Abstract
Src is the prototypic protein tyrosine kinase and is critical for controlling diverse cellular functions. Regions in Src define structural and functional domains conserved in many cell signaling proteins. Src also contains a region of low sequence conservation termed the unique domain, the function of which has until now remained enigmatic. Here, we show that the unique domain of Src is a protein–protein interaction region and we identify NADH dehydrogenase subunit 2 (ND2) as a Src unique domain-interacting protein. ND2 is a subunit of complex I in mitochondria, but we find that ND2 interacts with Src outside this organelle at excitatory synapses in the brain. ND2 acts as an adapter protein anchoring Src to the N-methyl-d-aspartate (NMDA) receptor complex, and is crucial for Src regulation of synaptic NMDA receptor activity. By showing an extramitochondrial action for a protein encoded in the mitochondrial genome, we identify a previously unsuspected means by which mitochondria regulate cellular function, suggesting a new paradigm that may be of general relevance for control of Src signaling.
The nonreceptor protein tyrosine kinase Src is a ubiquitous enzyme with key roles in diverse developmental, physiological and pathological processes (1, 2). Domains identified in Src, the Src homology 3 (SH3) domain, the SH2 domain, and the SH1, or catalytic, domain, are signature regions that define highly conserved protein modules found in a wide variety of signaling proteins (3). In addition to the highly conserved regions, Src also contains a region of low sequence conservation and unknown function termed the unique domain.
The CNS is a major site of expression of Src, and numerous functions are ascribed to Src in the developing and adult CNS. A growing body of evidence indicates that in the CNS a key function of Src is to regulate glutamatergic neurotransmission and synaptic plasticity (4). At glutamatergic synapses, Src modulates the activity of N-methyl-d-aspartate receptors (NMDARs) (5–7), a principal subtype of glutamate receptor (8). NMDARs are crucial for CNS development, neuroplasticity, and pathophysiology (9, 10). NMDARs are multiprotein complexes comprised of core channel subunits together with associated scaffolding and regulatory proteins (11, 12) that control receptor localization, ionic flux through the receptor, and downstream signaling events. The function of NMDARs is regulated by multiple factors, including regulation by dynamic cycling of protein phosphorylation and dephosphorylation at serine/threonine or tyrosine residues (13, 14). Src represents a point through which multiple signaling cascades from G protein-coupled receptors (15), Eph receptors (16–18), and integrins (19, 20) converge to up-regulate NMDAR channel activity. The up-regulation of NMDAR activity by Src is necessary for long-term potentiation (LTP) of synaptic transmission at Schaffer collateral–CA1 synapses in the hippocampus (4), the predominant cellular model for learning and memory (21). Src-mediated up-regulation of NMDAR activity is prevented by peptide fragments of the Src unique domain and by a unique domain-binding antibody (5, 6) leading to the hypothesis that up-regulation of NMDAR function requires an interaction between a region in the unique domain of Src and an unknown protein within the NMDAR complex (4). Thus, in the present study, we set out to search for proteins that may interact with the Src unique domain and may thereby mediate the interaction between Src and NMDARs.
Materials and Methods
Detailed methods are found in ref. 22 and Supporting Text, which is published as supporting information on the PNAS web site.
Yeast Two-Hybrid Screen. cDNAs encoding amino acids 4–82 (the Src unique domain) and amino acids 4–150 (the Src unique and SH3 domains) of murine n-Src were ligated into pEG202 (23) to create two expression vectors encoding in frame LexA fusions containing the Src unique domain. To create the selection strains for screening, each bait plasmid was individually transformed into the yeast strain EGY48. The selection strains were transformed with a representative activation-tagged cDNA prey fusion library constructed by using ≈1-kb EcoRI fragmented poly(A)(+) RNA from human fetal brain. Yeast transformed with the prey library (≈1.1 × 106 clones) were screened by double selection on 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) Leu- medium. Prey cDNAs encoding proteins that interacted with the bait were isolated and sequenced.
Src, Fyn, and NADH Dehydrogenase Subunit 2 (ND2) Recombinant Proteins. The cDNAs encoding the SH3 and SH2 domains of mouse n-Src and Fyn were PCR subcloned, ligated in frame into pGEX4T-1 (Amersham Pharmacia). These plasmids, as well as plasmids encoding the unique domains of Src and Fyn in pGEX2T′6, were transformed into BL21 bacteria, and GST fusion proteins were purified by glutathione affinity chromatography. To create the ND2.1, ND2.2, and ND2.3 GST fusion proteins, cDNAs encoding amino acids 239–321 (ND2.1–GST), amino acids 189–238 (ND2.2–GST), and amino acids 1–188 (ND2.3–GST) of human ND2 were PCR subcloned and ligated into pGEX4T-1. Using PCR-based single-nucleotide mutagenesis, all cDNAs encoding ND2 fusion proteins were corrected for differences between mitochondrial and nuclear codons to prevent premature translation termination and protein truncation. The plasmids were transformed into bacteria, and GST fusion proteins were purified as above.
