PK (DNA-PKcs). We found that fibroblasts derived from SCID mice (mutant in DNA-PKcs) were resistant to cisplatin compared with SCID fibroblasts that had been complemented by chromosome transfer with the human DNA-PKcs gene (Fig. 1D). The lack of DNA-PKcs expression in the SCID cells and the expression of DNA-PKcs in the complemented cells was confirmed by Western blot analysis (Fig. 6B). To directly test the role of the kinase function of the Ku/DNA-PK complex, we used the DNA-PKcs kinase inhibitor wortmannin. We found that wortmannin protected wild-type MEFs from cisplatin-induced cell death (Fig. 1E), suggesting that DNA-PKcs kinase activity, specifically, participates in cisplatin killing. Because wortmannin can also inhibit the ATM kinase, we compared cisplatin survival of a matched pair of ataxia telangiectasia mutated (ATM)-deficient and proficient human cell lines (20); no differences were seen under the same experimental conditions that had revealed the effect of DNA-PK (data not shown).
Because prior studies had not detected such resistance to cisplatin in Ku80- or DNA-PKcs-deficient cells (21–23), we carefully examined a number of experimental parameters that might have an influence on the cisplatin survival response. We discovered that the density at which monolayer cell cultures are exposed to cisplatin has a dramatic effect on cell survival (Fig. 2). At low density (500 cells per cm2), there was no difference in clonogenic survival between Ku80-deficient and wild-type cells (Fig. 2 A). However, at high cell density (30,000 cells per cm2), the survival of the wild-type cells was substantially decreased whereas that of the Ku80-deficient cells was unchanged, remaining similar to that of both the Ku80-deficient and -proficient cells at low density. This striking density dependence of the cisplatin response in the Ku80+/+ cells can be further visualized in the analysis of cell survival after treatment with 5 μg/ml cisplatin over a range of cell densities (Fig. 2B). As shown, the survival of the Ku80+/+ cells decreased with increasing density; the survival of the Ku80-/- cells was not affected by density. A similar difference in density dependence was seen in a comparison of DNA-PKcs-proficient and -deficient cells (data not shown). Therefore, there is a density-dependent sensitivity of wild-type cells to cisplatin that is mediated by Ku80 and DNA-PKcs.
The density dependence of the cisplatin response in the wild-type cells suggested a possible role for intercellular communication. A major pathway for such communication occurs by means of gap junctions. These structures are gated intercellular channels that allow the passage of ions and other small molecules (up to 1 kDa) involved in cell-to-cell signaling or propagation of action potentials (24, 25). Gap junctions are formed by docking together of two hemichannels (connexons) from adjacent cells, each composed of hexameric arrays of transmembrane proteins (connexins) arranged around a central pore.
To test the role of gap junction intercellular communication (GJIC) in cisplatin sensitivity, we used several methods to manipulate gap junction expression and function, including chemical inhibitors, mutant cell lines, RNA interference, and forced gene expression. Pretreatment of wild-type MEFs with either of two GJIC inhibitors, lindane or oleamide, protected high-density cells from cisplatin toxicity, yielding substantially increased survival, as visualized by monolayer growth (Fig. 3A). We verified that the concentrations of lindane and oleamide used in this assay do inhibit GJIC in these cells, based on a dye transfer assay (Fig. 8, which is published as supporting information on the PNAS web site). In clonogenic survival assays, lindane altered the cisplatin survival dramatically when the cells were plated at high density (Fig. 3B); but, at low cell density, there was very little effect of lindane on cisplatin response. In control experiments, we determined that lindane has no significant effect on the plating efficiency of the wild-type MEFs at the concentration used (Fig. 9, which is published as supporting information on the PNAS web site). [In a more extensive density-dependence analysis (Fig. 10, which is published as supporting information on the PNAS web site), the small effect of lindane at low cell density can be better visualized. The persistence of a small effect even at low density is consistent with the presence of occasional cell clusters even among sparsely seeded cells. Such clusters can carry out some GJIC, which can be inhibited by lindane.] Overall, the effect of lindane, and, in particular, its much greater impact at high density, supports a mode of cell killing that is mediated by GJIC.
