Abstract
The aim of this study is to identify general properties of emerging infectious agents that determine the likely success of two simple public health measures in controlling outbreaks, namely (i) isolating symptomatic individuals and (ii) tracing and quarantining their contacts. Because these measures depend on the recognition of specific disease symptoms, we investigate the relative timing of infectiousness and the appearance of symptoms by using a mathematical model. We show that the success of these control measures is determined as much by the proportion of transmission occurring prior to the onset of overt clinical symptoms (or via asymptomatic infection) as the inherent transmissibility of the etiological agent (measured by the reproductive number R0). From published studies, we estimate these quantities for two moderately transmissible viruses, severe acute respiratory syndrome coronavirus and HIV, and for two highly transmissible viruses, smallpox and pandemic influenza. We conclude that severe acute respiratory syndrome and smallpox are easier to control using these simple public health measures. Direct estimation of the proportion of asymptomatic and presymptomatic infections is achievable by contact tracing and should be a priority during an outbreak of a novel infectious agent.
Keywords: epidemiology, severe acute respiratory syndrome, HIV, smallpox, influenza
The global spread of severe acute respiratory syndrome (SARS) in early 2003 caused at least 800 deaths and substantial morbidity and had a significant economic cost for the worse affected countries (1–4). Despite rapid early spread, the epidemic eventually was contained, reflecting in part a highly effective global public health response. However, containment was aided also by specific epidemiological and biological characteristics of the SARS virus. Evaluating whether the methods used to control SARS are likely to be equally effective for future outbreaks of other emerging infectious diseases requires a more detailed understanding of the factors that make containment feasible even when effective vaccines or treatment are not available.
In the first instance, two basic public health policy options exist for controlling the spread of an infectious disease in the absence of effective vaccines or treatment: (i) effective isolation of symptomatic individuals and (ii) tracing and quarantining of the contacts of symptomatic cases. Both measures rely on rapid dissemination of information to facilitate accurate diagnosis of the symptoms of the disease based on a clear and precise case definition.
For SARS, the timing of the onset of symptoms relative to peak infectivity is likely to have been a crucial factor in the success of simple public health interventions aimed at reducing transmission. In SARS patients, viremia (as measured in both fecal material and respiratory tract exudates) seems to peak between 5 and 10 days after the onset of illness and overt clinical symptoms such as elevated temperature (5). Although viremia does not always predict infectivity, the very low levels measured in the days immediately after the onset of symptoms suggest that peak infectivity occurs somewhat later. Also, no confirmed cases of transmission from asymptomatic patients have been reported to date in detailed epidemiological analyses of clusters of SARS cases (6, 7), which suggests that, for SARS, there is a period after symptoms develop during which people can be isolated before their infectiousness increases. Actions taken during this period to isolate or quarantine ill patients can effectively interrupt transmission.
Modeling Infectious Disease Outbreaks
We develop a mathematical model of infectious disease outbreak dynamics that captures the distributions of times to symptoms and infectiousness for the etiological agent concerned and provides an alternative approach to earlier theoretical studies (8). This model can be used to evaluate the impact of simple public health control measures. By exploring different distributions and different intervention strategies, we aim to establish a general quantitative framework that can help predict whether simple control measures can be successful in reversing epidemic growth if applied efficaciously at an early stage of an outbreak.
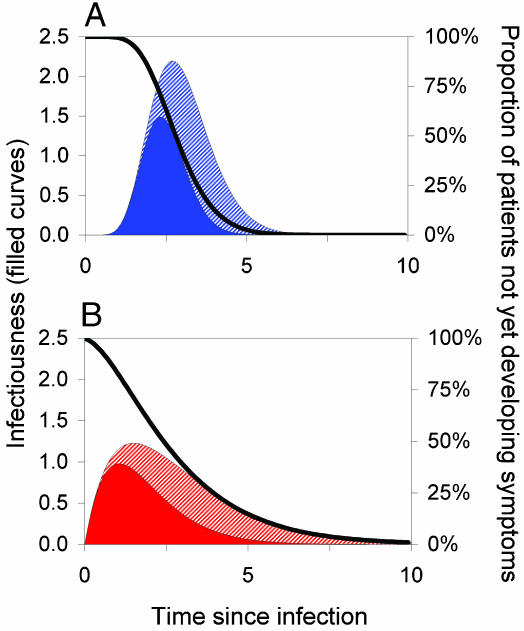
In our analyses, we focus on an infectious disease outbreak in its early stages within a community. We assume that the people in the community mix homogeneously; i.e., all susceptible individuals are equally likely to become infected. We characterize individuals in terms of their infectiousness as a function of the time (τ) since they were infected, β(τ), and also the probability that they have not yet developed symptoms, S(τ); example distributions are illustrated in Fig. 1. [Note that in the examples we illustrate, all patients eventually develop symptoms, because S(τ) tends to zero as the time since infection τ becomes large. More generally, if S(τ) tends to a fixed value S∞ > 0, then a proportion S∞ of infections are totally asymptomatic.]