Cellular Fractionation. Postsynaptic density (PSD) proteins (24) were prepared from rat brain as described (22). Cellular fractionation of rat brain tissue into nuclear, heavy mitochondrial, light mitochondrial, microsomal, and cytosolic fractions was performed by differential centrifugation of tissue homogenate in 0.25 M sucrose/10 mM Hepes-NaOH/1 mM EDTA, pH 7.4, with 2 μg each of aprotinin, pepstatin A, and leupeptin (Sigma) at 4°C. All pellets from individual fractions were resuspended in RIPA buffer (50 mM Tris, pH 7.6/150 mM NaCl/1 mM EDTA/1% Nonidet P-40/2.5 mg/ml NaDOC/1 mM Na3VO4/1 mM PMSF/2 μg/ml each of protease inhibitors). The light mitochondrial fraction was used in subsequent experiments. For immunoblots 50 μg of total protein was loaded per lane, resolved by SDS/PAGE, transferred to nitrocellulose membranes, and probed with anti-ND2, anti-Cyto1 (cytochrome c oxidase subunit 1), and anti-ND4 (mouse monoclonals, Molecular Probes), anti-PSD95 (mouse monoclonal clone 7E3-1B8, Oncogene Research Products, Cambridge, MA), anti-NR1 (mouse monoclonal clone 54.1, Pharmingen), anti-Src, or anti-synaptophysin (mouse monoclonal, Sigma).
Dot Blotting. Src40–58 and scrambled Src40–58 peptides were biotinylated by incubating with Sulfo-NHS-Biotin (Pierce) for 30 min at room temperature. The biotinylation reaction was then quenched by the addition of Tris·HCl (pH 8.0) to a final concentration of 20 mM. Purified recombinant fusion proteins (≈20 μg each) were dotted onto nitrocellulose and dried overnight. Membranes were blocked with 5% BSA in PBS (pH 7.5) for 1 h, after which biotinylated peptides (30 μg/ml) diluted 1:1,000 in fresh 5% BSA in PBS were added. The membranes were incubated with the peptides for 1 h, washed, and probed with streptavidin–horseradish peroxidase conjugate (SA-HRP). Bound probe was then detected on film by using an enhanced chemiluminescence kit.
Cultured Hippocampal Neurons. Fetal hippocampal neurons were prepared, cultured, and used for electrophysiological recordings 12–17 days after plating.
Results
ND2 Is a Src Unique Domain-Binding Protein. To search for proteins that interact with the Src unique domain, we did a yeast two-hybrid screen using bait constructs containing the Src unique domain. In two independent screens, we isolated cDNA fragments encoding overlapping regions within ND2 (Fig. 1A), a 347-aa protein that is a subunit of the inner mitochondrial membrane enzyme NADH dehydrogenase (complex I). ND2 is one of a group of seven oxidoreductase proteins that are encoded in the mitochondrial genome and which coassemble with 35 nuclear encoded subunits to form complex I. ND2 on its own lacks enzymatic activity (25–27).
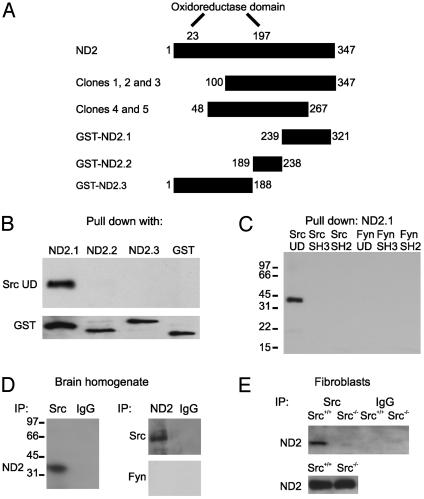
Fig. 1.
ND2 is a Src unique domain-interacting protein. (A) Schematic diagram illustrating the domain structure of ND2, clones isolated from the yeast two-hybrid screen, and recombinant GST-tagged fusion proteins. (B) Blot of ND2–GST fusion proteins probed with biotinylated Src unique domain followed by SA-HRP. (C) Blot of ND2.1–GST probed with biotinylated domains of Src and Fyn followed by SA-HRP. (D) Immunoblots of coimmunoprecipitates from brain homogenate probed with anti-ND2, anti-Src, or anti-Fyn. Nonspecific IgG was used as a negative control for immunoprecipitation. In these experiments, the cell lysates were prepared by using nondenaturing conditions; under denaturing conditions, we found no coimmunoprecipitation of Src by anti-ND2 or of ND2 by anti-Src (not shown). (E) Immunoblot of coimmunoprecipitates from cultured Src+/+ and Src-/- fibroblasts probed with anti-ND2. Nonspecific IgG was used as a negative control for immunoprecipitation, and immunoblotting of ND2 protein from both cell lines was used as a positive control.
