Abstract
The utility of adenovirus (Ad) vectors for gene transduction can be limited by receptor specificity. We developed a gene-delivery vehicle in which the potent Ad5 vector was genetically reengineered to display the mucosal-targeting σ1 protein of reovirus type 3 Dearing (T3D). A σ1 construct containing all but a small virion-anchoring domain was fused to the N-terminal 44 aa of Ad5 fiber. This chimeric attachment protein Fibtail-T3Dσ1 forms trimers and assembles onto Ad virions. Fibtail-T3Dσ1 was recombined into the Ad5 genome, replacing sequences encoding wild-type fiber. The resulting vector, Ad5-T3Dσ1, expresses Fibtail-T3Dσ1 and infects Chinese hamster ovary cells transfected with human or mouse homologs of the reovirus receptor, junctional adhesion molecule 1 (JAM1), but not the coxsackievirus and Ad receptor. Treatment of Caco-2 intestinal epithelial cells with either JAM1-specific antibody or neuraminidase reduced transduction by Ad5-T3Dσ1, and their combined effect decreased transduction by 95%. Ad5-T3Dσ1 transduces primary cultures of human dendritic cells substantially more efficiently than does Ad5, and this transduction depends on expression of JAM1. These data provide strong evidence that Ad5-T3Dσ1 can be redirected to cells expressing JAM1 and sialic acid for application as a vaccine vector.
Adenovirus (Ad) vectors are potent gene-delivery vehicles capable of eliciting both mucosal and systemic immune responses (1). Human Ad serotypes 2 and 5 (Ad2 and Ad5) bind and enter cells by using the combined interactions of the fiber and penton base proteins with cellular receptors. The fiber protein is an elongated trimer with an N-terminal fibrous tail domain (shaft) and a C-terminal globular head domain (knob). Ad2 and Ad5 engage the coxsackievirus and Ad receptor (CAR) (2, 3) via a binding site located in the knob (4). CAR is a member of the Ig superfamily (2, 3) expressed at regions of cell–cell contact (5). After fiber-mediated attachment, the penton base binds to cell surface αv integrins, which mediate internalization (6).
Although Ad5 vectors transduce many types of cells, the efficiency of these vectors is limited if cells lack one or more of its receptors (7). For example, dendritic cells (DCs) do not express CAR and are poorly transduced by Ad5 (8). This relatively poor transduction of DCs can be enhanced by reengineering the vector to target alternative receptors (9, 10). Ad serotypes that bind to other receptors [e.g., CD46 (11)] mediate increased transduction of immunologically relevant cells (12), but these vectors are more promiscuous than Ad5 and deliver genes into cells that may not contribute to vaccination and thus may increase toxicity. Therefore, although potent, current Ad vectors lack sufficient specificity to function in some applications.
Mammalian reoviruses are nonenveloped, double-stranded RNA viruses with a broad host range (13). Reovirus infections are common, but most are asymptomatic. Reovirus enters the host by either the respiratory or enteric routes and infects epithelium and associated lymphoid tissue (14). The reovirus attachment protein, σ1, plays a key role in targeting the virus to distinct cell types, including those at mucosal surfaces (15–18). Similar to the Ad fiber, reovirus σ1 is an elongated trimer with head-and-tail morphology (19–21). A domain in the fibrous tail of serotype 3 Dearing (T3D) σ1 binds to α-linked sialic acid (22–25), whereas the head binds to junctional adhesion molecule 1 (JAM1) (26). JAM1 is an Ig-superfamily member expressed by a variety of cells including DCs (27) and epithelial and endothelial barriers (28–30).
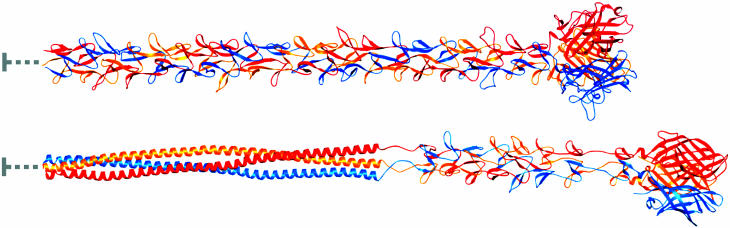
The structures of the Ad fiber (31) and reovirus σ1 (32) proteins are strikingly similar (Fig. 1). The two proteins are the only structures known to date to form trimers by using triple β-spiral motifs. The fiber shaft most likely is composed entirely of β-spiral repeats (31), whereas the σ1 tail is predicted to also contain an α-helical coiled-coil N-terminal to the β-spiral region (32). The head domains of both proteins are formed by eight antiparallel β-strands with identical interstrand connectivity. Therefore, although Ad and reovirus belong to different virus families and have few overall properties in common, the observed similarities between the attachment proteins and receptors of these viruses suggest a conserved mechanism of binding.
Fig. 1.