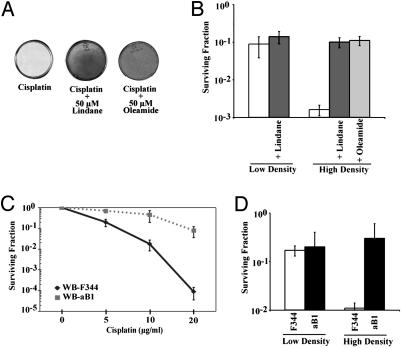
Fig. 3.
Inhibition of gap junction intercellular communication reduces cisplatin-induced cell death. (A) Cell monolayers stained with crystal violet 6 days posttreatment showing rescue from cisplatin killing by gap junction inhibitors, either lindane or oleamide. (B) Clonogenic survival of wild-type MEFs treated with 5 μg/ml cisplatin at low and high cell density and coincubated with either 50 μM lindane or 50 μM oleamide. (C) Clonogenic survival of rat liver epithelial cells, WB-F344 (GJIC-competent; solid line), and WB-aB1 (GJIC-deficient; dashed line), exposed to increasing doses of cisplatin at high cell density. (D) Clonogenic survival of WB-F344 and WB-aB1 cells treated with 10 μg/ml cisplatin at low and high cell density.
In addition, we found that lindane must be added to the cell culture medium before or simultaneous with cisplatin to achieve full protection (data not shown). If it is added >30 min after cisplatin treatment, no protective effect is seen, suggesting that the cytotoxic signal is generated and transmitted through gap junctions within minutes after cisplatin exposure.
We also compared clonogenic survival of a pair of rat liver epithelial cell lines either proficient or deficient in GJIC (WB-F344 and WB-aB1, respectively; the latter was derived from the former in a mutagenesis screen for loss of GJIC) (17). At high cell density, the GJIC-proficient cells (WB-F344) were much more sensitive to cisplatin (Fig. 3C), with survival 25-fold lower than that of the GJIC-deficient cells at a cisplatin dose of 10 μg/ml and 800-fold lower at a dose of 20 μg/ml. At low cell density, in contrast, there was minimal difference in cisplatin survival between the GJIC-proficient and -deficient cells (Fig. 3D); surviving fractions in both cases were in the same range as that of the GJIC-deficient cells at high density. These results further indicate that a substantial portion of cisplatin toxicity depends on GJIC.
To broaden these observations to other cell lines, we used RNAi to inhibit expression of connexin43, a component of gap junctions in many cell types (Fig. 4A). Killing by cisplatin was substantially reduced in wild-type MEFs in which connexin43 levels had been reduced by RNAi (Fig. 4B). Conversely, we tested the effect of forced expression of connexin43 (by means of a cDNA-expression vector) on cisplatin response in the human breast cancer cell line MCF-7. The MCF-7 cells have no detectable endogenous expression of connexin43 (Fig. 4C). As measured by dye transfer after scrape loading, the parental MCF-7 cells show minimal GJIC, but the connexin43-expressing subline has been made GJIC competent (Fig. 4D). In survival assays, we found that connexin43 expression sensitized the MCF-7 cells to cisplatin at high density (Fig. 4E).
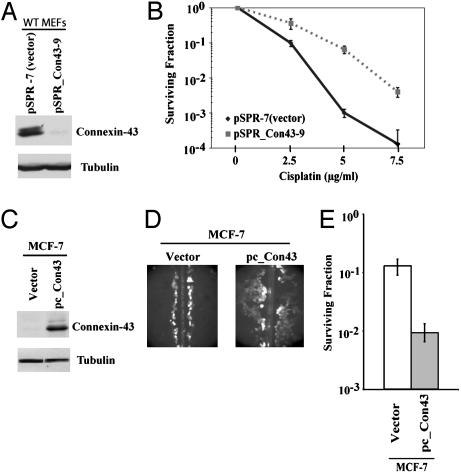
Fig. 4.
