Abstract
Impaired apoptosis is now recognized to be central to tumor development. Bcl2, activated by chromosomal translocation in human follicular lymphoma, promotes oncogenesis by inhibiting apoptosis. Bim, a distant proapoptotic relative, is emerging as a major physiologic antagonist of Bcl2. Here, we show that loss of Bim is oncogenic. Bim protein levels were elevated in the apoptosis-prone B lymphoid cells of Eμ-Myc-transgenic mice, and Bim-mutant Eμ-Myc mice had increased numbers of IgM-bearing B cells. Eμ-Myc-expressing B lymphoid cells deficient in Bim were refractory to apoptosis induced in vitro by cytokine deprivation or antigen receptor cross-linking. Thus, Bim is induced by Myc in B cells and mediates apoptosis. Remarkably, inactivation of even a single allele of Bim accelerated Myc-induced development of tumors, particularly acute B cell leukemia. None of the primary tumors from Bim+/- Eμ-Myc mice displayed loss of the second allele of Bim. These findings indicate that Bim is a tumor suppressor, at least in B lymphocytes, and is haploinsufficient. Whereas the p19Arf/p53 pathway is frequently mutated in tumors arising in Bim+/+ Eμ-Myc mice, it was unaffected in most Bim-deficient tumors, indicating that Bim reduction is an effective alternative to loss of p53 function.
One of the shields against cancer development is the intrinsic ability of cells to undergo apoptosis in response to various stresses, including activation of oncogenes such as Myc, cytokine deprivation, or genotoxic damage (1, 2). Hence, mutations that prevent cells from undergoing apoptosis can be oncogenic.
The link between impaired apoptosis and tumorigenesis was first appreciated when Bcl2, the gene found linked to the Ig heavy-chain locus by chromosomal translocation in human follicular lymphoma (3) was found to promote cell survival (4), and to act synergistically with Myc in promoting lymphomagenesis in mice (5). Whereas Bcl2 and several homologs enhance cell survival during development and stress, other close relatives such as Bax and Bak instead promote apoptosis (6, 7). A group of distant relatives attracting increasing interest are the Bcl2-homology 3 (BH3)-only proteins; so named because the only homology they bear to Bcl2 or to each other is the 9- to 16-amino acid BH3 domain. BH3-only proteins appear to be essential triggers of apoptosis. Once activated in response to intracellular damage signals, they bind to and neutralize prosurvival family members by means of the BH3 domain, which is necessary and probably sufficient for their death-promoting capacity. One unusual BH3-only protein, Bid, may instead function by interacting (transiently) with Bax or Bak, thereby triggering their pro-apoptotic activity.
The BH3-only protein Bim (8) is a major physiological antagonist of Bcl2, particularly in the hematopoietic system. Multiple defects in Bcl2-/- mice can be prevented by loss of Bim, including severe lymphopenia (9). Furthermore, like mice with enforced pan-hematopoietic expression of Bcl2 (10), mice lacking Bim accumulate excess lymphoid and myeloid cells (11). Bim regulates apoptosis of T cells in response to certain cytotoxic stimuli but not others (11), and is essential for elimination of autoreactive thymocytes (12) and B cells (13).
Because Bcl2 and Bim oppose each other and Bcl2 is oncogenic, we conjectured that Bim is a tumor suppressor. To test this hypothesis, we analyzed the impact of loss of Bim in Eμ-Myc-transgenic mice, which develop clonal pre-B or B cell lymphomas (14, 15) after a polyclonal expansion of pre-B cells (16) that is limited by increased apoptosis (17). Because constitutive expression of Myc induces apoptosis when cytokines are limiting (18, 19), curtailing apoptosis is a critical step toward malignancy in these mice. Although elevated Bcl2 can serve that role (5), a more common route is inactivation of the p53 tumor suppressor pathway. Loss of p53 (1, 20, 21) or its upstream regulator, p19Arf (21, 22), accelerates Eμ-Myc-induced lymphomagenesis, and lymphoid tumors induced by Eμ-Myc frequently harbor mutations that inactivate the p19Arf/Mdm2/p53 pathway (22). To determine whether Bim also plays a role in suppressing Myc-induced lymphomagenesis, we assessed the impact of loss of Bim on tumor development in Eμ-Myc mice.
Materials and Methods
Mice. Eμ-Myc-transgenic mice (15) were congenic with C57BL/6J. Bim-/- (266/266del) mice (11) had been backcrossed to C57BL/6J mice for eight or more generations; they were no longer susceptible to the autoimmune kidney disease found in the initial mixed (C57BL/6J × 129/Sv) strain background. Eμ-Myc males were crossed with Bim+/- females, and then Bim+/- Eμ-Myc male offspring were crossed with Bim-/- females to generate Bim-/- Eμ-Myc mice. The expected genotypes were obtained in normal Mendelian ratios. The transgene was detected by PCR amplification of an 830-bp product from the pUC vector backbone using the primers pUC-1: 5′-CAGCTGGCGTAATAGCGAAGAG-3′ and pUC-2: 5′-CTGTGACTGGTGAGTACTCAACC-3′. Genotyping for Bim by PCR used the primers PB20 (common): 5′-CATTCTCGTAAGTCCGAGTCT-3′; PB335 (wild-type allele): 5′-GTGCTAACTGAAACCAGATTAG-3′; and PB65 (targeted allele): 5′-CTCAGTCCATTCATCAACAG-3′.