Fig. 1.
Key epidemiological determinants. These determinants describe the pattern of typical disease progression for an individual patient as a function of time since infection (measured in arbitrary units). Filled curves represent infectiousness through time (left axis). The black curve represents S(τ), the probability of a person not having developed symptoms by a certain time (right axis). The basic reproduction number R0 is the area under the infectiousness curve (solid color plus cross-hatched section). The solid-colored area represents transmission arising prior to symptoms such that θ, the proportion of presymptomatic transmission, is the proportion of the total area under the infectiousness curve that is solid-colored. (A and B) Low- and high-variance incubation and infectiousness distributions, respectively. Both cases have R0 = 5, Tg = 3 (in arbitrary time units), andθ = 0.5; A shows a low variance of 0.1 × mean2, whereas B shows a high variance of 0.5 × mean2.
From this description of the course of infection in the individual, illustrated in Fig. 1, we identify three important parameters:
The basic reproduction number (9), R0, defined as the number of secondary infections generated by a primary infection in a susceptible population and which thus measures the intrinsic transmissibility of an infectious agent; it can be calculated as the area under the infectiousness curve (see Fig. 1 and Eq. 3). For an epidemic to expand in the early stages of spread, more than one secondary case has to be generated by the primary case, and hence we need R0 > 1.
The disease generation time Tg, which is the mean time interval between infection of one person and infection of the people that individual infects; together with R0, Tg sets the time scale of epidemic growth and thereby the speed with which intervention measures need to be put in place to avoid a large outbreak. Specifically, the doubling time for the number of cases in a growing outbreak is of order ln (2) Tg/(R0 - 1).
The proportion of transmission occurring prior to symptoms (or asymptomatically), θ, which determines the potential for symptom-based public health control measures to reduce the number of infections.
We base the analysis on an idealized optimal intervention, without delays in implementation of isolation and quarantining, so that Tg does not play an important role in our analysis. However, the framework can account for a distributed delay between onset of clinical symptoms and admission to hospital for isolation (in other words, delays in implementation), within the definition of θ. The effect of delays is always to increase θ. In the SARS epidemic, for example, there were significant delays between onset of symptoms and isolation in settings such as Hong Kong. These delays shortened over the course of the epidemic because of public health announcements to encourage early reporting to a health care setting (4). Obviously the definition of θ also depends on the clinical definition of symptoms: for example, for smallpox, different values will be obtained depending on whether prodromal fever or overt rash are used to determine isolation measures. Such uncertainties need to be incorporated into the estimation of θ.
The choice of parameter θ has the key advantage that at the start of an outbreak it can readily be estimated by using contact tracing since it is the proportion of infections occurring with an asymptomatic or presymptomatic infector.
Once public health interventions are implemented, a person is isolated immediately after symptoms with an efficacy εI, and a proportion of the people he or she infected prior to isolation are contact-traced and quarantined with efficacy εT. The two parameters εI and εT together determine the efficacy of implementation of the public health measures.
By analysis and simulation of the mathematical formulation of this model (discussed in detail in the next section), we find that the interventions are sufficient to control outbreaks of infections for combinations of values of parameters R0 and θ falling below a certain critical line. Estimated ranges of the parameters for four infections we consider here are shown as shaded areas in Fig. 2. The critical values of R0 and θ will depend on the intervention efficacies, as well as other parameters, and are shown for some selected cases in Fig. 3. Additional assumptions about the interpatient variability of the time to symptoms and the variance of the infectiousness function are made as appropriate and are color-coded into the three figures.
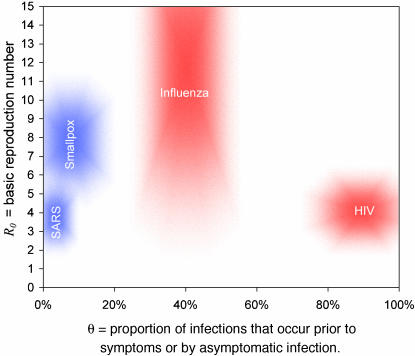
Fig. 2.
Parameter estimates. Plausible ranges for the key parameters R0 and θ (see main text for sources) for four viral infections of public concern are shown as shaded regions. The size of the shaded area reflects the uncertainties in the parameter estimates. The areas are color-coded to match the assumed variance values for β(τ) and S(τ) of Fig. 1 appropriate for each disease, for reasons that are apparent in Fig. 3.