Connexin43 expression mediates sensitivity to cisplatin. (A) Western blot showing inhibition of connexin43 expression in wild-type MEFs by RNA interference. Wild-type MEFs were stably transfected with either empty vector, pSPR7, or vector containing an insert designed to express siRNA targeted to connexin43, pSPR_Con43-9. (B) Clonogenic survival after cisplatin treatment of MEFs with reduced levels of connexin43 vs. control cells. (C) Western blot demonstrating forced expression of connexin43 in the human MCF-7 cells after stable transfection with the human connexin43 cDNA driven by the CMV immediate early promoter (pc_Con43). Endogenous expression is not detected in an MCF-7 subclone transfected with an empty vector. (D) Demonstration of functional GJIC in MCF-7 cells expressing recombinant connexin43 but not in vector control cells, as visualized by lucifer yellow dye transfer after scrape loading. (E) Clonogenic survival after 20 μg/ml cisplatin treatment of MCF-7 cells expressing connexin43 vs. control cells.
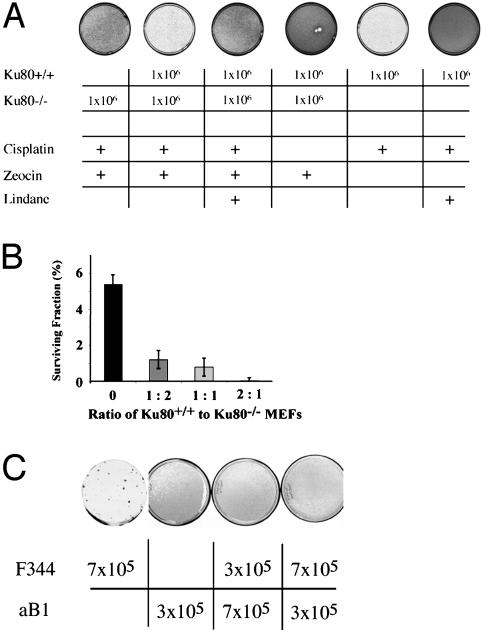
The above results suggested a model in which cisplatin damage triggers signal transduction by the DNA-PK complex, ultimately leading to gap junction transmission of an intercellular death signal. Based on this model, we hypothesized (i) that DNA-PK is required to send the intercellular signal but not necessarily to receive it and (ii) that functional gap junctions are needed for cells both to send and receive the signal. To test these hypotheses, we examined cisplatin response in mixed cell populations (Fig. 5). We found that the presence of Ku80+/+ MEFs in contact with Ku80-/- MEFs during cisplatin exposure decreased the survival of the Ku80-/- cells. This effect was determined both by visualization of monolayer growth (Fig. 5A) and by quantification of clonogenic survival of the Ku80-/- cells (Fig. 5B). In these experiments, the Ku80-/- cells were made zeocin resistant by prior gene transfer to allow specific detection in both the monolayer growth assay and the clonogenic survival assay. Note that Fig. 5A includes a control showing that killing of the Ku80+/+ cells by zeocin does not cause cell death in neighboring Ku80-/- cells; it is only when the Ku80+/+ cells are treated with cisplatin that an intercellular death signal is produced affecting the Ku80-/- cells. As further evidence that the death signal transmitted from the Ku80+/+ to the Ku80-/- cells passes through gap junctions, we found that treatment with lindane blocked killing of the Ku80-/- cells by the Ku80+/+ cells after cisplatin treatment of the mixed cell populations (Fig. 5A). To support our interpretation of the data, we confirmed that the Ku80+/+ and the Ku80-/- MEFs both express connexin43 and both have functional gap junctions (Fig. 11, which is published as supporting information on the PNAS web site).
Fig. 5.
Intercellular transduction of a cytotoxicity signal after cisplatin treatment in mixed cell populations. (A) Admixture of Ku80+/+ cells with Ku80-/- MEFs decreases the survival of the latter after cisplatin exposure (5 μg/ml). The number of cells of each genotype initially plated in the mixed cell populations are as indicated. The Ku80-/- cells were made zeocin resistant by prior gene transfer to allow detection of growth and colony formation specifically by these cells in the mixed populations. Cell monolayers were stained 6 days after cisplatin treatment. Note that transmission of the death signal from Ku80+/+ MEFs to Ku80-/- MEFs was blocked by lindane exposure. (B) Clonogenic survival after cisplatin exposure of Ku80-/- cells in the presence of increasing proportions of Ku80+/+ cells. (C) Lack of transduction of an intercellular death signal from gap junction-proficient (F344) to gap junction-deficient cells (aB1) after cisplatin treatment in mixed cell populations.