Mice of each genotype were monitored daily for tumor development. Tumors, peripheral blood, and tissue samples were collected immediately after the killing of sick mice. Nucleated peripheral blood cell counts and mean nuclear volume measurements were performed with a Coulter particle count and size analyzer Z2 and Zap-oglobin II lytic reagent (Beckman Coulter). Histopathological analysis was performed on representative samples of tumor tissue, lymph nodes, spleen, thymus, kidney, lung, heart, and liver fixed in Bouin's solution and on sternum fixed in formalin. Blood films were stained with Diff Quik (Lab Aids Pty, Narrabeen, Australia). statview software (SAS Institute, Cary, NC) was used for generating Kaplan–Meier plots and for performing statistical analysis. All mice used for breeding were censored from the analysis at the time of mating to exclude any effect of breeding on tumor development. Clonality of tumors was assessed by Ig heavy-chain CDR3 spectratyping as described (23). Sequence analysis of Bim exon 5, which encodes the BH3 region, was performed directly on purified PCR products amplified from genomic DNA extracted from lymphoma tissue; tail DNA served as a control.
Antibodies. Antibodies used for flow cytometry were as described (5). Antibodies used for immunoblotting were: rabbit anti-p19Arf (Abcam, Cambridge, U.K.), sheep Ab-7 anti-p53 (Oncogene Research Products, San Diego), rat 3C5 monoclonal anti-Bim (kindly given by D. Huang and L. O'Reilly, The Walter and Eliza Hall Institute of Medical Research), rabbit anti-Bim (Stressgen), hamster 3F11 monoclonal anti-Bcl2 (Pharmingen), mouse AC-40 monoclonal anti-actin (Sigma), and mouse N6 monoclonal anti-heat shock protein 70 (generous gift from R. Anderson, Peter MacCallum Cancer Institute, Melbourne).
Fluorescence-Activated Cell Sorter Analysis and Cell Sorting. Cells (106 per analysis) were stained with relevant antibodies (5) labeled with fluorochromes (FITC, phycoerythrin, or Cy5) or biotin, using 1% normal rat serum to block Fc receptors. Streptavidin conjugated to FITC or phycoerythrin (Caltag, Burlingame, CA) was used as a secondary reagent for biotinylated antibodies. Analyses were performed on a Becton Dickinson LSR flow cytometer. To isolate pre-B and B cells, single-cell suspensions were prepared from bone marrow and spleen, erythrocytes were removed by lysis in NH4Cl solution, and the leukocytes were stained with Cy5-labeled anti-B220 and phycoerythrin-labeled anti-IgM antibody. After washing, the cells were filtered through cotton wool to remove aggregates and were stained with propidium iodide to exclude dead cells. Bone marrow pre-B cells (B220+ IgM-) and splenic B cells (B220+ IgM+) were sorted on a modified MoFlo cell sorter (DakoCytomation, Fort Collins, CO).
Cell Death Assays. Cells were cultured in DMEM plus 10% FCS/100 μM l-asparagine/50 μM 2-mercaptoethanol. To monitor cell death, 8 × 105 sorted cells were incubated in 200 μl of medium and stained with annexin V-FITC and propidium iodide, followed by flow cytometric analysis. Viable cells were negative for both annexin V and propidium iodide.
Immunoblotting. Whole-cell protein extracts were prepared from primary tumors or sorted pre-B and B cells. Briefly, cells were lysed in 20 μl per 106 cells of ice-cold radioimmunoprecipitation assay buffer (150 mM NaCl/50 mM Tris·HCl, pH 8.0/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS) containing 1 μg/ml each of leupeptin, aprotinin, soybean trypsin inhibitor, and pepstatin (Sigma, and Roche Diagnostics Australia, Castle Hill, NSW, Australia) for 30 min. After lysates were cleared by centrifugation, proteins were size-fractionated by SDS/PAGE and were electroblotted onto poly(vinylidene difluoride) membranes (Immobilon-P, Millipore). Membranes were incubated in 5% skim milk in PBS with 0.05% Tween 20 to block nonspecific binding, then probed with relevant antibodies in blocking solution, followed by HRP-conjugated secondary antibodies and enhanced chemiluminescence detection (Amersham Pharmacia Biosciences, Little Chalfont, U.K.).
Results
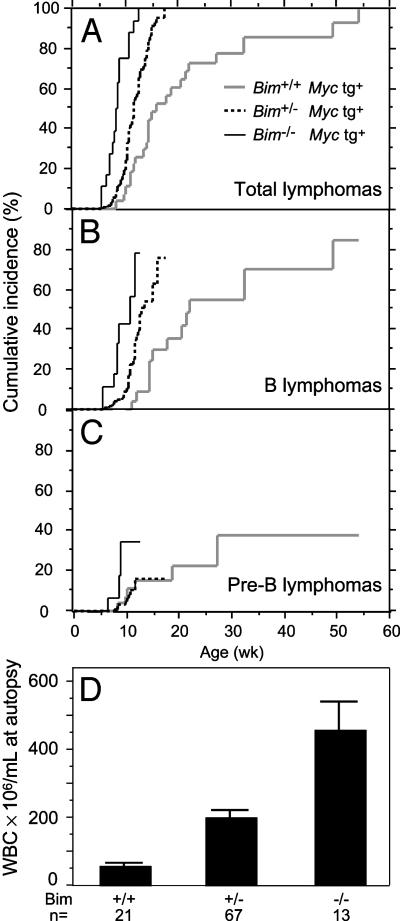
Loss of Bim Accelerates Morbidity in Eμ-Myc Mice by Promoting Acute B Cell Leukemia. By itself, loss of Bim does not appear to be markedly tumorigenic because only 1 of 30 Bim-/- mice monitored for at least 12 months developed a tumor: a follicular lymphoma at 37 wk of age. When we combined Bim deficiency with a lymphomagenic Eμ-Myc transgene, however, we saw a striking acceleration of morbidity (Fig. 1A). Whereas control (Bim+/+) Eμ-Myc mice developed tumors stochastically over a period of >12 months, with a median lifespan of ≈15 wk, all of the Bim-/- Eμ-Myc mice studied (11 of 11) became terminally ill between 7 and 12 wk of age (median 8.2 wk). Loss of one allele of Bim was almost as effective as loss of both alleles; the median lifespan for the 39 Bim+/- Eμ-Myc mice was 11 wk and 90% of the mice became terminally ill before 50% of Bim+/+ mice were affected.