Because yeast two-hybrid screens may reveal false positive protein–protein interactions, we investigated whether the interaction between Src and ND2 was observed with an independent methodological approach. We tested for direct binding in vitro between ND2 and Src by using recombinant proteins. We made a series of GST fusion proteins comprised of portions of ND2 that spanned the overlapping region found with the yeast two-hybrid screen (Fig. 1 A). Importantly, cDNAs encoding ND2 fusion proteins were corrected for differences between mitochondrial and nuclear codons so that the sequence of the ND2 portion of the fusion proteins was that which would be produced by mitochondrial translation. We tested each of these proteins individually for interaction with the Src unique domain. We found that a GST fusion protein containing amino acids 239–321 of ND2 (ND2.1–GST) bound to the unique domain of Src (Fig. 1B). In contrast, fusion proteins containing amino acids 189–238 (ND2.2–GST) or 1–188 (ND2.3–GST) of ND2 did not bind to the Src unique domain. These results, together with those from the yeast two-hybrid screen, show that ND2 is a Src unique domain-binding protein. Our results indicate further that the Src-binding portion of ND2 is contained within the region of amino acids 239–321. This region of ND2 has little conservation among mitochondrially encoded oxidoreductase proteins and is outside the so-called “oxidoreductase domain,” a signature region present in all mitochondrially encoded subunits of NADH dehydrogenase (25–27) and some antiporters (28).
We wondered whether binding of ND2 might generalize to other domains of Src or to other Src family tyrosine kinases. However, we found that ND2.1–GST did not bind to either of the prototypic protein–protein interaction domains of Src, the SH2 or SH3 domains (Fig. 1C). To examine the potential interaction of ND2 with other kinases of the Src family, we tested recombinant domains of Fyn. Although Fyn is closely related to Src, it has low sequence conservation in the unique domain (1–3). We found that ND2.1–GST did not interact in vitro with the Fyn unique domain; nor did it bind to the SH2 or SH3 domains of Fyn. Thus, the ND2.1 region does not interact with the SH2 or SH3 domains of Src or Fyn, nor does it generally bind to the unique domain of Src family tyrosine kinases.
To investigate the possibility that Src and ND2 may interact in vivo, we immunoprecipitated brain lysates with antibodies directed against ND2 (anti-ND2) or against Src (anti-Src). We found that immunoprecipitating Src led to coimmunoprecipitation of ND2 and, conversely, immunoprecipitating with anti-ND2 resulted in coimmunoprecipitation of Src (Fig. 1D). In contrast, anti-ND2 did not coimmunoprecipitate Fyn and neither ND2 nor Src was immunoprecipitated with nonspecific IgG. As an independent immunoprecipitation control, we found that ND2 was coimmunoprecipitated by anti-Src from Src+/+ fibroblasts but not from Src-/- fibroblasts (Fig. 1E). Thus, in addition to finding the ND2-Src unique domain interaction in the yeast two-hybrid screens and in vitro binding assays, we found that ND2 and Src coimmunoprecipitated with each other, leading us to conclude that ND2 is a Src unique domain binding protein that may interact with Src in vivo.
ND2 Is Present in PSDs in Brain. In the CNS a prominent subcellular location for Src is in the PSD (5), a subsynaptic specialization at glutamatergic synapses comprised of α-amino-3-hydroxy-5-methylisoxazolepropionic acid (AMPA-) and NMDA-type glutamate receptors together with scaffolding, signaling, and regulatory proteins (29). Because Src regulates subsynaptic NMDARs (5), we predicted that if ND2 is the protein mediating the interaction between NMDARs and the Src unique domain, then ND2 would be present in the PSD. Therefore, we prepared PSD proteins from rat brain homogenate by sequential fractionation and determined whether ND2 was present in this fraction. Characteristic of a bona fide PSD fraction, this fraction contained postsynaptic proteins including PSD-95 and NMDAR subunit proteins, but lacked the presynaptic protein synaptophysin (Fig. 2A). We found that ND2 was present in the PSD fraction, and we estimated the amount of ND2 in this fraction was ≈15% of that in total brain homogenate. In contrast to ND2, neither the oxidoreductase protein ND4, another mitochondrially encoded component of complex I (25–27), nor Cyto1, an inner mitochondrial membrane protein component of complex IV (30), was detectable in the PSD fraction. Conversely, Cyto1 and ND4, as well as ND2, were readily detected in proteins from brain mitochondria (Fig. 2B). It is possible that the protein detected in the PSD fraction was not ND2, but a protein of the same molecular size that was spuriously recognized by anti-ND2. However, incubating anti-ND2 with the antigen used to derive the antibody prevented the immunoblot signal (Fig. 2C). Moreover, a second antibody directed toward a distinct epitope in a region of ND2 remote from that of the anti-ND2 epitope also detected ND2 in both PSD and mitochondrial preparations (Fig. 2D). Thus, ND2 was found in the PSD fraction by two separate antibodies, and this was not caused by a general contamination with mitochondrial proteins because neither Cyto1 nor ND4 were detected in the PSD.
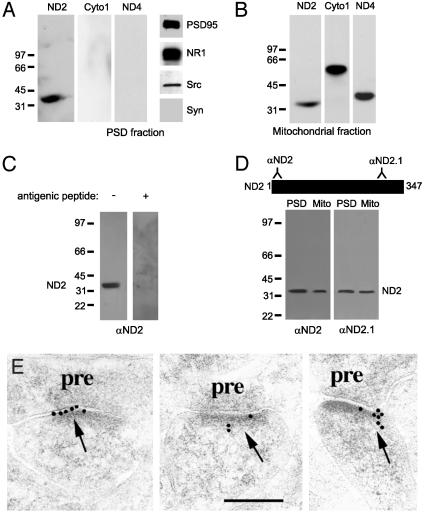
Fig. 2.