When GJIC-proficient cells (WB-F344) were mixed with GJIC-deficient cells (WB-aB1), the presence of the proficient cells did not alter the cisplatin survival of the deficient cells regardless of the ratio of the two (Fig. 5C), indicating that GJIC-deficient cells cannot receive the death signal even at high density.
We also tested whether culture medium from cisplatin-treated Ku80+/+ cells could transmit a toxicity signal to separately cultured Ku80-/- cells. There was no cytotoxic effect of this extracellular medium transfer (data not shown), again suggesting a requirement for direct cell-to-cell communication. Because of the limited sensitivity of this sort of medium transfer experiment, we cannot absolutely rule out that some signal transfer might occur between non-neighboring cells by means of open connexon hemi-channels. However, our cell-mixing experiments would indicate that open hemi-channels would be needed both to send and to receive the signal, still an interesting (although unlikely) result.
In testing other DNA damage and repair pathways, we found that the previously reported cisplatin resistance of cells deficient in DNA MMR (11, 12) occurs at both low and high cell densities (data not shown), indicating that the MMR-associated damage response pathway is distinct from the density-dependent, DNA-PK- and GJIC-mediated pathway that we have identified here. In addition, with respect to other types of DNA damage, we found no differences between Ku80- and GJIC-proficient and -deficient cells treated with UV, mitomycin C, or 1-methyl-3-nitro-1-nitrosoguanidine at high density (data not shown), indicating that the cell-interdependent cisplatin response pathway is damage-specific. In the case of IR, opposite results are seen: the Ku80-deficient MEFs are extremely sensitive to IR, not resistant as they are to cisplatin (Fig. 1B).
Because differential uptake of cisplatin and altered cisplatin sensitivity have been associated in some cell lines with the function of a copper transporter (7, 8), we tested whether either Ku80 or connexin43 expression or lindane-mediated inhibition of GJIC can affect cisplatin uptake (as measured by formation of platinum DNA-adducts in treated cells). By using mass spectrometry to analyze hydrolyzed cell DNA, we found no significant differences in adduct formation (data not shown), ruling out differences in cisplatin uptake as an explanation for the effects we have observed.
Discussion
Taken together, our results demonstrate that there is a cell-interdependent pathway of cisplatin toxicity that requires Ku/DNA-PK signaling and intercellular communication through gap junctions. By careful attention to cell growth conditions, we found that treatment of monolayer cells at high density with cisplatin results in greater cell killing than when the same cells are treated at low density with the same dose of drug (and when the cells are handled in the same way), consistent with a cell-interdependent mechanism of cell death. In comparisons of wild-type, Ku80-deficient, and DNA-PKcs-deficient cells, we found that this cell-interdependent killing depends on a functional Ku/DNA-PK signaling complex and on DNA-PKcs kinase activity. The pathway was also found to depend on gap junction communication because it was absent in GJIC-deficient cells and could be abrogated by pretreatment of wild-type cells with gap junction inhibitors. Down-regulation of connexin43 by RNAi conferred increased resistance to cisplatin in wild-type cells, and forced expression of connexin43, in a human cancer cell line that does not otherwise express it, produced increased sensitivity to cisplatin. In all cases, experimental manipulation of connexin expression was shown to correlate with the expected changes in gap junction function, as determined by dye transfer assays.
In cell-mixing experiments, we established that wild-type cells can transmit a cytotoxicity signal to neighboring cells that are deficient in Ku80 but not to cells deficient in GJIC. These results are consistent with a signal transduction pathway in which the Ku/DNA-PK complex produces a signal within cisplatin-damaged cells that is subsequently propagated through gap junctions into neighboring cells, causing cell death.
Previously, the cytotoxicity of cisplatin was thought to occur only on a cell-autonomous basis, primarily from the ability of unrepaired platinum-DNA adducts to block transcription and replication (1). Factors described as playing a role in the cisplatin response, including high-mobility group proteins, DNA MMR factors, signaling molecules such as c-jun, c-abl, and p73 (4, 10, 26, 27), and alterations in uptake and inactivation (1, 7, 8, 28), were all thought to act within individual cells.