Fig. 1.
Loss of Bim accelerates lymphoma development initiated by Eμ-Myc transgene. (A) Cumulative incidence of all tumors in mice of the indicated genotype. Tumors occurred earlier in Bim+/- mice (P < 0.0001) and Bim-/- mice (P < 0.0001) than in Bim+/+ mice. (B) B cell lymphoma development was accelerated in Eμ-Myc mice by the loss of one allele (P = 0.0008) or two alleles of Bim (P < 0.0001). (C) Pre-B lymphoma development was not altered in Bim+/- mice (P = 1), and a trend toward earlier onset in Bim-/- mice was not statistically significant (P = 0.1). (D) Frequency of total leukocytes in the blood of sick Eμ-Myc mice of indicated Bim genotype. Bim+/- mice had higher leukocyte counts than Bim+/+ mice (P < 0.0001), and Bim-/- mice had significantly higher counts than in either Bim+/- (P < 0.0001) or in Bim+/+ mice (P < 0.0001). The Bim-/- and Bim+/- mice had much higher numbers of B cells than did Bim+/+ mice (P < 0.0001 and P = 0.003, respectively). Pre-B cell numbers were somewhat higher in the Bim-/- mice than in the Bim+/+ mice (P = 0.01), but were comparable in the Bim+/- mice (P = 0.9). Leukocyte counts were 7.9 ± 0.3 × 106 per ml in 45 healthy wild-type C57BL/6 mice and 16 ± 3 × 106 per ml in 39 healthy 3-wk-old Eμ-Myc Bim+/+ mice.
As in Bim+/+ mice, the tumors induced by Myc in Bim-deficient mice were either B cell lymphomas (70%) or pre-B lymphomas (Table 1). All those examined were monoclonal by PCR analysis of rearranged Ig heavy-chain variable region genes (ref. 23; 11 were tested from Bim+/+,10from Bim+/-, and 6 from Bim-/- mice) and transplantable in C57BL/6 mice (four tested for each genotype). Importantly, most of the accelerated morbidity was due to early onset of surface IgM-positive B cell tumors. The median time to terminal illness for these tumors was shortened from 23 wk in Bim+/+ Eμ-Myc-transgenic mice to 12 wk in Bim+/- Eμ-Myc mice, and 10 wk in Bim-/- Eμ-Myc mice (Fig. 1B). In contrast, the kinetics of pre-B cell tumor development was relatively unaffected by Bim deficiency (Fig. 1C).
Table 1. Frequency of pre-B and B cell tumors induced by the Eμ-Myc transgene.
| Pre-B lymphoma*
|
B lymphoma*
|
|||
|---|---|---|---|---|
| Genotype | Frequency, % | Incidence at 11 wk,† % | Frequency, % | Incidence at 11 wk,† % |
| Bim+/+ | 6 of 18 (33) | 11 | 12 of 18 (66) | 0 |
| Bim+/— | 9 of 39 (23) | 10 | 30 of 39 (77)‡ | 22 |
| Bim—/— | 3 of 11 (27) | 34 | 9 of 11 (73) | 57 |
Lymphomas were typed by flow cytometry of surface immunofluorescence: pre-B lymphoma (B220+ CD19+ IgM—); and B lymphoma (B220+ CD19+ IgM+)
Cumulative incidence at 11 wk of age. The other tumor type was censored, so the incidence shown is independent of the competing risk of the other type
The frequency of B lymphoma in Bim+/— versus Bim+/+ Eμ-Myc mice was not significantly different (P = 0.41)
Remarkably, loss of Bim favored leukemia development. Most terminally ill Bim-/- Eμ-Myc mice lacked the disseminated lymphadenopathy characteristic of Bim+/+ Eμ-Myc mice (15). Instead, they displayed very high levels of circulating lymphoblasts; on average, 46-fold greater than normal and 8-fold greater than tumor-bearing Bim+/+ Eμ-Myc mice (Fig. 1D). The leukemia was more pronounced for B cell tumors than for pre-B cell tumors. At autopsy, the mean nucleated blood cell count in Bim-/- Eμ-Myc mice bearing such tumors was twice that in animals having pre-B cell tumors (490 ± 120 × 106 versus 240 ± 100 × 106 per ml), and in heterozygotes, the difference was even greater (240 ± 30 × 106 versus 78 ± 16 × 106 per ml).
Lack of Bim Results in B Cell Excess. To investigate the influence of loss of Bim before the onset of malignancy, we analyzed peripheral blood from weanling mice of each genotype (Fig. 2A). The increase in leukocytes previously observed in adult Bim-/- mice (11) was apparent even at 3 wk of age, and indeed was comparable to that induced by the Myc transgene in a wild-type (Bim+/+) background (compare columns 3 and 4 with column 1). Notably, the two types of mutation were synergistic: Bim-/- Eμ-Myc mice had almost 8-fold more white blood cells than normal mice and almost 4-fold more than Bim+/+ Eμ-Myc animals. Significantly, loss of Bim altered the relative proportions of pre-B and B cells in Eμ-Myc mice (Fig. 2B). Whereas the pre-B cell population was elevated in all genotypes of Eμ-Myc mice, this increase was more pronounced for conventional than Bim mutant Eμ-Myc mice. In contrast, the increase in IgM-bearing B cells was more pronounced in the Bim mutant mice, particularly the homozygous mutants. A similar increase in B cells was observed in the spleen of Bim-deficient transgenic animals (data not shown). Thus, even in the preleukemic phase, loss of Bim favors accumulation of B cells in Eμ-Myc-transgenic mice.