ND2 is present at the PSD. (A) Immunoblots of PSD proteins probed with anti-ND2, anti-cytochrome c oxidase I (Cyto1), anti-ND4, anti-PSD95, anti-NR1, anti-Src, and anti-synaptophysin. The PSD preparation contained PSD95, NR1, and Src, but lacked the presynaptic marker synaptophysin. (B) Immunoblots of mitochondrial proteins probed with anti-ND2, anti-Cyto1, and anti-ND4. Neither NR1 nor NR2A/B was detected in the mitochondrial fraction (not shown). (C) Immunoblots of PSD proteins showing the specificity of the N-terminal ND2 antibody by preadsorption with the antigenic peptide used to derive the antibody. (D) Immunoblots of PSD and mitochondrial proteins probed with two independent rabbit polyclonal antibodies directed against two disparate regions of ND2. The N-terminal ND2 antibody was used for all subsequent experiments shown. (E) Postembedding immunogold electron microscopy images of rat hippocampus CA1 synapses. ND2 immunoreactivity in PSDs is visualized by secondary antibody conjugated to 10-nm gold particles. pre, presynaptic. (Scale bar is 200 nm.)
We also tested for the presence of ND2 in PSDs by postembedding immunogold electron microscopy in the CA1 stratum radiatum of rat hippocampus (31, 32). In this approach, the tissue is fixed immediately after death to preserve protein localization before sectioning. We found ND2 labeling, as visualized by secondary antibody conjugated to 10-nm gold particles, in the PSD and the postsynaptic membrane in dendritic spines of CA1 neurons (Fig. 2E), as well as over mitochondria (see Fig. 6, which is published as supporting information on the PNAS web site). ND2 labeling was enriched in the postsynaptic membrane ≈30-fold as compared with the plasma membrane in the remainder of the dendritic spine (0.37 particles per PSD/section versus 0.012, P < 0.05) and there was no obvious accumulation of ND2 labeling along the plasma membrane of the dendritic shaft. Because mitochondria are excluded from dendritic spines (33), the ND2 labeling observed in the PSD and postsynaptic membrane was not caused by mitochondrial labeling. Thus, these results show that ND2 is present in the biochemically defined PSD protein fraction and is localized at PSDs in CA1 neurons.
ND2 Interacts with Src at the NMDA Receptor Complex in PSDs. Because our results show that ND2 is present in PSDs from brain, we examined whether ND2 interacts with Src in PSDs. We found that immunoprecipitating ND2 from the PSD fraction led to coimmunoprecipitation of Src and vice versa (Fig. 3A), showing that ND2 and Src interact postsynaptically at glutamatergic synapses. Moreover, Src was pulled from the PSD fraction by the fusion protein ND2.1–GST, but not by either ND2.2– or ND2.3–GST (Fig. 3B). Thus, as we found with Src–ND2 binding in vitro, these results indicate that amino acids 239–321 of ND2 are both necessary and sufficient for ND2 to interact with Src in the PSD.
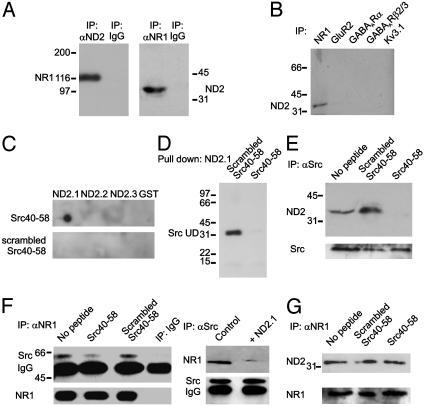
Fig. 3.
ND2 interacts with Src at the postsynaptic density. (A) Immunoblots of coimmunoprecipitates from PSD preparations probed with anti-ND2 or anti-Src as indicated. Nonspecific IgG (either rabbit or mouse) was used as a negative control for both antibodies. (B) Recombinant ND2.1–GST fusion protein, but not ND2.2–GST, ND2.3–GST, or GST alone, pulls Src from PSD preparations.
The hypothesis that ND2 is the protein mediating the interaction between Src and NMDARs requires that, in addition to being present in the PSD and interacting there with Src, ND2 is part of the NMDAR protein complex. To determine whether ND2 is a component of the NMDAR complex, we used an antibody directed against the core NMDAR subunit NR1 (8) to immunoprecipitate NMDAR complexes from the PSD fraction and probed the coimmunoprecipitates with anti-ND2. We found that ND2 coimmunoprecipitated and, conversely, immunoprecipitating with anti-ND2 led to coimmunoprecipitation of NR1 (Fig. 4A). Neither ND2 nor NR1 was immunoprecipitated by nonspecific IgG, and ND2 did not coimmunoprecipitate with the potassium channel Kv3.1 (Fig. 4B), a negative control for nonspecific immunoprecipitation of postsynaptic proteins, therefore we concluded that ND2 is an NMDAR complex protein. Importantly, neither ND4 nor Cyto1 was detected in coimmunoprecipitates of NR1 (not shown) indicating that mitochondrial proteins in general are not part of the NMDAR complex. Moreover, ND2 did not coimmunoprecipitate with GluR2, γ-aminobutyric acid type A receptor α (GABAARα), or GABAARβ2/3 (Fig. 4B) showing that ND2 is not a detectable component of AMPA receptor or GABA receptor complexes. Thus, although ND2 is a part of NMDAR complexes, it is not generally a component of neurotransmitter receptor complexes.