However, our work demonstrates that there is also a cell-interdependent mechanism by which cisplatin kills cells. For example, at a dose of 10 μg/ml cisplatin, there is 95% killing of wild-type rat liver cells (Fig. 3C). Of this, ≈2/3 can be attributed to cell-autonomous effects (based on the extent of killing of the GJIC-deficient subclone) and 1/3 to the effects of intercellular signaling. Hence, the cell-interdependent pathway can make a substantial contribution to overall cell killing when there is sufficient cell-to-cell contact (as in high-density cell monolayers or in 3D solid tumors) and when DNA-PK and GJIC are functional. In analyzing the mechanism(s) of cell death after cisplatin treatment of the wild-type cells, we found evidence for both necrosis (loss of membrane integrity) and apoptosis (characteristic DNA fragmentation; data not shown), consistent with the activation of more than one pathway of cytotoxicity.
The Ku80-dependent sensitivity to cisplatin was not identified in previous studies (21–23). This Ku80 dependence may have been missed due to lack of gap junctions in the tested cells because many cell lines lose connexin expression on adaptation to in vitro culture conditions. In the absence of gap junctions, the role of the Ku/DNA-PK pathway in cisplatin response could not have been detected. In addition, the prior studies were conducted without knowledge of the importance of cell density.
The role of gap junctions in mediating cisplatin killing also had not been previously detected. However, prior work had suggested that forced connexin expression could suppress cell growth under certain conditions, indicating a possible role for connexins as tumor suppressors (29–31). This growth suppression was also reported to be additive to the toxic effects of certain chemotherapy agents, including cisplatin (32). Hence, connexins may be important factors in cancer both because of a regulatory effect on cell growth, as previously described, and because of a key role in cisplatin response, as reported here.
Gap junctions are also known to be required for the bystander effect seen with low-dose IR. In this phenomenon, IR-damaged cells pass a signal to unirradiated cells, triggering certain signaling pathways and producing genomic instability and cytotoxicity in the untreated cells (33–36). It is also known that toxic metabolites of pro-drugs can also be passed between cells through gap junctions [for example, 5-fluorouracil that has been converted from 5-fluorocytosine by cytosine deaminase (37)]. However, what we have found is a true signal transduction pathway that requires the kinase activity of the Ku/DNA-PK complex, not just sharing of a toxic metabolite in a combined pro-drug/gene therapy protocol.
We propose a model in which the cellular response to cisplatin damage occurs by means of several pathways, both cell-interdependent and cell-autonomous. In cells that have functional DNA-PK complex and that have established gap junction communication, our results suggest that the DNA-PK-mediated cytotoxic signal is triggered rapidly and is transmitted between cells in <1 hr. The means by which DNA-PK is activated and the intermediate signal transduction steps that follow this activation have yet to be established, but these steps must lead to the production of a signaling molecule of <1,000 daltons to allow transmission through gap junctions.
Because the DNA-PK- and gap junction-mediated cell-interdependent pathway can therefore play a major role in the effects of cisplatin, the activity of this pathway and the status of its components in human cancers are likely to be important determinants of the clinical response to platinum-based chemotherapy. Clearly, solid tumors grow in vivo at high density in three dimensions, with a high level of cell-to-cell contact, both among the malignant cells themselves and between malignant cells and normal ones. However, the ability of cells within solid tumors to carry out gap junction communication can be variable. In fact, distinct variations in connexin expression have been reported in human cancers (38). Consequently, further examination of gap junction expression in human tumor samples may yield insight into the variability in clinical response to cisplatin. Such variability occurs both within tumor types (for example, sensitive and resistant ovarian cancers) and among tumor types (for example, the high curability of testicular cancers by cisplatin-containing regimens). In addition, the work reported here suggests that pharmacologic strategies designed to stimulate DNA-PK activity, to increase connexin expression, or to enhance gap junction communication could sensitize malignant cells to cisplatin, providing the basis for new combined approaches to cancer therapy.