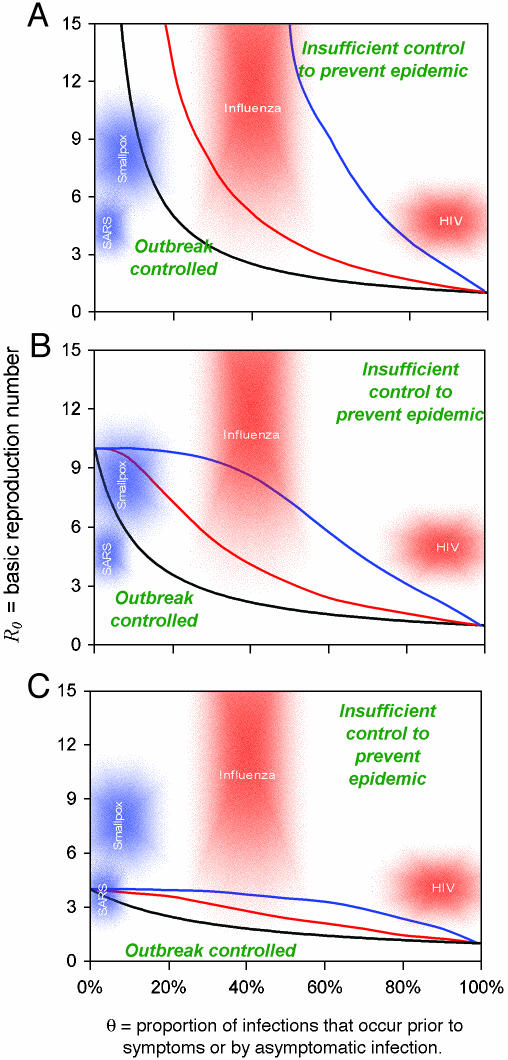
Fig. 3.
Criteria for outbreak control. Each curve represents a different scenario, consisting of a combination of interventions and a choice of parameters. For each scenario, if a given infectious agent is below the R0–θ curve, the outbreak is always controlled eventually. Above the curve, additional control measures (e.g., movement restrictions) would be required to control spread. Black lines correspond to isolating symptomatic individuals only. Colored lines correspond to the addition of immediate tracing and quarantining of all contacts of isolated symptomatic individuals. The black (isolation only) line is independent of distributional assumptions made (low or high variance), whereas the colored (isolation + contact tracing) lines match the variance assumptions made in Fig. 1 (red = high variance; blue = low variance). The efficacy of isolation of symptomatic individuals is 100% in A, 90% in B, and 75% in C. Contact tracing and isolation is always assumed 100% effective in the scenarios in which it is implemented (colored lines). Curves are calculated by using a mathematical model of outbreak spread incorporating quarantining and contact tracing (see main text).
Mathematical Model Formulation and Analysis‡
The basic model of disease transmission determines the dynamics of Y (t,τ), the current number of people, at time t, who were infected time τ ago. The cumulative epidemic size by time t is given by Y(t) = ∫∞0 Y(t, τ) dτ, whereas the incidence of infection (i.e., rate of people acquiring infection) at time t is Λ(t) = Y(t, 0). The model predicts that an outbreak will be controlled if the incidence declines to zero, i.e., Λ(t) → 0 as t becomes large. The model of disease spread is determined by the Von Foerster equations (15),
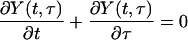 |
[1] |
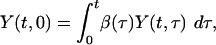 |
[2] |
together with the boundary conditions Y(0, 0) = Yi and Y(t, τ) = 0 when t < τ. Here β(τ) represents infectiousness at time τ since infection. The reproduction number for this model is defined by
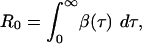 |
[3] |
whereas the generation time (or serial interval), denoted Tg,isgiven by the mean of the infectiousness distribution β(τ),
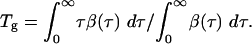 |
[4] |
The asymptotic behavior of the model (in the limit t → ∞)iseither exponential growth or decline: by substituting an exponential function Y(t, τ) = K(τ)exp(rt) into Eqs. 1 and 2, we see that r > 0, i.e., epidemic growth, occurs when R0 > 1, and r < 0, i.e., epidemic decline, occurs when R0 < 1, confirming the empirical definition of R0 in Eq. 3.