Fig. 2.
Bim deficiency in Eμ-Myc mice results in excess B lymphoid cells. (A) White blood cell counts in healthy 3-wk-old mice of indicated Bim genotype. Open bars, nontransgenic mice; black bars, Eμ-Myc-transgenic mice. Differences between all Bim genotypes were statistically significant for Eμ-Myc-transgenic mice (P < 0.0005), as were the differences between nontransgenic and transgenic mice for each Bim genotype (P = 0.03, P < 0.0001 and P < 0.0001 for Bim+/+, Bim+/-, and Bim-/- mice, respectively). (B) Frequency of pre-B cells (B220+ sIgM-) and B cells (B220+ sIgM+) per ml of peripheral blood of healthy 3-wk-old wild-type C57BL/6 mice and Eμ-Myc mice of the indicated genotype, determined by flow cytometric analysis. B cells were more numerous in Bim-/- Eμ-Myc mice (P = 0.03) than in wild-type C57BL/6, whereas in Bim+/+ Eμ-Myc and Bim+/- Eμ-Myc mice, the difference from Bim+/+ Eμ-Myc mice was not significant (P = 0.44 and 0.24, respectively). The number of pre-B cells was significantly different between Bim+/+ Eμ-Myc mice and wild-type C57BL/6 (P < 0.003), but not between wild-type C57BL/6 and either Bim+/- Eμ-Myc (P = 0.26) or Bim-/- Eμ-Myc (P = 0.29) mice. Bim+/+ Eμ-Myc mice had significantly more pre-B cells than did Bim+/- Eμ-Myc or Bim-/- Eμ-Myc mice (P < 0.02 for each comparison).
Bim Is Induced by Myc and Mediates Myc-Induced Apoptosis. The enlargement of the B lymphoid compartment in Bim+/- and Bim-/- Myc-transgenic mice suggested that loss of Bim may have countered the apoptosis normally induced by Myc when cytokines become limiting (18, 19). To test this hypothesis, we assessed the survival of B lymphoid cells cultured in the absence of cytokines. Whereas the pre-B and B cells from Bim+/+ Eμ-Myc mice died rapidly, as reported (24), those from Bim-/- Eμ-Myc mice survived almost as well as cells from wild-type littermates (Fig. 3A). Inactivation of even just one Bim allele afforded significant protection (Fig. 3A).
Fig. 3.
Bim levels are elevated in preleukemic Eμ-Myc–expressing B lymphoid cells, and loss of Bim enhances their survival. (A) Loss of Bim enhances survival of B lymphoid cells deprived of cytokines. Sorted splenic B cells (B220+ IgM+; Left) and bone marrow pre-B cells (B220+ IgM-; Right) from healthy young Eμ-Myc mice were cultured and their viability was analyzed by flow cytometry at the indicated intervals, as described in Materials and Methods. The data are pooled for three Bim-/- Eμ-Myc-transgenic mice and four mice of each of the other genotypes. Error bars, SEM. (B) Loss of Bim protects against apoptosis induced by crosslinking surface IgM. B cells were sorted from the spleen of nontransgenic C57BL/6 animals of the indicated genotype and were incubated in the presence of 0.5 μg/ml anti-IgM Fab fragment. The survival of treated cells is shown, corrected for the spontaneous apoptosis that occurred in culture (see A). Loss of one or both alleles of Bim protected B cells significantly (Fisher's protected least significant difference for 40-h values, P = 0.0025 and P < 0.0001, respectively). (C)Eμ-Myc transgene modulates Bcl2 and Bim expression. Western blot analysis of Bim and Bcl2 protein in sorted pre-B and B cells from healthy 3-wk-old control and Eμ-Myc-transgenic mice. Samples were adjusted for total protein content, and actin staining was used as a loading control. Extracts from two independent sorts are shown in each group and are representative of the results from five experiments. BimEL and BimL are the two most abundant Bim isoforms.
As a further test of the impact of loss of Bim on survival capacity, B cells were treated with an anti-IgM Fab fragment to crosslink their antigen receptors. Once again, loss of a single Bim allele inhibited cell death, and loss of both alleles markedly increased survival (Fig. 3B; see also ref. 13). (The comparable experiment with Eμ-Myc-transgenic cells could not be performed because apoptosis induced by Myc proceeds faster in vitro than that induced by IgM cross-linking.)
To explore why B-lymphoid cells are so susceptible to Myc-induced apoptosis, we assessed Bim and Bcl2 protein levels in lysates of bone marrow pre-B cells and splenic B cells from healthy young Eμ-Myc animals and normal littermates. Both major isoforms of Bim (BimEL and BimL) (25) were markedly increased in transgenic B cells and, more variably, in pre-B cells (Fig. 3C). As reported (26), the transgenic cells also had lower levels of Bcl2 (Fig. 3C). Thus, Myc appears to promote apoptosis by inducing Bim and suppressing a major antagonist.