Supplementary Material
Acknowledgments
We thank S. Deschenes, R. Bindra, H. Sun, J. Sweasy, M. Caplan, R. Franklin, S. Baserga, and L. Cabral for their help and C. Kirchgessner, E. Azzam, J. Trosko, and G. Li for reagents. This work was supported by National Institutes of Health Grant ES05775 and the Yale Cancer Center.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DNA-PK, DNA-dependent protein kinase; DNA-PKcs, DNA-PK catalytic subunit; GJIC, gap junction intercellular communication; MMR, mismatch repair; MEF, mouse embryonic fibroblast; SCID, severe combined immunodeficient; CHO, Chinese hamster ovary; IR, ionizing radiation; RNAi, RNA interference.
References
- 1.Kartalou, M. & Essigmann, J. M. (2001) Mutat. Res. 478, 23-43. [DOI] [PubMed] [Google Scholar]
- 2.Kartalou, M. & Essigmann, J. M. (2001) Mutat. Res. 478, 1-21. [DOI] [PubMed] [Google Scholar]
- 3.Zdraveski, Z. Z., Mello, J. A., Marinus, M. G. & Essigmann, J. M. (2000) Chem. Biol. 7, 39-50. [DOI] [PubMed] [Google Scholar]
- 4.Huang, J. C., Zamble, D. B., Reardon, J. T., Lippard, S. J. & Sancar, A. (1994) Proc. Natl. Acad. Sci. USA 91, 10394-10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aloyz, R., Xu, Z. Y., Bello, V., Bergeron, J., Han, F. Y., Yan, Y., Malapetsa, A., Alaoui-Jamali, M. A., Duncan, A. M. & Panasci, L. (2002) Cancer Res. 62, 5457-5462. [PubMed] [Google Scholar]
- 6.Cohen, S. M. & Lippard, S. J. (2001) Prog. Nucleic Acid Res. Mol. Biol. 67, 93-130. [DOI] [PubMed] [Google Scholar]
- 7.Ishida, S., Lee, J., Thiele, D. J. & Herskowitz, I. (2002) Proc. Natl. Acad. Sci. USA 99, 14298-14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katano, K., Kondo, A., Safaei, R., Holzer, A., Samimi, G., Mishima, M., Kuo, Y. M., Rochdi, M. & Howell, S. B. (2002) Cancer Res. 62, 6559-6565. [PubMed] [Google Scholar]
- 9.Duckett, D. R., Bronstein, S. M., Taya, Y. & Modrich, P. (1999) Proc. Natl. Acad. Sci. USA 96, 12384-12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong, J. G., Costanzo, A., Yang, H. Q., Melino, G., Kaelin, W. G., Jr., Levrero, M. & Wang, J. Y. (1999) Nature 399, 806-809. [DOI] [PubMed] [Google Scholar]
- 11.Fink, D., Nebel, S., Aebi, S., Zheng, H., Cenni, B., Nehme, A., Christen, R. D. & Howell, S. B. (1996) Cancer Res. 56, 4881-4886. [PubMed] [Google Scholar]
- 12.Drummond, J. T., Anthoney, A., Brown, R. & Modrich, P. (1996) J. Biol. Chem. 271, 19645-19648. [DOI] [PubMed] [Google Scholar]
- 13.Featherstone, C. & Jackson, S. P. (1999) Mutat. Res. 434, 3-15. [DOI] [PubMed] [Google Scholar]
- 14.Turchi, J. J., Henkels, K. M., Hermanson, I. L. & Patrick, S. M. (1999) J. Inorg. Biochem. 77, 83-87. [DOI] [PubMed] [Google Scholar]
- 15.Nussenzweig, A., Chen, C., da Costa Soares, V., Sanchez, M., Sokol, K., Nussenzweig, M. C. & Li, G. C. (1996) Nature 382, 551-555. [DOI] [PubMed] [Google Scholar]
- 16.Ross, G. M., Eady, J. J., Mithal, N. P., Bush, C., Steel, G. G., Jeggo, P. A. & McMillan, T. J. (1995) Cancer Res. 55, 1235-1238. [PubMed] [Google Scholar]