Other quantities are of practical importance in determining the extent and rapidity of control measures needed to control an outbreak. In any novel outbreak, a detailed description of clinical symptoms should be publicized as soon as possible to facilitate the rapid isolation (self-imposed or otherwise) of symptomatic individuals. We define εI as the efficacy of this isolation measure, which could equally be thought of as the fraction of people isolated or the degree by which their infectiousness is reduced. Isolation of symptomatic individuals modifies the model by changing Eq. 2 to
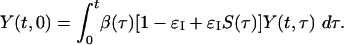 |
[5] |
Here, S(τ) is the proportion of people not having symptoms by time τ, i.e., the cumulative density function of the incubation period distribution (see Fig. 1).
It follows that isolation reduces the reproduction number from R0 to
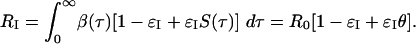 |
[6] |
Here, the parameter θ is the proportion of transmission occurring before symptoms develop or by asymptomatic infection:
 |
[7] |
Isolation will lead to the control of the outbreak when RI < 1. If isolation is perfect (i.e., εI = 1) and instantaneous (after symptoms), this measure can contain outbreaks when θ < 1/R0. Conditions of this general form are familiar in other contexts of infectious disease control (see ref. 9). This sets a biological upper bound for the efficacy of isolation. An intuitive interpretation of this condition is that isolating symptomatic individuals can only control an outbreak for which, on average, each person infects less than one person prior to symptoms appearing.
A more intuitive definition θ is obtained by noting that we can define the contributions of symptomatic and asymptomatic infection to R0 as 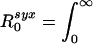 β(τ)[1 - S(τ)]dτ and
β(τ)[1 - S(τ)]dτ and 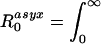 β(τ)S(τ)dτ, respectively. θ then is just the proportion
β(τ)S(τ)dτ, respectively. θ then is just the proportion 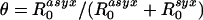 . This definition may also make it easier to generalize our analysis to models in which there is greater granularity or heterogeneity in contact processes and can be used directly for empirical estimators of
. This definition may also make it easier to generalize our analysis to models in which there is greater granularity or heterogeneity in contact processes and can be used directly for empirical estimators of 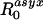 and
and 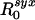 .
.
To capture the additional effect of tracing and quarantining the contacts of an isolated individual, we extend the model and define Y(t, τ, τ′) as the total number of people, at time t, who were infected time τ ago by people who themselves were infected time τ' ago. We define εT as the efficacy of contact tracing. An approximate model for Y(t, τ, τ′) is obtained by assuming that isolation and contact tracing are independent events, which will overestimate the efficacy of contact tracing, because more realistically it is performed on isolated individuals, creating a correlation between the events. To derive the equations for the model, we first define I(t, τ, τ′) as the number of infected people who are neither quarantined nor isolated [note that Y(t, τ, τ′) includes both quarantined and isolated individuals]. The hazard of being isolated then is
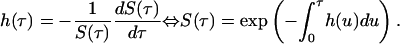 |
[8] |
We subdivide I(t, τ, τ′) into four groups of individuals: there are IĪT̄(t, τ, τ′) individuals who will never be isolated or contact-traced; IIT̄(t, τ, τ′) individuals who will be isolated but never contact-traced; IĪT(t, τ, τ′) individuals who will never be isolated but will be contact-traced; and IIT(t, τ, τ′) individuals who will be either isolated or contact-traced (competing hazards). If we define the differential operator Δ= ∂t + ∂τ + ∂τ′, then the equations of state are
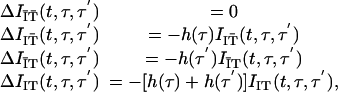 |
[9] |
which represents the removal of people by isolation or contact tracing, and
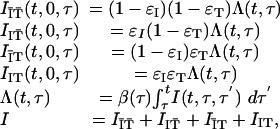 |
[10] |
which represents the efficacy of isolation and contact tracing in terms of the proportions of people entering into each subgroup, occurring at a total rate proportional to the incidence Λ(t, τ) of people infected by people who themselves have been infected for a time τ. The dynamics of Y(t, τ, τ′) then are recovered by the following transformation:
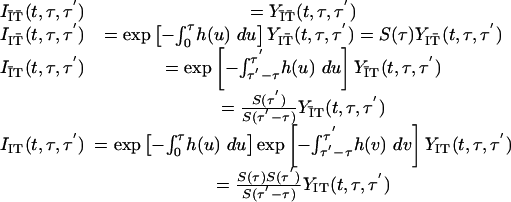 |
[11] |
such that
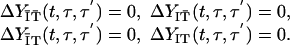 |
[12] |
The incidence Λ (t, τ) can be rewritten
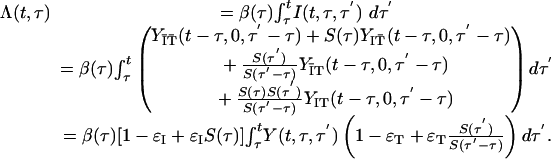 |
[13] |
The final model for the number of infected people with isolation and contact tracing then is given simply by
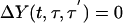 |
[14] |
and
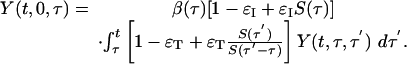 |
[15] |
The threshold condition that separates exponential growth from decline is found when the eigenvalue of the next generation operator is 1. The eigenvector then is the stationary distribution of infection generation times. From Eq. 15, we see that the operator is
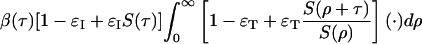 |
[16] |
Eq. 16 can be solved for simple functions β(τ) and S(τ).