Fig. 4.
ND2 interacts with Src at the NMDAR complex. (A) Immunoblots of coimmunoprecipitates from PSD preparations probed with anti-ND2 or with anti-NMDA receptor subunit 1 (NR1) as indicated. Nonspecific IgG (either rabbit or mouse) was used as a negative control for both antibodies. (B) Immunoblot of coimmunoprecipitates from PSD preparations using anti-GluR2, anti-GABAARα, anti-GABAARβ2/3, and anti-Kv3.1 antibodies to immunoprecipitate and probed with anti-ND2. (C) Dot blot of ND2–GST fusion proteins probed with biotinylated Src40–58 or scrambled Src40–58 peptides followed by SA–HRP. (D) Blot of ND2.1–GST probed with biotinylated Src unique domain in the presence of either Src40–58 or scrambled Src40–58 followed by SA-HRP. (E) Immunoblots of coimmunoprecipitates obtained from PSD proteins in the presence of either Src40–58 or scrambled Src40–58 probed with anti-ND2 or stripped and reprobed with anti-Src. (F) Immunoblots of coimmunoprecipitates obtained from PSD proteins in the presence of either Src40–58 or scrambled Src40–58 (Left) or in the presence of ND2.1–GST fusion protein (Right) probed with anti-Src or anti-NR1. (G) Immunoblots of coimmunoprecipitates obtained from PSD proteins in the presence of either Src40–58 or scrambled Src40–58 probed with anti-ND2 or stripped and reprobed with anti-NR1.
ND2 Acts as an Adapter Protein for Src. Amino acids 40–58 within the Src unique domain were implicated in the binding of Src to the interacting protein in the NMDAR complex (5–7) and thus, ND2 was predicted to bind to this region of Src. We tested this prediction in vitro by using a peptide with the sequence of amino acids 40–58 (Src40–58) that we found to bind directly to ND2.1–GST (Fig. 4C). In contrast, a peptide with identical amino acid composition, but a scrambled sequence (scrambled Src40–58), did not bind to ND2.1–GST. Neither Src40–58 nor scrambled Src40–58 bound to ND2.2–GST, ND2.3–GST, or GST alone (Fig. 4C). We next examined the effect of Src40–58 on the interaction between Src and ND2 and found that incubating ND2.1-GST with Src40–58 prevented pull down of the Src unique domain protein in vitro (Fig. 4D). Conversely, scrambled Src40–58 did not affect the interaction between ND2.1-GST and the Src unique domain. Incubating PSD proteins with Src40–58 prevented the coimmunoprecipitation of ND2 by anti-Src but this was unaffected by scrambled Src40–58 (Fig. 4E). Importantly, Src40–58 did not affect the immunoprecipitation of Src from PSDs. Thus, we conclude that amino acids 40–58 of Src interact with the region spanned by ND2.1, thereby mediating the binding between the Src unique domain and ND2.
Because ND2 alone is not catalytically active (25–27), we investigated its functional role in the NMDAR complex. ND2 might be a phosphorylation target for Src, but we found that ND2 immunoprecipitated from PSD protein fractions was not detectably phosphorylated on tyrosine. Moreover, inclusion of ND2.1–GST did not alter the catalytic activity of Src in vitro (not shown), consistent with binding of ND2 to the unique domain rather than to the regulatory or catalytic domains. Thus, it is unlikely that ND2 is a target of Src or a regulator of Src kinase activity.
These results led us to consider a role for ND2 in the association of Src with the NMDAR complex. Antibody directed against the core NMDAR subunit NR1 was used to immunoprecipitate NMDAR complexes from PSDs, and the coimmunoprecipitates were probed with anti-Src. We found that the coimmunoprecipitation of Src with NMDARs (Fig. 4F Left) was suppressed by Src40–58, but not scrambled Src40–58, and by ND2.1 (Fig. 4F Right), indicating that the association of Src with NMDAR complexes depends on the interaction with ND2. In contrast, the coimmunoprecipitation of ND2 with NMDARs was not affected by Src40–58 (Fig. 4G), implying that binding ND2 to Src is not required for ND2 to associate with NMDAR complexes. Taking these results together, we conclude that ND2 may function as an adapter protein that anchors Src in the NMDAR complex.
Loss of ND2 in Neurons Prevents the Regulation of NMDA Receptor Activity by Src. We reasoned that if ND2 is a Src adapter protein then loss of ND2 should prevent the up-regulation of NMDAR activity by endogenous Src (5). We tested this by recording miniature excitatory postsynaptic currents (mEPSCs) from cultured hippocampal neurons (34). In these neurons, the NMDAR-mediated component of mEPSCs is increased by activating endogenous Src via intracellular administration of a high-affinity activating phosphopeptide EPQ(pY)EEIPIA (35) and is reduced by applying the Src40–58 peptide (see ref. 5 and Fig. 5). We predicted that if ND2 acts as an adaptor protein for Src in the NMDAR complex, then blocking the expression of ND2 would cause the loss of each of these effects. To suppress ND2 expression, we treated hippocampal cultures with chloramphenicol (CAP) to selectively inhibit translation of mitochondrially encoded proteins but not translation of proteins encoded in the nucleus (36). After 48-h treatment with CAP, we found that the level of ND2 in the cultures was reduced by >95%, whereas there was no significant change in the levels of the nuclear encoded proteins examined (Fig. 5A). Importantly, CAP did not affect the level of Src or of the NMDAR subunit NR1, but suppressed the coimmunoprecipitation of Src with the NMDAR complex (Fig. 5B) as predicted if ND2 is an adapter protein linking Src to the complex.