- 17.Oh, S. Y., Dupont, E., Madhukar, B. V., Briand, J. P., Chang, C. C., Beyer, E. & Trosko, J. E. (1993) Eur. J. Cell Biol. 60, 250-255. [PubMed] [Google Scholar]
- 18.Kirchgessner, C. U., Patil, C. K., Evans, J. W., Cuomo, C. A., Fried, L. M., Carter, T., Oettinger, M. A. & Brown, J. M. (1995) Science 267, 1178-1183. [DOI] [PubMed] [Google Scholar]
- 19.van Putten, J. W. G., Groen, H. J. M., Smid, K., Peters, G. J. & Kampinga, H. H. (2001) Cancer Res. 61, 1585-1591. [PubMed] [Google Scholar]
- 20.Peretz, S., Jensen, R., Baserga, R. & Glazer, P. M. (2001) Proc. Natl. Acad. Sci. USA 98, 1676-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arrington, E. D., Caldwell, M. C., Kumaravel, T. S., Lohani, A., Joshi, A., Evans, M. K., Chen, H. T., Nussenzweig, A., Holbrook, N. J. & Gorospe, M. (2000) Free Radical Biol. Med. 29, 1166-1176. [DOI] [PubMed] [Google Scholar]
- 22.Caldecott, K. & Jeggo, P. (1991) Mutat. Res. 255, 111-121. [DOI] [PubMed] [Google Scholar]
- 23.Muller, C., Calsou, P., Frit, P., Cayrol, C., Carter, T. & Salles, B. (1998) Nucleic Acids Res. 26, 1382-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans, W. H. & Martin, P. E. (2002) Mol. Membr. Biol. 19, 121-136. [DOI] [PubMed] [Google Scholar]
- 25.Simon, A. M. & Goodenough, D. A. (1998) Trends Cell Biol. 8, 477-483. [DOI] [PubMed] [Google Scholar]
- 26.Nehme, A., Baskaran, R., Aebi, S., Fink, D., Nebel, S., Cenni, B., Wang, J. Y., Howell, S. B. & Christen, R. D. (1997) Cancer Res. 57, 3253-3257. [PubMed] [Google Scholar]
- 27.Zamble, D. B., Mu, D., Reardon, J. T., Sancar, A. & Lippard, S. J. (1996) Biochemistry 35, 10004-10013. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, K., Chew, M., Yang, E. B., Wong, K. P. & Mack, P. (2001) Mol. Pharmacol. 59, 837-843. [DOI] [PubMed] [Google Scholar]
- 29.Hirschi, K. K., Xu, C. E., Tsukamoto, T. & Sager, R. (1996) Cell Growth Differ. 7, 861-870. [PubMed] [Google Scholar]
- 30.Mehta, P. P., Bertram, J. S. & Loewenstein, W. R. (1986) Cell 44, 187-196. [DOI] [PubMed] [Google Scholar]
- 31.Yamasaki, H. & Naus, C. C. (1996) Carcinogenesis 17, 1199-1213. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka, M. & Grossman, H. B. (2001) Hum. Gene Ther. 12, 2225-2236. [DOI] [PubMed] [Google Scholar]
- 33.Azzam, E. I., de Toledo, S. M. & Little, J. B. (2003) Oncogene 22, 7050-7057. [DOI] [PubMed] [Google Scholar]
- 34.Morgan, W. F. (2002) Mil. Med. 167, 44-45. [PubMed] [Google Scholar]
- 35.Mothershill, C. & Seymour, C. B. (2004) Nat. Rev. Cancer 4, 158-164. [DOI] [PubMed] [Google Scholar]
- 36.Prise, K. M., Belyakov, O. V., Folkard, M. & Michael, B. D. (1998) Int. J. Radiat. Biol. 74, 793-798. [DOI] [PubMed] [Google Scholar]
- 37.Aghi, M., Hochberg, F. & Breakefield, X. O. (2000) J. Gene Med. 2, 148-164. [DOI] [PubMed] [Google Scholar]
- 38.Laird, D. W., Fistouris, P., Batist, G., Alpert, L., Huynh, H. T., Carystinos, G. D. & Alaoui-Jamali, M. A. (1999) Cancer Res. 59, 4104-4110. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.