A similar result to Eq. 16 was derived by Müller et al. (8), although for a different set of assumptions regarding the contact-tracing process; in ref. 8, contact tracing was assumed to be an immediately recursive process, such that contacts of infected contacts would be screened and traced before symptoms develop, in a continuing chain until all infected contacts have been isolated. Their approach is mathematically convenient but perhaps not as realistic for when no screening tools are available or for pathogens with short infectious periods.
The calculation simplifies dramatically when the distribution of time to symptoms is exponential, i.e., S(τ) = exp(-ντ), because in that case S(ρ + τ)/S(ρ) = S(τ), and Eq. 16 reduced to the algebraic equation
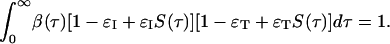 |
[17] |
If, in addition, we make the unrealistic but simplifying assumption that the infectiousness distribution is exponential, i.e., β(τ) = R0e-τ, such that the proportion of presymptomatic or asymptomatic transmission is θ = 1/(ν + 1), then the critical line dividing outbreak control from epidemic growth is determined by
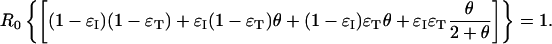 |
[18] |
We investigated the validity of the approximation underlying the model defined by Eqs. 14 and 15, namely that quarantining and contact tracing are separate independent processes determined by the same distribution S (τ), by investigating the dynamics of an individual-based, discrete-time simulation defined by the following, more realistic rules:
-
At time t = 0, a number Yi of people are infected with τ = 0.
Thereafter, at each time step of size δ:
Each τ value is incremented by δ.
If an individual is not isolated, he or she infects a random number of new people, drawn from a Poisson distribution of mean β (τ) δ.
Newly infected people are assigned a τ value of 0 and a time to onset of symptoms of S-1 (ρ), where ρ is a uniform random number between 0 and 1.
When an individual becomes symptomatic, he or she is isolated with probability εI. Symptomatic individuals who have been isolated have their contacts traced, and the people they have infected are themselves quarantined with probability εT. Contacts of individuals who are nonsymptomatic when quarantined are only themselves traced after symptoms develop.
The time step δ was reduced until convergence of model results was achieved, at which point the model can be expected to reproduce the dynamics of the comparable continuous time model. In the absence of contact tracing, this stochastic individual-based model was exactly equivalent in its mean behavior (dynamic as well as steady-state) to the deterministic model defined by Eqs. 14 and 15.
Once contact tracing was introduced, the analytic solution given above overestimated (sometimes substantially) the efficacy of contact tracing when isolation is <100% effective (i.e., εI < 1). To understand the mismatch, we note that the analytical solution agreed exactly with a modified form of the individual-based simulation in which in step 4, newly infected people are instead assigned two independent times to symptoms, both drawn from the same distribution S, the first time being the time at which a person is isolated and the second time being the time at which their contacts are traced and quarantined. The distribution of time to isolation and time to contact tracing are unchanged for this modified model, but the correlation between the two events is removed. We leave the development of an analytically tractable approach to capturing this correlation for future study.
When isolation is 100% effective, i.e., εI = 1, the correlation no longer becomes important, so that once again agreement between the deterministic model and the individual-based simulation was exact (at least for the exponential and simple gamma distributions we tested).
Parameter Estimates for a Selection of Viral Infections
We estimated R0 and θ values from published studies for four viral infections of interest. These estimates are plotted as shaded regions in Fig. 2 to compare with the scenario analysis created from the model, plotted in Fig. 3.
SARS. The basic reproduction number R0 for SARS has been estimated in a number of ways. For the Hong Kong outbreak, we fitted a detailed transmission model to the incidence time series, which gave estimates of 2–4 (2). Lipsitch et al (3) estimated R0 from exponential doubling times of several epidemics, which resulted in a wider range of just in excess of 1–7. To estimate θ, we first looked at the time to symptoms and infectiousness distributions, S(τ) and β(τ). S(τ) was determined from detailed analysis of clinical patient records (4). Up-to-date estimates based on Donnelly et al. (4) indicate a mean of 4.25 days and a variance of 14.25 days2. β(τ) can be inferred from viral shedding data (5), which peaks 5–10 days after onset of symptoms. To maximize θ, we chose a low variance distribution (var = 0.1 × mean2) and a peak at 9.25 days after infection, yielding θ < 11%. Because there is no evidence of presymptomatic transmission having occurred, no minimum value is given.