Full-length models of Ad5 fiber (Upper) and reovirus σ1 (Lower). The three monomers within each trimer are shown in red, orange, and blue. Both proteins have head-and-tail morphology, with an eight-stranded β-barrel domain forming the head. The Ad5 fiber shaft is predicted to consist of 21 β-spiral repeats (31). The Ad5 fiber model was generated by adding 17 β-spiral repeats to the four present in the crystal structure of an Ad2 fragment, which also has 21 β-spiral repeats (31). Sequence predictions suggest that σ1 contains an N-terminal ≈135-residue α-helical coiled coil followed by eight β-spiral repeats and the globular head domain (32, 49). The σ1 model was generated by first adding five β-spiral repeats to the N terminus of the crystallized fragment (32). This model then was joined with a 135-residue trimeric coiled coil formed by elongating an existing coiled-coil structure (50). The N-terminal 45 and 39 residuesof fiber and σ1, respectively, are not included in the model, because they form a virion-anchoring structure (indicated by gray lines). The overall lengths of the fiber and σ1 models are ≈325 and 385 Å, respectively, which is consistent with data from electron microscopy studies. This figure was prepared by using ribbons (51).
Based on the structural similarities between Ad fiber and reovirus σ1, we engineered chimeric fiber-σ1 attachment proteins to exploit the JAM1- and sialic acid-binding properties of σ1. Of those tested, only a near-full-length version of σ1 grafted onto the virion-insertion domain of Ad fiber (Fibtail-T3Dσ1) formed trimers and assembled onto Ad particles. We show here that when the fiber gene in the Ad5 genome is replaced with Fibtail-T3Dσ1, the resulting virus, Ad5-T3Dσ1, is capable of infecting intestinal epithelial cells expressing JAM1 and sialic acid and primary human DCs expressing JAM1. These data provide proof of principle for the development of chimeric Ad vectors encoding reovirus σ1 for gene delivery to mucosal surfaces. This work also establishes a foundation for the use of Ad-σ1 chimeric viruses as a template to enable facile reverse genetic manipulation of the reovirus attachment protein for studies of virus–cell and virus–host interactions.
Methods
Cells, Antibodies, and Viruses. 293A (Q-BIOgene, Carlsbad, CA) and Chinese hamster ovary (CHO) cells (American Type Culture Collection) were maintained as described (10). 633 cells, a derivative of A549 cells expressing E1, E2A, and Ad5 fiber, were provided by D. Von Seggern (The Scripps Research Institute, La Jolla, CA) and maintained as described (33). Caco-2 cells (American Type Culture Collection) were maintained in Alpha minimum essential medium (GIBCO) with 20% FBS. Primary human DCs (NHDC, Cambrex, Baltimore) were maintained according to vendor protocol.
The human (h)CAR-specific mAb RmcB was purified from CRL-2379 hybridoma cells (American Type Culture Collection). The hJAM1-specific mAb J10.4 was provided by Chuck Parkos (Emory University School of Medicine, Atlanta). Rabbit polyclonal serum 1561 was raised against the N-terminal region of Ad5 fiber (peptide ARPSEDTFNPVY). The c-Myc-specific mAb was purchased from PharMingen.
Ad vectors used in this study are based on the AdEasy system (Q-BIOgene) and carry the full E1- and E3-deleted Ad5 genome with the firefly luciferase gene, an internal ribosome entry site, and the humanized Renilla GFP expressed from a cytomegalovirus (CMV) immediate-early promoter in the E1 region.
Generation of Chimeric Fiber-σ1 Attachment Proteins. Fiber-σ1 fusion constructs were generated by using λ phage red recombinase (34) expressed in Escherichia coli strain BW25113/pKD46 (35) obtained from the E. coli Genetic Stock Center (http://cgsc.biology.yale.edu) as follows: Fibshaft-T3Dσ1, consisting of the N-terminal 396 aa of Ad5 fiber fused to amino acid 292 of T3D σ1; Fib8-T3Dσ1, consisting of the N-terminal 170 aa of Ad5 fiber fused to amino acid 167 of T3D σ1; and Fibtail-T3Dσ1, consisting of the N-terminal 44 aa of Ad5 fiber fused to amino acid 18 of T3D σ1. Sequences encoding the reovirus T3D σ1 protein flanked by a bovine growth hormone polyadenylation signal and a zeocin-resistance gene were amplified by using Pfu polymerase (Stratagene) and primers containing 39-nt overhangs homologous to the pCMVfiber plasmid. The pCMVfiber plasmid, containing the Ad5 fiber gene expressed from a CMV immediate-early promoter, was cotransformed with the PCR product into the λ phage red strain BW25113/pKD46. Recombinants were selected by using zeocin-containing agar plates.
Fibtail-T3Dσ1 was subcloned into a plasmid containing sequences homologous to E4 and then recombined into the Ad5 genome to replace the fiber gene using red recombinase. To aid in detection of the chimeric protein, two c-Myc tags (C2), and one hexahistidine tag (H6) were added to the C terminus of the chimera (Fibtail-T3Dσ1C2H6) before recombination. The recombinants were screened for loss of the fiber gene by restriction endonuclease mapping and sequencing.