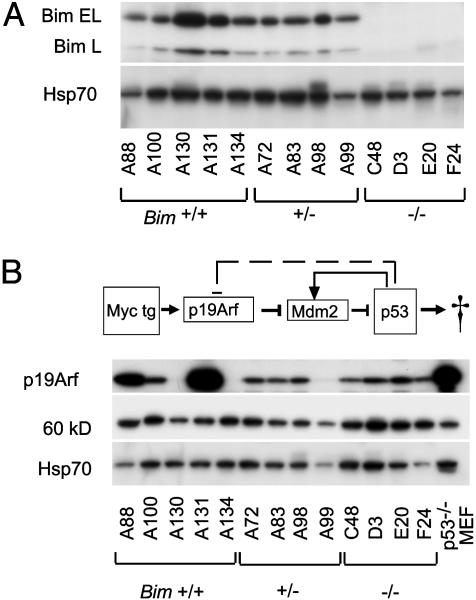
Tumor Development Does Not Involve Loss of the Second Bim Allele. The 1- to 3-month latency before tumors became manifest in Bim+/- and Bim-/- Eμ-Myc mice (Fig. 1 A–C) implicated somatic mutation in their genesis. For the tumors arising in Bim+/- Eμ-Myc mice, loss of the second Bim allele seemed a likely contributor. Surprisingly, however, Bim was readily detected in all 24 tumors analyzed (5 pre-B and 19 B lymphomas; e.g., Fig. 4A). Because the BH3 region of Bim is the only domain thought to be required for its proapoptotic activity (8, 11), we tested whether that region was mutated in the tumors. In 12 randomly selected tumors from Bim+/- Eμ-Myc mice, the BH3-encoding exon 5 sequence (27) was readily amplified from genomic DNA and yielded only wild-type sequence (data not shown). These results indicate that there is no strong selective pressure in vivo to further reduce Bim levels or Bim proapoptotic activity. The implication is that there is a critical concentration threshold below which Bim is unable to serve as a tumor suppressor during B lymphopoiesis.
Fig. 4.
Bim and p19Arf expression in Eμ-Myc tumors. (A) Western blot analysis of Bim protein in individual tumors from Eμ-Myc-transgenic mice of the indicated Bim genotype. BimEL and BimL are the two most abundant Bim isoforms. Heat shock protein 70 protein served as loading control. (B) Western blot analysis of p19Arf in the same tumors. Heat shock protein 70 and a 60-kDa protein detected by nonspecific staining with the anti-p19 antibody served as loading control, and p53-null mouse embryo fibroblasts (MEF) provided a positive control for p19Arf expression. p19Arf was detected with rabbit antibody. The diagram depicts Myc regulation of the p19Arf/Mdm2/p53 axis (modified from ref. 22).
Loss of Bim Relieves Pressure for Inactivation of p53 Downstream of Myc. We next investigated whether the p19Arf/Mdm2/p53 apoptotic pathway had been inactivated in the Eμ-Myc-induced tumors arising in Bim mutant mice, as is common in a wild-type background (21, 22). The level of p19Arf is a sensitive indicator for the functionality of this pathway (22). Myc up-regulates p19Arf expression, probably indirectly, and p19Arf sequesters Mdm2, thereby preventing inhibition and degradation of p53 (28).
Mutations that inactivate the p19 protein, induce overexpression of Mdm2, or inactivate p53 result in overexpression of p19Arf, due to the absence of the p53-induced negative-feedback loop (see diagram at the top of Fig. 4B). Whereas ≈30% of the tumors (5 of 16) arising in Bim+/+ mice overexpressed p19Arf (Fig. 4B and Table 2), which is similar to the incidence reported by others (22), none of the tumors from Bim-/- mice (0 of 12), and only one from Bim+/- mice (1 of 21) did so. The p53 response can also be crippled by loss of p19 expression, and Arf was undetectable in 3 of 21 (14%) Eμ-Myc tumors from Bim+/- mice, compared with 3 of 16 (19%) from Bim+/+ mice.
Table 2. Frequency of p19Arf alterations in Eμ-Myc lymphomas.
| P19Arf overexpressed
|
p19Arf undetectable
|
|||
|---|---|---|---|---|
| Genotype | Frequency, % | χ2; P* | Frequency, % | χ2; P* |
| Bim+/+ | 5 of 16 (31) | 3 of 16 (19) | ||
| Bim+/- | 1 of 21 (5) | 4.7; 0.03 | 3 of 21 (14) | 0.1; 0.71 |
| Bim-/- | 0 of 12 (0) | 4.6; 0.03 | 0 of 12 (0) | 2.5; 0.11 |
Lymphomas were assayed by immunoblotting for expression of p19Arf (see representative blot in Fig. 4).
Significance was determined by log-rank test in comparison with values for Eμ-Myc Bim+/+ mice
If these results are considered together (Table 2), expression of p19Arf was normal in 100% (12 of 12) of the assessable Bim-/- Eμ-Myc tumors and in 80% (17 of 21) of the heterozygous tumors, which is in marked contrast to the Bim+/+ Eμ-Myc tumors where p19 levels were perturbed (either increased or lost) in 50% (8 of 16) of those analyzed. Thus, loss of Bim removes the strong selection pressure for functional inactivation of the p53 apoptotic pathway during Myc-induced lymphomagenesis.
Discussion
In view of the oncogenic potential of Bcl2 (5, 29, 30) and antiapoptotic relatives such as BclxL (31) and Mcl1 (32), we reasoned that their antagonists, the BH3-only proteins, might function as tumor suppressors. To test this hypothesis, we focused on Bim, a major regulator of lymphoid and myeloid homeostasis (9, 11, 12, 33). Here, we have shown that loss of Bim accelerated Myc-induced lymphomagenesis (Fig. 1 A). Notably, loss of a single allele of Bim accelerated tumorigenesis almost as much as did the loss of both alleles (Fig. 1 A). Furthermore, none of the primary tumors from Bim+/- Myc-transgenic mice displayed loss of the second allele or evidence of its mutation (Fig. 4A). These results establish that Bim is a tumor suppressor, at least in B lymphoid cells. The striking impact of the loss of a single allele of Bim on tumorigenesis mirrors previous observations that Bim is haploinsufficient, most notably in the setting of the Bcl2 knockout mouse, in which Bim heterozygosity completely prevented polycystic kidney disease and the ensuing runting and early death, and improved the survival of lymphoid cells (9).