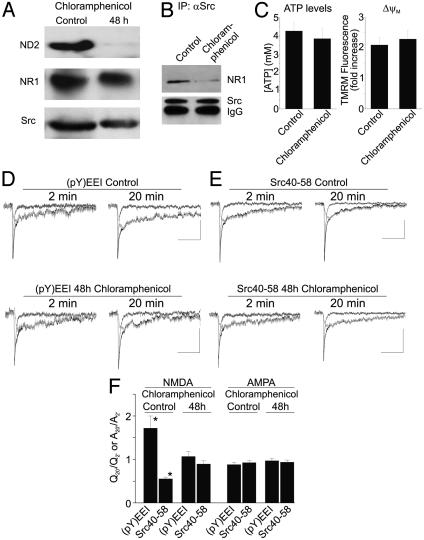
Fig. 5.
Blocking expression of ND2 prevents Src-dependent regulation of NMDA receptor activity. (A) Immunoblots of total soluble protein obtained from cultured rat hippocampal neurons treated with 50 μg/ml chloramphenicol (CAP) for 48 h probed with anti-ND2, anti-NR1, and anti-Src as indicated. (B) Immunoblot of coimmunoprecipitates obtained from cultured hippocampal neurons, either untreated or treated with CAP for 48 h, probed with anti-NR1 or anti-Src. (C) Summary histograms (Left) of ATP level or mitochondrial membrane potential (▵ψM), as assessed by TMRM fluorescence dequenching (Right), in cultured hippocampal neurons either untreated or treated with CAP for 48 h. (D) The up-regulation of NMDAR activity by the Src activator peptide EPQ(pY)EEIPIA, labeled (pY)EEI, is prevented in CAP-treated neurons. (E) The reduction of NMDAR activity by Src40–58 peptide is also prevented in neurons treated with CAP for 48 h. Composite traces are shown in black, the NMDAR component is shown in dark gray, and the AMPA receptor (AMPAR) component is shown in light gray. (Scale bars are 50 ms/10 pA.) (F) Summary histogram of electrophysiology data. NMDA component data were calculated as Q20′/Q2′, and AMPA component data were calculated as A20′/A2′. CAP treatment for 48 h prevents modulation of NMDAR function by the Src activator and Src40–58 peptides, whereas neither of these reagents affected the AMPAR component of mEPSCs. Asterisks indicate a significant difference, Student's t test, P < 0.05.
We tested the effect of 48-h treatment with CAP on ATP levels, mitochondrial membrane potential, viability, and general functioning of the hippocampal neurons in culture. We found that CAP did not significantly affect ATP levels in the neurons (Fig. 5C), consistent with the lack of effect of CAP treatment for up to 55 h on ATP levels in other cell types in culture (37). To examine the effect of CAP on mitochondrial membrane potential (▵ψM), we monitored the dequenching of tetramethylrhodamine methyl ester fluorescence evoked by bath applying the mitochondrial uncoupler carbonyl cyanide p-trifluoromethoxyphenylhydrazone (38) in individual neurons from CAP-treated or control cultures. We found that the dequenching response of neurons treated with CAP was not different from that of untreated neurons (Fig. 5C), indicating that ▵ψM was unaffected by CAP. Moreover, CAP-treated neurons were indistinguishable from control neurons in terms of cell number and morphology, resting membrane potential, resting intracellular [Ca2+], action potential amplitude, or mEPSC frequency (data not shown). Thus, from these data together we concluded that treatment with chloramphenicol for 48 h did not detectably compromise the functioning of the neurons. We note, nevertheless, that the intracellular solution used for whole-cell recording contained 2 mM Mg-ATP, so that the level of intracellular ATP was equal in all cells throughout the experiments.
In neurons treated with CAP for 48 h, we found that the NMDAR component of mEPSCs was unaffected by administering either the EPQ(pY)EEIPIA peptide or the Src40–58 peptide (Fig. 5 D–F). In contrast, in control experiments, administering EPQ(pY)EEIPIA increased the NMDAR component of mEPSCs by 172 ± 28% and application of Src40–58 decreased the NMDAR component to 56 ± 4% (Fig. 5 D–F). CAP was present during the recording periods of the control experiments, and thus loss of effect of the activator and Src40–58 peptides cannot be attributed to an acute effect of CAP. Taking our results together, we conclude that Src-dependent regulation of NMDAR activity depends on expression of ND2 through its anchoring of Src to the NMDAR complex.
Discussion
The principal goal of the present study was to identify the protein mediating the interaction between NMDARs and the unique domain of Src. The criteria for identifying this protein, as inferred from previous work (4, 5), are as follows: the protein must bind directly to the Src unique domain through amino acids 40–58; this binding must be prevented by the Src40–58 peptide; the protein must be present at excitatory synapses and must be a component of the NMDAR complex; and lack of the protein must prevent the up-regulation of NMDAR activity by endogenous Src. In the present study, we provide multiple, converging lines of evidence leading to the conclusion that ND2 is the protein mediating the interaction between NMDARs and the Src unique domain.