Protein Expression and Characterization. CHO cells were transfected with plasmids encoding fiber-σ1 chimeras by using Lipofectamine-PLUS (Invitrogen), and cell extracts were harvested for SDS/PAGE. Immunoblots were performed as described (10).
Generation of a Chimeric Ad Vector. Linearized Ad genome encoding the Fibtail-T3Dσ1C2H6 chimera was transfected into 633 cells and maintained in the presence of 0.3 μM dexamethasone and 4 μg/ml polybrene. Virus was propagated, purified by CsCl gradient centrifugation, and quantitated as described (36). The resultant recombinant virus, Ad5-T3Dσ1, was amplified for a final round by using 293A cells to remove any residual fiber from newly assembled virions.
CsCl-banded Ad5, CAR-ablated biotinylated Ad [Ad5-BAP-TR (10)], and Ad5-T3Dσ1 were precipitated with trichloroacetic acid. Pellets were resuspended in loading buffer, and 4 × 1010 particles per lane were resolved by SDS/PAGE and immunoblotting. For total protein analysis, precipitated virus (1.5 × 1011 particles per lane) was resolved by SDS/PAGE, and gels were stained with Coomassie blue.
Transduction of CHO Cells Transfected with Receptor Constructs. CHO cells were transfected with plasmids expressing hCAR, hJAM1, or murine (m)JAM1 (37, 38). After 48 h, the cells were washed once with Hanks' balanced salt solution (GIBCO) with 1% BSA (HBSS-BSA) and adsorbed with 5,000 particles per cell of Ad5-T3Dσ1 at 4°C for 30 min. Cells were washed twice with HBSS-BSA, and fresh medium was added. After incubation at 37°C for 24 h, cells were lysed, and luciferase activity (in lumens) was measured as described (10).
Transduction of Caco-2 Cells and Primary DCs After Receptor Blockade. Cells were harvested, washed with HBSS-BSA, and incubated in suspension with 10 μg/ml of either hCAR-specific mAb RmcB or hJAM1-specific mAb J10.4 at 4°C for 30 min. Alternatively, cells were treated with 333 milliunits/ml of Clostridium perfringens neuraminidase type X (Sigma) at 37°C for 30 min to remove cell-surface sialic acid, followed by two washes with HBSS-BSA. Cells then were adsorbed with 5,000 particles per cell of Ad5-T3Dσ1 at 4°C for an additional 30 min, washed twice, and seeded onto 24-well plates in fresh medium. After incubation at 37°C for 24 h, cells were harvested for determination of luciferase activity.
Results
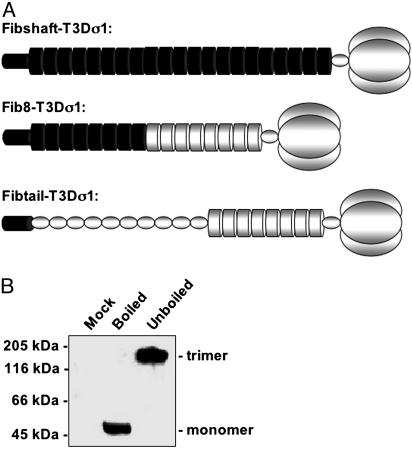
Design and Characterization of a Functional Fiber-σ1 Chimera. Based on the structural similarities between Ad5 fiber and reovirus σ1 (Fig. 1), we engineered three Ad fiber-reovirus σ1 chimeras with increasingly larger portions of σ1 protein replacing structurally homologous regions of fiber (Fig. 2A). Fibshaft-T3Dσ1 contains the N-terminal 21 β-spiral repeats of fiber fused to the head domain of T3D σ1. Fib8-T3Dσ1 contains the N-terminal eight β-spiral repeats of fiber fused to the T3D σ1 β-spiral and head domains. Fibtail-T3Dσ1 contains the N-terminal 44 aa virion-anchoring domain (39) fused to T3D σ1 lacking only the N-terminal 17 amino acids. After transfection of CHO cells, each of the chimeric attachment proteins was expressed, but only Fibtail-T3Dσ1 formed trimers (Fig. 2B and data not shown), suggesting that only this chimera maintains native folding.
Fig. 2.
Design and expression of chimeric fiber-σ1 attachment proteins. (A) Schematic diagram of the chimeric fiber-σ1 attachment proteins described in the text. Regions corresponding to fiber and σ1 in the diagrams are shaded black and gray, respectively (not drawn to scale). Fiber tail, which mediates virion anchoring, is represented as a small cylinder, the α-helical coiled coils as small ovals, the β-spiral repeats as large cylinders, and the head domain as three large ovals. (B) Immunoblots of denatured (boiled) and native (unboiled) lysates of CHO cells transfected with plasmid expressing Fibtail-T3Dσ1 probed with a serum (1561) that recognizes the N-terminal region of Ad5 fiber.