Smallpox. R0 and θ have been determined from a detailed analysis of an outbreak in Nigeria by Eichner and Dietz (10). They concluded 4 < R0 < 10 and 0 < θ < 20% (defining symptoms as the appearance of rash). The reported incubation distributions (10) suggest that our low variance model is appropriate for smallpox.
Pandemic Influenza. A maximum bound for R0 can be obtained by analyzing the case data from an outbreak of the 1978 H1N1 flu in a boys boarding school (11), yielding an upper bound of R0 < 21. No lower bound can be defined for a novel recombinant influenza strain. β (τ) can be estimated from experimental infections. Rvachev and Longini (12) suggest, by analyzing an unpublished experiment from a Soviet laboratory, a mean of 3 days (when variance = 0.5 × mean2), whereas viral shedding peaking at 2 days suggests that S (τ) has an estimated mean of 2 days (13). This results in a range of θ estimates of 30% < θ < 50%.
HIV. In populations for which spread into the general population has been seen (e.g., sub-Saharan Africa), R0 is by definition >1. We are not aware of published estimates of R0 for these generalized heterosexual epidemics; however, based on the formulas shown by Anderson and May (9), an upper bound of ≈5 can be obtained. If most transmission occurs during primary infection, then θ ≈ 100%. If transmission is more uniform, then the distribution of time to AIDS (14) leads to a lower bound: θ > 80%.
Intervention Strategies
We investigated six intervention strategies chosen to demonstrate the impact of the biology of the etiological agent, as characterized by R0 and θ, on the efficacy of the public health intervention. First, we considered three scenarios with no contact tracing: one with 100% effective isolation of symptomatic patients (i.e., εI = 1); one with 90% effective isolation (i.e., εI = 0.9); and one with 75% effective isolation (i.e., εI = 0.75) (Fig. 3). To these, we added three more scenarios by adding 100% effective contact tracing, which results in effective isolation of all the infected contacts of those who have been identified as symptomatic cases.
The model required two more assumptions to be made to be fully parametrized, namely characterization of the variance of the key distributions β(τ) and S(τ). We considered two cases that qualitatively matched the viral infections we describe below: a low variance case illustrated in Fig. 1A, for which the distributions are derived from gamma distributions with variance = 0.1 × mean2, chosen to match SARS and smallpox, and a high variance case illustrated in Fig. 1B, for which the distributions are derived from gamma distributions with variance = 0.5 × mean2, chosen to match influenza and (very approximately) HIV.
To summarize the predictions of the model, we illustrated for each scenario the critical line of R0 and θ values that separates epidemic growth (above the line) from outbreak control (below the line) (Fig. 3). In the absence of contact tracing, the line was determined analytically by Eq. 6, i.e., R0(1 - εI + εIθ) = 1, and was independent of which variances were chosen (i.e., Fig. 1 A or B). For the cases with contact tracing, a range of R0 and θ values was explored with the stochastic simulation repeated 100 times to determine critical parameter combinations for which, on average, infection incidence neither grew nor declined. The results depended on the variances of the distributions, and thus the analysis was repeated for the two cases of interest. In total we have nine critical lines, corresponding to six possible public health measures, and two possible variances for the distributions β(τ) and S(τ). The lines are plotted in Fig. 3, which is color-coded to match the assumed variances for β(τ) and S(τ) of Figs. 1 and 2.