ND2 is mitochondrially encoded and translated, yet we find it within PSDs of glutamatergic synapses in the brain. We did not detect other mitochondrial proteins (ND4 or Cyto1) in the PSD fraction, implying that this fraction is not contaminated nonspecifically by mitochondrial proteins. Furthermore, we find ND2 immunoreactivity by immunogold electron microscopy within identified PSDs in dendritic spines of CA1 neurons. In this preparation, immobilization of proteins by tissue fixation precludes the possibility that ND2 could have relocated from mitochondria to the PSD during processing. Moreover, because dendritic spines are devoid of mitochondria (33), ND2 immunoreactivity cannot be accounted for by mitochondria abutting the PSD. Taken together, these findings indicate that ND2, but not the entire complex I, is normally present within the PSD. Many enzymes in the PSD may be involved in regulating synaptic function (39), including glycolytic enzymes capable of generating ATP (40). However, without the remaining components of complex I, it is unlikely that ND2 functions catalytically in the PSD.
Thus, in addition to its localization in mitochondria and function as a component of complex I, the present results indicate that ND2 has a second location and function outside mitochondria. Mitochondria are intimately linked to overall cellular functioning through generation of ATP by oxidative phosphorylation, are key for sequestration of intracellular calcium (41, 42), and participate in programmed cell death (43, 44). Some mitochondrial proteins are present at extramitochondrial sites (45, 46), but our evidence reveals a function for a mitochondrial protein outside this organelle; that is, ND2 acts as an adapter protein that anchors Src within the NMDAR complex, where it thereby allows Src to up-regulate NMDAR activity.
Up-regulating the activity of NMDARs is a major function of Src in neurons in the adult CNS (6, 22, 47), and this mediates the induction of long-term potentiation of excitatory synaptic transmission in CA1 hippocampal neurons (4). Our present findings imply that the ND2-Src interaction is essential for LTP induction because LTP in CA1 neurons is prevented by Src40–58 and by anti-Src1, an antibody that recognizes this amino acid sequence within the Src unique domain and prevents binding of the Src unique domain to ND2.1 in vitro (J.R.G. and M.W.S., unpublished data). LTP at Schaffer collateral-CA1 synapses is the prototypic example of NMDAR-dependent enhancement of excitatory synaptic transmission observed at numerous types of glutamatergic synapses throughout the CNS (48). In addition, Src is implicated in NMDAR-dependent seizures (49), chronic pain (50), and neurotoxicity (51). Thus, our discovery of the Src–ND2 interaction at NMDARs defines a protein–protein interaction of general relevance to regulation of neuronal function, synaptic plasticity, and pathophysiology in the CNS.
Supplementary Material
Acknowledgments
We thank R. F. Doolittle, J. Bolen, S. A. Courtneidge, R. Brent, Y. T. Wang, and P. S. Pennefather for reagents; J. L. Hicks, R. Diaz, B. Owen, C. Tsang, and W. Ju for assistance and advice; and J. F. MacDonald and L. Y. Wang for comments on the manuscript. This work was generously supported by the Hospital for Sick Children Research Institute and by the Canadian Institutes of Health Research.
Abbreviations: SH3, Src homology 3; NMDA, N-methyl-d-aspartate; NMDAR, NMDA receptor; LTP, long-term potentiation; ND2, NADH dehydrogenase subunit 2; PSD, postsynaptic density; SA-HRP, streptavidin–horseradish peroxidase; AMPA, α-amino-3-hydroxy-5-methylisoxazolepropionic acid; Cyto1, cytochrome c oxidase subunit 1; CAP, chloramphenicol; mEPSCs, miniature excitatory postsynaptic currents.