Production and Characterization of an Ad Vector Expressing a Chimeric Fiber-σ1 Attachment Protein. The Fibtail-T3Dσ1 gene was recombined into an Ad5 genome lacking E1 and E3 to replace the fiber gene by using λ phage red recombinase (34). During the cloning process, two c-Myc tags (C2) and one hexahistidine tag (H6) were added to the C terminus of Fibtail-T3Dσ1 (Fibtail-T3Dσ1C2H6) to facilitate protein detection. The resulting virus, Ad5-T3Dσ1, was rescued by transfection and production in 633 fiber-expressing cells (33). After amplification in 633 cells, the virus was passaged in 293A cells to eliminate fiber from the virions and allow only Fibtail-T3Dσ1C2H6 to be encapsidated.
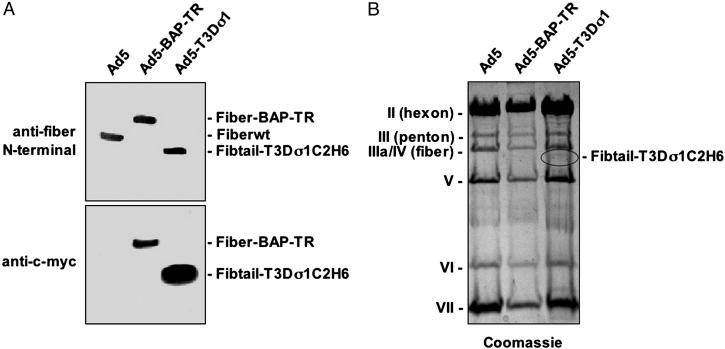
To determine whether Fibtail-T3Dσ1C2H6 was encapsidated onto Ad5 virions, CsCl-purified Ad5, Ad5-BAP-TR, which displays biotinylated fibers (10), and Ad5-T3Dσ1 were analyzed by immunoblotting with antibodies specific for either the fiber N terminus or the c-Myc epitope tag (Fig. 3A). Comparison of the immunoblots demonstrated that Fibtail-T3Dσ1C2H6 was encapsidated onto Ad5 virions at levels similar to those of fiber on Ad5 and Ad5-BAP-TR. As anticipated, the anti-c-Myc antibody recognized both Ad5-BAP-TR and Ad5-T3Dσ1, which contain c-Myc tags but not wild-type fiber. Coomassie blue staining demonstrated that relative amounts of the capsid proteins of wild-type Ad5 and Ad5-T3Dσ1 were indistinguishable (Fig. 3B). Thus, Fibtail-T3Dσ1C2H6 is encapsidated onto Ad virions and enables normal virion maturation.
Fig. 3.
Characterization of Ad5-T3Dσ1. Ad5 virions expressing wild-type fiber (Fiberwt), CAR-ablated biotinylated fiber (Fiber-BAP-TR) (10), and Fibtail-T3Dσ1C2H6 were precipitated with trichloroacetic acid. (A) Precipitated particles (4 × 1010 per lane) were resolved by SDS/PAGE and immunoblotted with anti-c-Myc mAb 9E10 or antiserum 1561, which recognizes the N-terminal region of Ad5 fiber. (B) Precipitated particles (1.5 × 1011 per lane) were resolved by SDS/PAGE and stained with Coomassie blue.
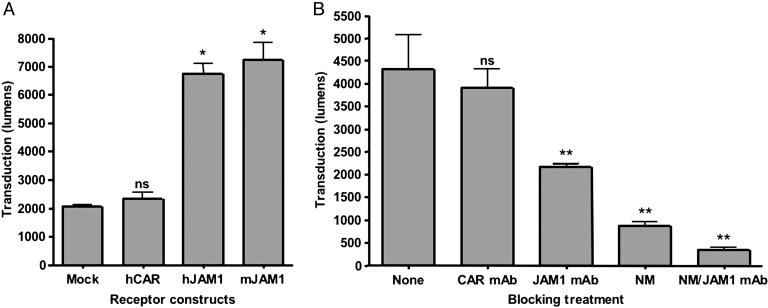
Transient Transfection of CHO Cells with JAM1 Rescues Infection by Ad5-T3Dσ1. To determine whether the chimeric Fibtail-T3Dσ1 attachment protein could bind to JAM1, CHO cells were transfected with plasmids expressing hCAR, hJAM1, and mJAM1 and tested for infection by luciferase-expressing Ad5-T3Dσ1. CHO cells were chosen for these studies, because they lack both CAR and JAM1 and are poorly infected by both Ad and reovirus (38). Transduction of CHO cells by Ad5-T3Dσ1 was increased substantially by expression of either hJAM1 or mJAM1 but not by expression of hCAR (Fig. 4A), the receptor for Ad5 (2, 3). These data indicate that the JAM1-binding domain of Ad5-T3Dσ1 is functional and can target JAM1-expressing cells in a species-independent fashion.
Fig. 4.