Effect of Delays in Isolation
In reality, delays will occur between a patient becoming ill and being isolated. This delay obviously will reduce the efficacy of the control measures. Defining patient isolation in this context may not be straightforward, because self-isolation may arise prior to a patient presenting to the hospital and being isolated formally. This will depend on the nature of symptoms, the concomitant severity of illness, and also on the time scales involved. For influenza, for example, a person with flu-like symptoms at a workplace may not self-isolate before the end of the working day, which will be a substantial delay on influenza's rapid time scale of development and spread. For smallpox, on the other hand, the main rash is preceded by prodromal fever, and in an outbreak situation, it is plausible that most people would isolate themselves prior to or very near the start of the infectious period. For SARS, our analysis of patient reports in Hong Kong has shown that substantial delays of ≥2 days before patient hospitalization persisted during the outbreak, but these delays were substantially shorter than in the start of the outbreak (4). The effectiveness of self-isolation after fever is not known in that context. It is straightforward to implement these delays in our modeling framework by replacing the distribution S(τ) by a new distribution Σ(τ), which we define as the probability that a person has not been isolated by time τ since infection. Mathematically, Σ(τ) then can be expressed as the convolution of the time from infection to symptoms distribution S(τ) and the time from symptoms to isolation distribution F(τ). Specifically, if σ(τ) = -dΣ(τ)/dτ, s(τ) = -dS(τ)/dτ, and f(τ) = -dF(τ)/dτ are the corresponding probability density functions, then they are related by σ(τ) = ∫τ0 f(τ - u)s(u) du. The parameter θ then must be interpreted as the proportion of infections that arise prior to a person being isolated, defined by the equation θ =∫+∞0 β(τ)Σ(τ)dτ. Implementation of additional delays between a patient being isolated and his or her contacts being traced and quarantined also may be important and can be implemented in the individual-based model. The effect of delays in isolation can be envisaged in Fig. 3 by displacing the shaded areas to the right, whereas delays between isolation and contact tracing will simply bring the isolation-plus-contact-tracing lines closer to the isolation-only line. In this article, we focus on providing an estimate of what may be achieved in the best-case scenario.
Results and Conclusions
We propose that the proportion of transmission that occurs before the onset of symptoms or via asymptomatic transmission, which we call θ, is a useful new statistic for summarizing the likely feasibility of isolation- or contact-tracing-based intervention measures in controlling an epidemic outbreak. For control through isolation alone, we need θ < 1/R0. For diseases in which θ > 1/R0, contact tracing needs to be added to the set of control measures used. Fig. 3 shows how the two key parameters R0 and θ can be used to predict whether control policies involving isolation and contact tracing will lead to outbreak containment. In general, the curves show that for very high values of θ, neither contact tracing nor isolation make any impact in preventing an epidemic; for low values of θ, only isolation is effective. Contact tracing can be important, however, to counter the effect of delays in implementation of patient isolation, because these would effectively increase θ. For intermediate values of θ,the impact of contact tracing depends on the efficacy of reporting and isolation of symptomatic cases. For efficient (i.e., >90% effective) isolation, Fig. 3 shows that contact tracing can give substantial (up to 4-fold) additional reductions in transmission.
One key advantage of using R0 and θ as summary statistics for emerging pathogens is that, in principle, they can be readily estimated from detailed contact tracing and data collected from the first few hundred people infected in a novel disease outbreak. In addition, once the pathogen has been identified, θ can be inferred from longitudinal data on clinical symptoms and pathogen load within the infected patient. The framework presented here could be used to assess the likely success or failure of simple public health measures earlier than might be possible otherwise.
Comparing R0 and θ estimates for SARS, smallpox, “pandemic” influenza, and HIV (Fig. 2), it is clear that SARS is the easiest of the four infections to control because of its low R0 and θ values. Indeed, for SARS, our analysis indicates that effective isolation of symptomatic patients is sufficient to control an outbreak. The second most readily controlled infection of the four examined is smallpox. Here isolation even at the 90% level is insufficient to guarantee control, but effective contact tracing together with isolation of symptomatic cases is predicted to readily control an outbreak even for the highest feasible R0 values. This prediction contradicts other recent modeling studies of smallpox control (16, 17), largely because those studies assumed unrealistically high values of θ for smallpox (18). Influenza, on the other hand, is predicted to be very difficult to control even with 90% quarantining and contact tracing because of the high level of presymptomatic transmission. In addition, quarantining and contact tracing for influenza would probably be unfeasible because of the very short incubation (2 days) and infectious (3–4 days) periods of that disease. Last, despite its relatively low transmissibility (outside high-risk groups), our analysis predicts that effective “self-isolation” (i.e., cessation of risk behaviors) and contact tracing of AIDS patients would have done little to control the early stages of the HIV pandemic, again because of the high level of presymptomatic transmission. However, we note that we have not considered backward contact tracing for HIV in which infectious individuals may be identified by those they have infected as well as by those who infected them, which may be important for HIV because of the high mean and variance of the incubation-time distribution (8).
The analysis presented here highlights the need for the development of a more robust analytical framework for capturing the impact of contact tracing and other reactive, locally targeted control policies on disease-transmission dynamics, particularly when realistic infectiousness and incubation distributions are being modeled. Naive analytical approaches fail to capture the correlation structure between disease generations induced by contact tracing and thus can tend to overestimate its efficacy at reducing transmission. Additional development of the model structure introduced here and alternative approaches (8, 19) is needed, together with investigation of the impact of heterogeneity of transmission. Such heterogeneity could be important in diseases for which infectiousness and time to symptoms are strongly correlated. In the case of SARS, a few superspread events resulted in a large proportion of all cases (2): early isolation of these cases would have had a dramatic impact on the course of the epidemic. Similarly, for a sexually transmitted infection, reductions in risk-taking by the most sexually active members of the population can have a disproportionately large impact on an outbreak.