The acceleration of lymphomagenesis induced in Eμ-Myc mice by loss of Bim was comparable with that produced by gain of Bcl2 (5). Strikingly, however, the tumor phenotype was very different. Whereas constitutive overexpression of Bcl2 uniformly resulted in the development of lymphomyeloid progenitor cell tumors in Eμ-Myc mice (5, 24), loss of Bim favored the development of acute B cell leukemia (Fig. 1). Thus, loss of Bim markedly increases the vulnerability of IgM-bearing B cells to malignant transformation rather than more immature cells in the B lymphoid lineage (see further below). The impact of loss of Bim on lymphopoiesis was apparent before tumor onset. Peripheral blood cell counts in healthy Bim-/- Eμ-Myc weanlings were 8-fold higher than normal and almost 4-fold higher than in transgenic Bim+/+ littermates. Significantly, whereas pre-B cells were elevated in both, most of the excess cells in the young Bim-/- Eμ-Myc mice were mature B cells. Thus, the preleukemic state presages the major tumor phenotype.
Lymphoid cells lacking Bim are more refractory to apoptosis (this paper and ref. 11). Pre-B and B cells from Bim-deficient Eμ-Myc mice survived cytokine deprivation in culture (Fig. 3A), and presumably also in vivo. The Myc-driven population can thus continue to expand, increasing the probability of acquiring further oncogenic mutations. It is striking that lack of Bim impacts primarily on the IgM-bearing B cell compartment. This result may reflect variation in the expression patterns of Bcl2 family members during B cell development. Bim may be the principal BH3-only protein expressed by IgM-bearing B cells, but progenitor and pre-B cells may express sufficiently high levels of other BH3-only proteins to compensate for lack of Bim. Alternatively, or in addition, the B cell increase may indicate specificity among the BH3-only proteins, in response to different physiologic cues. Pertinently, our data suggest that apoptosis at the B cell receptor checkpoint, in particular, depends on Bim (Fig. 3B and ref. 13).
The elevated level of Bim in B lymphoid cells from Eμ-Myc mice (Fig. 3C) suggests that Bim is a proapoptotic target of Myc, either directly or indirectly. One pathway for Myc-induced apoptosis is through activation of p53; Myc up-regulates p19Arf expression, probably indirectly, and p19Arf sequesters Mdm2, thereby preventing inhibition and degradation of p53 (28). Once induced, p53 activates transcription of the genes encoding the BH3-only proteins Puma and Noxa (34, 35). Bim does not appear to be a p53 target, however, because Bim transcripts are not elevated in lymphocytes after irradiation (L. Coultas, P.B., and A. Strasser, unpublished observations), and Bim-/- lymphocytes retain nearly normal sensitivity to γ-irradiation and etoposide, which act by means of p53 up-regulation (ref. 11 and data not shown). Thus, Myc appears to induce Bim via a p53-independent pathway (Fig. 5). One possibility (Fig. 5, solid arrow) is that Myc induces transcriptional activation of Bim. Although there are two conserved canonical Myc binding sites within the Bim locus, we have been unable to demonstrate any direct activation of Bim by Myc (P.B., unpublished results), suggesting that Myc may act through induction of another transcription factor. Alternatively, Myc may act by means of p19Arf through a p53-independent mechanism (Fig. 5, broken arrow).
Fig. 5.

Model for Myc regulation of apoptosis. Data presented in this paper suggest that Bim is a p53-independent target of Myc, either directly or indirectly. Conceivably, as indicated by the broken line, p53-independent induction of apoptosis by p19Arf may also involve activation of Bim (see Discussion).
The importance of the p19Arf/Mdm2/p53 axis in mediating Myc-induced apoptosis has been demonstrated in both fibroblasts (36) and B lymphoid cells (21, 22). Pertinently, more than half of the lymphomas arising in Eμ-Myc mice were found to have sustained loss of function of either p53 or p19Arf (22), and tumorigenesis in these mice was markedly accelerated in a p19Arf-deficient background (21, 22). It was therefore surprising to find that the p19Arf/p53 pathway remained functional in 100% of the lymphomas induced by Myc in Bim-/- mice and in >80% of those from Bim+/- mice (Table 2). Thus, loss of Bim relieves the selection pressure for inactivation of the p19Arf/Mdm2/p53 pathway.
Our data suggest that Myc sensitizes cells for death by altering the balance between BH3-only proteins and Bcl2-like antiapoptotic proteins. Not only were Bim levels higher in B lymphoid cells expressing the Myc transgene than in their normal counterparts, but the levels of Bcl2 and BclxL were lower (Fig. 3C and ref. 26). The resulting increase in the ratio of pro- to antiapoptotic family members would lower the threshold for induction of apoptosis. The major impact of loss of a single allele of Bim suggests that the balance is finely poised in B cells, and that Bim is the major physiologically relevant BH3-only protein in these cells. Our hypothesis predicts that a similar outcome would be achieved by increasing the level of Bcl2-like proteins and, pertinently, >50% of tumors arising spontaneously in Bim+/+ Eμ-Myc mice displayed high expression of Bcl2 or BclxL (relative to nontransformed pre-B or B cells) rather than loss of p19Arf or p53 function (26).
Bim is expressed in a wide range of cell types, including many epithelial cells and neurons (25). Consequently, our observations encourage a search for Bim mutations, including single-allele loss, in a variety of human cancers. The human BIM gene (official name BCL2L11) is located at chromosome 2q12 or 2q13 (27), a region where alterations, primarily deletions, have been reported for 17 cases of human malignancy, nine of which were hematopoietic in origin (http://cgap.nci.nih.gov/Chromosomes/Mitelman).