Ad5-T3Dσ1 transduction is mediated by JAM1 and sialic acid. (A) CHO cells were transiently transfected with plasmids encoding hCAR, hJAM1, or mJAM1. After 48 h of incubation to permit receptor expression, cells were adsorbed with 5,000 particles per cell of Ad5-T3Dσ1 and harvested 24 h later for luciferase assay. Transduction was measured in lumens. (B) Caco-2 cells were either untreated or treated with 10 μg/ml hCAR-specific mAb RmcB (CAR mAb), 10 μg/ml hJAM1-specific mAb J10.4 (JAM1 mAb), 333 milliunits/ml C. perfringens neuraminidase (NM), or both JAM1 mAb and neuraminidase. Cells were adsorbed with 5,000 particles per cell of Ad5-T3Dσ1 and harvested 24 h later for luciferase assay. Transduction was measured in lumens. The results are presented as the means for three independent experiments. Error bars indicate SD. A paired Student's t test was performed to compare transduction of transfected or treated cells versus mock or untreated cells (*, P < 0.01; **, P < 0.05; ns, not significant).
Inhibition of Binding to JAM1 and Sialic Acid Blocks Ad5-T3Dσ1 Infection of Caco-2 Cells. We next tested the capacity of hJAM1-specific mAb J10.4 and C. perfringens neuraminidase to inhibit transduction by Ad5-T3Dσ1. Caco-2 intestinal epithelial cells, a model for enteric mucosal surfaces (40, 41), were used for these experiments, because these cells express CAR, JAM1, and sialic acid (26, 42). Transduction by Ad5-T3Dσ1 was inhibited 50% by JAM1-specific mAb J10.4 and 80% by neuraminidase (Fig. 4B). Combined treatment with both mAb J10.4 and neuraminidase reduced transduction nearly 95%. In contrast, isotype-matched hCAR-specific mAb RmcB, used as a negative control, did not diminish luciferase transduction (Fig. 4B).
To ensure that JAM1-dependent transduction by Ad5-T3Dσ1 depends on σ1 and not another Ad protein, we tested the capacity of the T3D σ1-specific mAb 9BG5 (24) to block infection of Caco-2 cells. In contrast to T1L σ1-specific mAb 5C6 (24), mAb 9BG5 inhibited transduction in a dose-dependent fashion (data not shown). We noted a similar decrease in transduction efficiency after incubation of Ad5-T3Dσ1 with sialoglycophorin, which is known to interact with reovirus T3D σ1 (22), before infection (data not shown). These results demonstrate that transduction by Ad5-T3Dσ1 requires σ1 and its receptors, JAM1 and sialic acid.
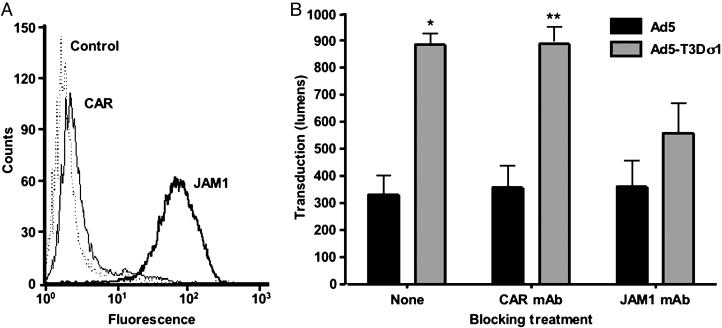
Ad5-T3Dσ1 Transduces Primary Human DCs. DCs play important roles in induction of adaptive immune responses (43). To determine whether Ad5-T3Dσ1 is capable of transducing DCs, we infected primary cultures of human DCs with Ad5 and Ad5-T3Dσ1. DCs express JAM1 but not CAR (Fig. 5A), which is consistent with previous observations (27). Transduction of DCs by Ad5-T3Dσ1 was substantially more efficient than by Ad5 (Fig. 5B). Moreover, transduction was eliminated almost completely by treatment with hJAM1-specific mAb J10.4 (Fig. 5B). These findings suggest that Ad5-T3Dσ1 may have utility for transducing CAR-negative DCs at mucosal and other sites.
Fig. 5.
Ad5 and Ad5-T3Dσ1 transduction of primary human DCs. (A) DCs were assessed for surface expression of CAR and JAM1 by flow cytometry by using hCAR-specific mAb RmcB and hJAM1-specific mAb J10.4, respectively (38). (B) DCs were either untreated or treated with 10 μg/ml hCAR-specific mAb RmcB (CAR mAb) or hJAM1-specific mAb J10.4 (JAM1 mAb) before adsorption with 5,000 particles per cell of either Ad5 or Ad5-T3Dσ1. Cells were harvested 24 h later for luciferase assay. Transduction was measured in lumens. The results are presented as the means for three independent experiments. Error bars indicate SD. A paired Student's t test was performed to compare transduction by Ad5 versus Ad5-T3Dσ1 (*, P < 0.01; **, P < 0.05).