Nonetheless, the lesson from the SARS outbreak is that θ, the proportion of transmission that occurs before the onset of clinical symptoms or in asymptomatic infection, may be equally as important for determining the ease of control of a novel outbreak as the intrinsic transmissibility, R0. Both need to be considered when assessing the risks posed by emerging infectious agents. We conclude that the control of SARS through the use of simple public health measures was achieved because of the efficacy with which those measures were introduced and the moderate transmissibility of the pathogen coupled with its low infectiousness prior to clinical symptoms.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: SARS, severe acute respiratory syndrome.
Footnotes
Reading this section is not essential to an understanding of the results in this article.
References
- 1.Pearson, H., Clarke, T., Abbott, A., Knight, J. & Cyranoski, D. (2003) Nature 424, 121-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riley, S., Fraser, C., Donnelly, C. A., Ghani, A. C., Abu-Raddad, L. J., Hedley, A. J., Leung, G. M., Ho, L. M., Lam, T. H., Thach, T. Q., et al. (2003) Science 300, 1961-1966. [DOI] [PubMed] [Google Scholar]
- 3.Lipsitch, M., Cohen, T., Cooper, B., Robins, J. M., Ma, S., James, L., Gopalakrishna, G., Chew, S. K., Tan, C. C., Samore, M. H., et al. (2003) Science 300, 1966-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnelly, C. A., Ghani, A. C., Leung, G. M., Hedley, A. J., Fraser, C., Riley, S., Abu-Raddad, L. J., Ho, L. M., Thach, T. Q., Chau, P., et al. (2003) Lancet 361, 1761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiris, J. S., Chu, C. M., Cheng, V. C., Chan, K. S., Hung, I. F., Poor, L. L., Law, K. I., Tang, B. S., Hon, T. Y., Chan, C. S., et al. (2003) Lancet 361, 1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ksiazek, T. G., Erdman, D., Goldsmith, C. S., Zaki, S. R., Peret, T., Emery, S., Tong, S., Urbani, C., Comer, J. A., Lim, W., et al. (2003) N. Engl. J. Med. 348, 1953-1966. [DOI] [PubMed] [Google Scholar]
- 7.Lee, N., Hui, D., Wu, A., Chan, P., Cameron, P., Joynt, G. M., Ahuja, A., Yung, M. Y., Leung, C. B., To, K. F., et al. (2003) N. Engl. J. Med. 348, 1986-1994. [DOI] [PubMed] [Google Scholar]
- 8.Müller, J., Kretzschmar, M. & Dietz, K. (2000) Math. Biosci. 164, 39-64. [DOI] [PubMed] [Google Scholar]
- 9.Anderson, R. M. & May, R. M. (1991) Infectious Diseases of Humans: Dynamics and Control (Oxford Univ. Press, Oxford).
- 10.Eichner, M. & Dietz, K. (2003) Am. J. Epidemiol. 158, 110-117. [DOI] [PubMed] [Google Scholar]
- 11.Communicable Disease Surveillance Centre (Public Health Laboratory Service) (1978) Br. Med. J., 587. [DOI] [PMC free article] [PubMed]
- 12.Rvachev, L. A. & Longini, I. M. (1985) Math. Biosci. 75, 3-23. [Google Scholar]
- 13.Hayden, F. G., Fritz, R. S., Lobo, M. C., Alvord, W. G., Strober, W. & Straus, S. E. (1998) J. Clin. Invest. 101, 643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babiker, A., Darby, S., DeAngelis, D., Kwart, D., Porter, K., Beral, V., Darbyshire, J., Day, N., Gill, N., Coutinho, R., et al. (2000) Lancet 355, 1131-1137.10791375 [Google Scholar]
- 15.Murray, J. D. (2001) Mathematical Biology. I. An introduction (Springer, New York).
- 16.Halloran, M. E., Longini, I. M., Jr., Nizam, A. & Yang, Y. (2002) Science 298, 1428-1432. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan, E. H., Craft, D. L. & Wein, L. M. (2002) Proc. Natl. Acad. Sci. USA 99, 10935-10940.12118122 [Google Scholar]
- 18.Ferguson, N. M., Keeling, M. J., Edmunds, W. J., Gani, R., Grenfell, B. T., Anderson, R. M. & Leach, S. (2003) Nature 425, 681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huerta, R. & Tsimring, L. S. (2002) Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 66, 056115. [DOI] [PubMed] [Google Scholar]