Other BH3-only proteins may also serve as tumor suppressors. Mice lacking Bid have an increased tendency to develop chronic myelomonocytic leukemia (37), and some aged Bad-deficient mice develop diffuse large cell lymphoma (38). Other BH3-only proteins that are attractive candidate tumor suppressors include Bmf, which triggers anoikis and therefore may protect against metastasis (39), and the p53 targets Noxa and Puma (34, 35, 40–42).
Finally, our studies suggest that small molecules that mimic the action of Bim are potential cancer therapeutic agents. The conserved BH3 domain is an amphipathic helix that targets the hydrophobic surface groove on Bcl2 and its prosurvival relatives (43). The search for BH3 mimetics is intensifying and some promising leads are beginning to emerge (7, 44, 45). Conventional therapies work mainly by activating the cell death program indirectly (46, 47), and it seems likely that drugs that activate the cell-suicide machinery directly would be more effective. Bcl2 is a prime target, particularly because it acts downstream of p53, and thus should remain vulnerable, even in human tumors in which p53 has been crippled by mutation.
Acknowledgments
We thank A. Wiegmans and V. Marshall for technical assistance; A. Strasser for antibodies; S. Mihajlovic for histologic processing; and J. M. Adams and A. Strasser for helpful discussions. This work was supported by National Health and Medical Research Council (Canberra) Grants 222099 and 257502, National Cancer Institute Grant CA43540, and Leukemia and Lymphoma Society Special Center of Research Grant 7015. A.E. was supported by Austrian Science Fund Grants J2129 and J1921, and P.B. was supported by a Sylvia and Charles Viertel Senior Medical Fellowship.
Abbreviation: BH3, Bcl2-homology 3.
References
- 1.Cory, S., Vaux, D. L., Strasser, A., Harris, A. W. & Adams, J. M. (1999) Cancer Res. 59, 1685s-1692s. [PubMed] [Google Scholar]
- 2.Hanahan, D. & Weinberg, R. A. (2000) Cell 100, 57-70. [DOI] [PubMed] [Google Scholar]
- 3.Tsujimoto, Y., Finger, L. R., Yunis, J., Nowell, P. C. & Croce, C. M. (1984) Science 226, 1097-1099. [DOI] [PubMed] [Google Scholar]
- 4.Vaux, D. L., Cory, S. & Adams, J. M. (1988) Nature 335, 440-442. [DOI] [PubMed] [Google Scholar]
- 5.Strasser, A., Harris, A. W., Bath, M. L. & Cory, S. (1990) Nature 348, 331-333. [DOI] [PubMed] [Google Scholar]
- 6.Cory, S. & Adams, J. M. (2002) Nat. Rev. Cancer 2, 647-656. [DOI] [PubMed] [Google Scholar]
- 7.Cory, S., Huang, D. C. S. & Adams, J. M. (2003) Oncogene 22, 8590-8607. [DOI] [PubMed] [Google Scholar]
- 8.O'Connor, L., Strasser, A., O'Reilly, L. A., Hausmann, G., Adams, J. M., Cory, S. & Huang, D. C. S. (1998) EMBO J. 17, 384-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouillet, P., Cory, S., Zhang, L.-C., Strasser, A. & Adams, J. M. (2001) Dev. Cell 1, 645-653. [DOI] [PubMed] [Google Scholar]
- 10.Ogilvy, S., Metcalf, D., Print, C. G., Bath, M. L., Harris, A. W. & Adams, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 14943-14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouillet, P., Metcalf, D., Huang, D. C. S., Tarlinton, D. M., Kay, T. W. H., Köntgen, F., Adams, J. M. & Strasser, A. (1999) Science 286, 1735-1738. [DOI] [PubMed] [Google Scholar]
- 12.Bouillet, P., Purton, J. F., Godfrey, D. I., Zhang, L.-C., Coultas, L., Puthalakath, H., Pellegrini, M., Cory, S., Adams, J. M. & Strasser, A. (2002) Nature 415, 922-926. [DOI] [PubMed] [Google Scholar]
- 13.Enders, A., Bouillet, P., Puthalakath, H., Xu, Y., Tarlinton, D. M. & Strasser, A. (2003) J. Exp. Med. 198, 1119-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams, J. M., Harris, A. W., Pinkert, C. A., Corcoran, L. M., Alexander, W. S., Cory, S., Palmiter, R. D. & Brinster, R. L. (1985) Nature 318, 533-538. [DOI] [PubMed] [Google Scholar]
- 15.Harris, A. W., Pinkert, C. A., Crawford, M., Langdon, W. Y., Brinster, R. L. & Adams, J. M. (1988) J. Exp. Med. 167, 353-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langdon, W. Y., Harris, A. W., Cory, S. & Adams, J. M. (1986) Cell 47, 11-18. [DOI] [PubMed] [Google Scholar]
- 17.Jacobsen, K. A., Prasad, V. S., Sidman, C. L. & Osmond, D. G. (1994) Blood 84, 2784-2794. [PubMed] [Google Scholar]
- 18.Askew, D. S., Ashmun, R. A., Simmons, B. C. & Cleveland, J. L. (1991) Oncogene 6, 1915-1922. [PubMed] [Google Scholar]