Discussion
In this study, we fused two structurally homologous viral attachment proteins, Ad fiber and reovirus σ1, to produce a functional chimeric virus, Ad5-T3Dσ1. Of the three fiber-σ1 chimeras tested, only Fibtail-T3Dσ1 bearing the Ad5 fiber virion-insertion domain fused to an almost-full-length version of T3D σ1 protein formed trimers and assembled onto Ad virions. The lack of trimerization of Fib8-T3Dσ1 and Fibshaft-T3Dσ1 was surprising, because both the head and tail regions of σ1 contain trimerization domains (44), whereas the fiber knob domain initiates and maintains trimerization (45). Because only Fibtail-T3Dσ1 formed trimers, it is likely that the C-terminal trimerization domain of σ1 is insufficient for trimerization of the fiber shaft. Alternatively, it is possible that the chimeric Fib8-T3Dσ1 and Fibshaft-T3Dσ1 proteins do not form trimers, because the fused β-spiral junctions are imperfectly matched.
In Ad5-T3Dσ1 virions, Fibtail-T3Dσ1 was encapsidated at levels comparable with wild-type fiber. Furthermore, the capsid protein profile of Ad5-T3Dσ1 is identical to that of wild-type Ad5. Most importantly, experiments using receptor-transfected cells, antibodies, and reagents that block σ1–sialic acid interactions provide compelling evidence that Ad5-T3Dσ1 displaying Fibtail-T3Dσ1 retains both the JAM1- and sialic acid-binding functions of the T3D σ1 protein.
We envision at least four applications for chimeric Ad vectors in which the CAR-binding functions of fiber have been replaced with the JAM1- and sialic acid-binding functions of σ1. First, Ad vectors based on fiber-σ1 chimeras may serve to efficiently target mucosal sites for enhanced induction of immune responses at mucosal surfaces. Second, because JAM1 and sialic acid are expressed on a variety of cells, Ad5-T3Dσ1 and its derivatives may have utility for transducing cells deficient in CAR (e.g., DCs and certain types of cancer cells). Third, because σ1 incorporates its own trimerization motifs, fiber-σ1 fusions may provide a trimeric scaffold for the display of other cell-targeting ligands in a manner analogous to fiber-fibritin chimeras (46). In support of this approach, we recently appended single-chain antibodies onto truncated forms of Fibtail-T3Dσ1 (unpublished data). Fourth, Ad vectors based on Ad5-T3Dσ1 can be used as a simple genetic platform for directed mutagenesis of σ1 for studies of reovirus tropism and receptor-linked signaling.
The opportunity to use Ad vectors encoding fiber-σ1 chimeras for mucosal targeting is especially appealing. Increased delivery of antigens to intestinal epithelial cells and Peyer's patch lymphocytes by such vectors might result in more potent and less toxic gene-based vaccines. Reovirus binds to murine microfold cells (15, 16, 18), and the σ1 protein plays an important role in conferring this tropism (18, 47). Interactions of Ad5-σ1 vectors with microfold cells may facilitate efficient delivery to underlying Peyer's patches for induction of immune responses in the gut. Alternatively, σ1-bearing Ad vectors may directly infect DCs at the luminal surface, which are known to shuttle bacteria across epithelial monolayers by opening tight junctions and sampling the intestinal lumen (48). DCs express tight junction proteins, including JAM1 (27), which are hypothe-sized to facilitate epithelial barrier penetration. Our finding that Ad5-T3Dσ1 transduces primary DCs more efficiently than wild-type Ad5 suggests that Ad5-σ1 vectors may be useful for antigen gene delivery to DCs in the intestine and other sites.
Findings described in this report indicate that Ad vectors can be efficiently retargeted to cells expressing JAM1 and sialic acid by the reovirus attachment protein σ1. By virtue of the capacity to infect both intestinal epithelial cells and DCs, Ad5-σ1 vectors may have utility in the induction of immune responses at mucosal surfaces and thus prevention of infection at the site of pathogen entry. These vectors also will allow a precise determination of the contribution of the JAM1- and sialic acid-binding properties of σ1 to interactions of σ1 with cells in vivo. This approach should lead to improved Ad vectors for gene delivery and enhance an understanding of σ1 biology.
Acknowledgments
We thank Mary E. Barry and Jared Abramian for excellent technical assistance; members of the Barry and Dermody laboratories for many useful discussions; Chuck Parkos for providing hJAM1-specific mAb J10.4; and Dan Von Seggern for providing the 633 cells. This work was supported by National Science Foundation IGERT Award DGE-0114264 (to G.T.M.), Public Health Service Awards T32 CA09385 (to J.A.C.), T32 HL07751 (to J.D.C.), R01 GM67853 (to T.S. and T.S.D.), R01 AI38296 (to T.S.D.), and R01 AI42588 (to M.A.B.), and the Elizabeth B. Lamb Center for Pediatric Research. Additional support was provided by Public Health Service Awards AI36211 for the Center for AIDS Research at Baylor College of Medicine, DK056338 for the Texas Gulf Coast Digestive Diseases Center (Baylor College of Medicine), CA68485 for the Vanderbilt Cancer Center, and DK20593 for the Vanderbilt Diabetes Research and Training Center.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ad, adenovirus; CAR, coxsackievirus and Ad receptor; DC, dendritic cell; T3D, type 3 Dearing; JAM1, junctional adhesion molecule 1; CHO, Chinese hamster ovary; h, human; CMV, cytomegalovirus; m, murine.