- 19.Evan, G. I., Wyllie, A. H., Gilbert, C. S., Littlewood, T. D., Land, H., Brooks, M., Waters, C. M., Penn, L. Z. & Hancock, D. C. (1992) Cell 69, 119-128. [DOI] [PubMed] [Google Scholar]
- 20.Hsu, B., Marin, M. C., El-Naggar, A. K., Stephens, L. C., Brisbay, S. & McDonnell, T. J. (1995) Oncogene 11, 175-179. [PubMed] [Google Scholar]
- 21.Schmitt, C. A., McCurrach, M. E., de Stanchina, E., Wallace-Brodeur, R. R. & Lowe, S. W. (1999) Genes Dev. 13, 2670-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eischen, C. M., Weber, J. D., Roussel, M. F., Sherr, C. J. & Cleveland, J. L. (1999) Genes Dev. 13, 2658-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egle, A., Harris, A. W., Bath, M. L., O'Reilly, L. & Cory, S. (2004) Blood 103, 2276-2283. [DOI] [PubMed] [Google Scholar]
- 24.Strasser, A., Elefanty, A. G., Harris, A. W. & Cory, S. (1996) EMBO J. 15, 3823-3834. [PMC free article] [PubMed] [Google Scholar]
- 25.O'Reilly, L. A., Cullen, L., Visvader, J., Lindeman, G., Print, C., Bath, M. L., Huang, D. C. S. & Strasser, A. (2000) Am. J. Pathol. 157, 449-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eischen, C. M., Woo, D., Roussel, M. F. & Cleveland, J. L. (2001) Mol. Cell. Biol. 21, 5063-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouillet, P., Zhang, L. C., Huang, D. C., Webb, G. C., Bottema, C. D., Shore, P., Eyre, H. J., Sutherland, G. R. & Adams, J. M. (2001) Mamm. Genome 12, 163-168. [DOI] [PubMed] [Google Scholar]
- 28.Sherr, C. J. (2001) Nat. Rev. Mol. Cell Biol. 2, 731-737. [DOI] [PubMed] [Google Scholar]
- 29.McDonnell, T. J. & Korsmeyer, S. J. (1991) Nature 349, 254-256. [DOI] [PubMed] [Google Scholar]
- 30.Strasser, A., Harris, A. W. & Cory, S. (1993) Oncogene 8, 1-9. [PubMed] [Google Scholar]
- 31.Naik, P., Karrim, J. & Hanahan, D. (1996) Genes Dev. 10, 2105-2116. [DOI] [PubMed] [Google Scholar]
- 32.Zhou, P., Levy, N. B., Xie, H., Qian, L., Lee, C. Y., Gascoyne, R. D. & Craig, R. W. (2001) Blood 97, 3902-3909. [DOI] [PubMed] [Google Scholar]
- 33.Villunger, A., Scott, C., Bouillet, P. & Strasser, A. (2002) Blood 101, 2393-2400. [DOI] [PubMed] [Google Scholar]
- 34.Oda, E., Ohki, R., Murasawa, H., Nemoto, J., Shibue, T., Yamashita, T., Tokino, T., Taniguchi, T. & Tanaka, N. (2000) Science 288, 1053-1058. [DOI] [PubMed] [Google Scholar]
- 35.Nakano, K. & Vousden, K. H. (2001) Mol. Cell 7, 683-694. [DOI] [PubMed] [Google Scholar]
- 36.Zindy, F., Eischen, C. M., Randle, D. H., Kamijo, T., Cleveland, J. L., Sherr, C. J. & Roussel, M. F. (1998) Genes Dev. 12, 2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zinkel, S. S., Ong, C. C., Ferguson, D. O., Iwasaki, H., Akashi, K., Bronson, R. T., Kutok, J. L., Alt, F. W. & Korsmeyer, S. J. (2003) Genes Dev. 17, 229-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ranger, A. M., Zha, J., Harada, H., Datta, S. R., Danial, N. N., Gilmore, A. P., Kutok, J. L., Le Beau, M. M., Greenberg, M. E. & Korsmeyer, S. J. (2003) Proc. Natl. Acad. Sci. USA 100, 9324-9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puthalakath, H., Villunger, A., O'Reilly, L. A., Beaumont, J. G., Coultas, L., Cheney, R. E., Huang, D. C. S. & Strasser, A. (2001) Science 293, 1829-1832. [DOI] [PubMed] [Google Scholar]
- 40.Yu, J., Wang, Z., Kinzler, K. W., Vogelstein, B. & Zhang, L. (2003) Proc. Natl. Acad. Sci. USA 100, 1931-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villunger, A., Michalak, E. M., Coultas, L., Müllauer, F., Böck, G., Ausserlechner, M. J., Adams, J. M. & Strasser, A. (2003) Science 302, 1036-1038. [DOI] [PubMed] [Google Scholar]
- 42.Jeffers, J. R., Parganas, E., Lee, Y., Yang, C., Wang, J., Brennan, J., MacLean, K. H., Han, J., Chittenden, T., Ihle, J. N., et al. (2003) Cancer Cell 4, 321-328. [DOI] [PubMed] [Google Scholar]
- 43.Sattler, M., Liang, H., Nettesheim, D., Meadows, R. P., Harlan, J. E., Eberstadt, M., Yoon, H. S., Shuker, S. B., Chang, B. S., Minn, A. J., et al. (1997) Science 275, 983-986. [DOI] [PubMed] [Google Scholar]
- 44.Rutledge, S. E., Chin, J. W. & Schepartz, A. (2002) Curr. Opin. Chem. Biol. 6, 479-485. [DOI] [PubMed] [Google Scholar]
- 45.Baell, J. B. & Huang, D. C. S. (2002) Biochem. Pharmacol. 64, 851-863. [DOI] [PubMed] [Google Scholar]
- 46.Fisher, D. E. (1994) Cell 78, 539-542. [DOI] [PubMed] [Google Scholar]
- 47.Brown, J. M. & Wouters, B. G. (1999) Cancer Res. 59, 1391-1399. [PubMed] [Google Scholar]