References
- 1.Shiver, J. W. & Emini, E. A. (2004) Annu. Rev. Med. 55, 355-372. [DOI] [PubMed] [Google Scholar]
- 2.Bergelson, J. M., Cunningham, J. A., Droguett, G., Kurt-Jones, E. A., Krithivas, A., Hong, J. S., Horwitz, M. S., Crowell, R. L. & Finberg, R. W. (1997) Science 275, 1320-1323. [DOI] [PubMed] [Google Scholar]
- 3.Tomko, R. P., Xu, R. & Philipson, L. (1997) Proc. Natl. Acad. Sci. USA 94, 3352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roelvink, P. W., Lizonova, A., Lee, J. G., Li, Y., Bergelson, J. M., Finberg, R. W., Brough, D. E., Kovesdi, I. & Wickham, T. J. (1998) J. Virol. 72, 7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, C. J., Shieh, J. T., Pickles, R. J., Okegawa, T., Hsieh, J. T. & Bergelson, J. M. (2001) Proc. Natl. Acad. Sci. USA 98, 15191-15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wickham, T. J., Mathias, P., Cheresh, D. A. & Nemerow, G. R. (1993) Cell 73, 309-319. [DOI] [PubMed] [Google Scholar]
- 7.Huang, S., Kamata, T., Takada, Y., Ruggeri, Z. M. & Nemerow, G. R. (1996) J. Virol. 70, 4502-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tillman, B. W., de Gruijl, T. D., Luykx-de Bakker, S. A., Scheper, R. J., Pinedo, H. M., Curiel, T. J., Gerritsen, W. R. & Curiel, D. T. (1999) J. Immunol. 162, 6378-6383. [PubMed] [Google Scholar]
- 9.Belousova, N., Korokhov, N., Krendelshchikova, V., Simonenko, V., Mikheeva, G., Triozzi, P. L., Aldrich, W. A., Banerjee, P. T., Gillies, S. D., Curiel, D. T. & Krasnykh, V. (2003) J. Virol. 77, 11367-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parrott, M. B., Adams, K. E., Mercier, G. T., Mok, H., Campos, S. K. & Barry, M. A. (2003) Mol. Ther. 8, 689-702. [DOI] [PubMed] [Google Scholar]
- 11.Gaggar, A., Shayakhmetov, D. M. & Lieber, A. (2003) Nat. Med. 9, 1408-1412. [DOI] [PubMed] [Google Scholar]
- 12.Vogels, R., Zuijdgeest, D., van Rijnsoever, R., Hartkoorn, E., Damen, I., de Bethune, M. P., Kostense, S., Penders, G., Helmus, N., Koudstaal, W., et al. (2003) J. Virol. 77, 8263-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nibert, M. L. & Schiff, L. A. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott-Raven, Philadelphia), pp. 1679-1728.
- 14.Tyler, K. L. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott-Raven, Philadelphia), pp. 1729-1945.
- 15.Wolf, J. L., Rubin, D. H., Finberg, R., Kauffman, R. S., Sharpe, A. H., Trier, J. S. & Fields, B. N. (1981) Science 212, 471-472. [DOI] [PubMed] [Google Scholar]
- 16.Wolf, J. L., Kauffman, R. S., Finberg, R., Dambrauskas, R., Fields, B. N. & Trier, J. S. (1983) Gastroenterology 85, 291-300. [PubMed] [Google Scholar]
- 17.Bodkin, D. K., Nibert, M. L. & Fields, B. N. (1989) J. Virol. 63, 4676-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amerongen, H. M., Wilson, G. A., Fields, B. N. & Neutra, M. R. (1994) J. Virol. 68, 8428-8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furlong, D. B., Nibert, M. L. & Fields, B. N. (1988) J. Virol. 62, 246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjea, A. C., Brechling, K. A., Ray, C. A., Erikson, H., Pickup, D. J. & Joklik, W. K. (1988) Virology 167, 601-612. [PubMed] [Google Scholar]
- 21.Fraser, R. D., Furlong, D. B., Trus, B. L., Nibert, M. L., Fields, B. N. & Steven, A. C. (1990) J. Virol. 64, 2990-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dermody, T. S., Nibert, M. L., Bassel-Duby, R. & Fields, B. N. (1990) J. Virol. 64, 5173-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chappell, J. D., Gunn, V. L., Wetzel, J. D., Baer, G. S. & Dermody, T. S. (1997) J. Virol. 71, 1834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chappell, J. D., Duong, J. L., Wright, B. W. & Dermody, T. S. (2000) J. Virol. 74, 8472-8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barton, E. S., Connolly, J. L., Forrest, J. C., Chappell, J. D. & Dermody, T. S. (2001) J. Biol. Chem. 276, 2200-2211. [DOI] [PubMed] [Google Scholar]
- 26.Barton, E. S., Forrest, J. C., Connolly, J. L., Chappell, J. D., Liu, Y., Schnell, F. J., Nusrat, A., Parkos, C. A. & Dermody, T. S. (2001) Cell 104, 441-451. [DOI] [PubMed] [Google Scholar]
- 27.Rescigno, M., Rotta, G., Valzasina, B. & Ricciardi-Castagnoli, P. (2001) Immunobiology 204, 572-581. [DOI] [PubMed] [Google Scholar]
- 28.Martin-Padura, I., Lostaglio, S., Schneemann, M., Williams, L., Romano, M., Fruscella, P., Panzeri, C., Stoppacciaro, A., Ruco, L., Villa, A., et al. (1998) J. Cell Biol. 142, 117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozaki, H., Ishii, K., Horiuchi, H., Arai, H., Kawamoto, T., Okawa, K., Iwamatsu, A. & Kita, T. (1999) J. Immunol. 163, 553-557. [PubMed] [Google Scholar]
- 30.Liu, Y., Nusrat, A., Schnell, F. J., Reaves, T. A., Walsh, S., Pochet, M. & Parkos, C. A. (2000) J. Cell Sci. 113, 2363-2374. [DOI] [PubMed] [Google Scholar]
- 31.van Raaij, M. J., Mitraki, A., Lavigne, G. & Cusack, S. (1999) Nature 401, 935-938. [DOI] [PubMed] [Google Scholar]
- 32.Chappell, J. D., Prota, A. E., Dermody, T. S. & Stehle, T. (2002) EMBO J. 21, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Von Seggern, D. J., Huang, S., Fleck, S. K., Stevenson, S. C. & Nemerow, G. R. (2000) J. Virol. 74, 354-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poteete, A. R. (2001) FEMS Microbiol. Lett. 201, 9-14. [DOI] [PubMed] [Google Scholar]
- 35.Datsenko, K. A. & Wanner, B. L. (2000) Proc. Natl. Acad. Sci. USA 97, 6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis, A. R., Wivel, N. A., Palladino, J. L., Tao, L. & Wilson, J. M. (2000) Methods Mol. Biol. 135, 515-523. [DOI] [PubMed] [Google Scholar]
- 37.Bewley, M. C., Springer, K., Zhang, Y. B., Freimuth, P. & Flanagan, J. M. (1999) Science 286, 1579-1582. [DOI] [PubMed] [Google Scholar]
- 38.Forrest, J. C., Campbell, J. A., Schelling, P., Stehle, T. & Dermody, T. S. (2003) J. Biol. Chem. 278, 48434-48444. [DOI] [PubMed] [Google Scholar]
- 39.Chroboczek, J., Ruigrok, R. W. & Cusack, S. (1995) Curr. Top. Microbiol. Immunol. 199, 163-200. [DOI] [PubMed] [Google Scholar]
- 40.Kerneis, S., Bogdanova, A., Kraehenbuhl, J. P. & Pringault, E. (1997) Science 277, 949-952. [DOI] [PubMed] [Google Scholar]
- 41.van der Lubben, I. M., van Opdorp, F. A., Hengeveld, M. R., Onderwater, J. J., Koerten, H. K., Verhoef, J. C., Borchard, G. & Junginger, H. E. (2002) J. Drug Target. 10, 449-456. [DOI] [PubMed] [Google Scholar]
- 42.Cheng, X., Ming, X. & Croyle, M. A. (2003) Pharm. Res. 20, 1444-1451. [DOI] [PubMed] [Google Scholar]
- 43.Banchereau, J., Briere, F., Caux, C., Davoust, J., Lebecque, S., Liu, Y. J., Pulendran, B. & Palucka, K. (2000) Annu. Rev. Immunol. 18, 767-811. [DOI] [PubMed] [Google Scholar]
- 44.Gilmore, R., Coffey, M. C., Leone, G., McLure, K. & Lee, P. W. (1996) EMBO J. 15, 2651-2658. [PMC free article] [PubMed] [Google Scholar]
- 45.Hong, J. S. & Engler, J. A. (1996) J. Virol. 70, 7071-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krasnykh, V., Belousova, N., Korokhov, N., Mikheeva, G. & Curiel, D. T. (2001) J. Virol. 75, 4176-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helander, A., Silvey, K. J., Mantis, N. J., Hutchings, A. B., Chandran, K., Lucas, W. T., Nibert, M. L. & Neutra, M. R. (2003) J. Virol. 77, 7964-7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rescigno, M., Urbano, M., Valzasina, B., Francolini, M., Rotta, G., Bonasio, R., Granucci, F., Kraehenbuhl, J. P. & Ricciardi-Castagnoli, P. (2001) Nat. Immunol. 2, 361-367. [DOI] [PubMed] [Google Scholar]
- 49.Bassel-Duby, R., Jayasuriya, A., Chatterjee, D., Sonenberg, N., Maizel, J. V., Jr., & Fields, B. N. (1985) Nature 315, 421-423. [DOI] [PubMed] [Google Scholar]
- 50.Weis, W., Brown, J. H., Cusack, S., Paulson, J. C., Skehel, J. J. & Wiley, D. C. (1988) Nature 333, 426-431. [DOI] [PubMed] [Google Scholar]
- 51.Carson, M. (1987) J. Mol. Graphics 5, 103-106. [Google Scholar]