References
- 1.Brown, M. T. & Cooper, J. A. (1996) Biochim. Biophys. Acta 1287, 121-149. [DOI] [PubMed] [Google Scholar]
- 2.Superti-Furga, G. & Courtneidge, S. A. (1995) BioEssays 17, 321-330. [DOI] [PubMed] [Google Scholar]
- 3.Pawson, T. (1995) Nature 373, 573-580. [DOI] [PubMed] [Google Scholar]
- 4.Ali, D. W. & Salter, M. W. (2001) Curr. Opin. Neurobiol. 11, 336-342. [DOI] [PubMed] [Google Scholar]
- 5.Yu, X. M., Askalan, R., Keil, G. J. & Salter, M. W. (1997) Science 275, 674-678. [DOI] [PubMed] [Google Scholar]
- 6.Lu, Y. M., Roder, J. C., Davidow, J. & Salter, M. W. (1998) Science 279, 1363-1368. [DOI] [PubMed] [Google Scholar]
- 7.Yu, X. M. & Salter, M. W. (1998) Nature 396, 469-474. [DOI] [PubMed] [Google Scholar]
- 8.Dingledine, R., Borges, K., Bowie, D. & Traynelis, S. F. (1999) Pharmacol. Rev. 51, 7-61. [PubMed] [Google Scholar]
- 9.McBain, C. J. & Mayer, M. L. (1994) Physiol. Rev. 74, 723-760. [DOI] [PubMed] [Google Scholar]
- 10.Sheng, M. & Kim, M. J. (2002) Science 298, 776-780. [DOI] [PubMed] [Google Scholar]
- 11.Scannevin, R. H. & Huganir, R. L. (2000) Nat. Rev. Neurosci. 1, 133-141. [DOI] [PubMed] [Google Scholar]
- 12.Sheng, M. & Pak, D. T. (2000) Annu. Rev. Physiol 62, 755-778. [DOI] [PubMed] [Google Scholar]
- 13.Wang, Y. T. & Salter, M. W. (1994) Nature 369, 233-235. [DOI] [PubMed] [Google Scholar]
- 14.Smart, T. G. (1997) Curr. Opin. Neurobiol. 7, 358-367. [DOI] [PubMed] [Google Scholar]
- 15.Luttrell, L. M. & Lefkowitz, R. J. (2002) J. Cell Sci. 115, 455-465. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, J. T., Georgiou, J., Jia, Z., Robertson, J., Elowe, S., Roder, J. C. & Pawson, T. (2001) Neuron 32, 1041-1056. [DOI] [PubMed] [Google Scholar]
- 17.Takasu, M. A., Dalva, M. B., Zigmond, R. E. & Greenberg, M. E. (2002) Science 295, 491-495. [DOI] [PubMed] [Google Scholar]
- 18.Murai, K. K. & Pasquale, E. B. (2002) Neuron 33, 159-162. [DOI] [PubMed] [Google Scholar]
- 19.Lin, B., Arai, A. C., Lynch, G. & Gall, C. M. (2003) J. Neurophysiol. 89, 2874-2878. [DOI] [PubMed] [Google Scholar]
- 20.Kramar, E. A., Bernard, J. A., Gall, C. M. & Lynch, G. (2003) J. Biol. Chem. 278, 10722-10730. [DOI] [PubMed] [Google Scholar]
- 21.Kandel, E. R. (2001) Science 294, 1030-1038. [DOI] [PubMed] [Google Scholar]
- 22.Pelkey, K. A., Askalan, R., Paul, S., Kalia, L. V., Nguyen, T. H., Pitcher, G. M., Salter, M. W. & Lombroso, P. J. (2002) Neuron 34, 127-138. [DOI] [PubMed] [Google Scholar]
- 23.Gyuris, J., Golemis, E., Chertkov, H. & Brent, R. (1993) Cell 75, 791-803. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy, M. B., Bennett, M. K. & Erondu, N. E. (1983) Proc. Natl. Acad. Sci. USA 80, 7357-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker, J. E. (1992) Q. Rev. Biophys. 25, 253-324. [DOI] [PubMed] [Google Scholar]
- 26.Sazanov, L. A. & Walker, J. E. (2000) J. Mol. Biol. 302, 455-464. [DOI] [PubMed] [Google Scholar]
- 27.Sazanov, L. A., Peak-Chew, S. Y., Fearnley, I. M. & Walker, J. E. (2000) Biochemistry 39, 7229-7235. [DOI] [PubMed] [Google Scholar]
- 28.Fearnley, I. M. & Walker, J. E. (1992) Biochim. Biophys. Acta 1140, 105-134. [DOI] [PubMed] [Google Scholar]
- 29.Walikonis, R. S., Jensen, O. N., Mann, M., Provance, D. W., Jr., Mercer, J. A. & Kennedy, M. B. (2000) J. Neurosci. 20, 4069-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marusich, M. F., Robinson, B. H., Taanman, J. W., Kim, S. J., Schillace, R., Smith, J. L. & Capaldi, R. A. (1997) Biochim. Biophys. Acta 1362, 145-159. [DOI] [PubMed] [Google Scholar]
- 31.Petralia, R. S., Esteban, J. A., Wang, Y. X., Partridge, J. G., Zhao, H. M., Wenthold, R. J. & Malinow, R. (1999) Nat. Neurosci. 2, 31-36. [DOI] [PubMed] [Google Scholar]
- 32.Sans, N., Petralia, R. S., Wang, Y. X., Blahos, J., Hell, J. W. & Wenthold, R. J. (2000) J. Neurosci. 20, 1260-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shepherd, G. M. & Harris, K. M. (1998) J. Neurosci. 18, 8300-8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacDonald, J. F., Mody, I. & Salter, M. W. (1989) J. Physiol. (London) 414, 17-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, X., Brodeur, S. R., Gish, G., Songyang, Z., Cantley, L. C., Laudano, A. P. & Pawson, T. (1993) Oncogene 8, 1119-1126. [PubMed] [Google Scholar]
- 36.Ibrahim, N. G. & Beattie, D. S. (1976) J. Biol. Chem. 251, 108-115. [PubMed] [Google Scholar]
- 37.Ramachandran, A., Moellering, D. R., Ceaser, E., Shiva, S., Xu, J. & Darley-Usmar, V. (2002) Proc. Natl. Acad. Sci. USA 99, 6643-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reers, M., Smith, T. W. & Chen, L. B. (1991) Biochemistry 30, 4480-4486. [DOI] [PubMed] [Google Scholar]
- 39.Siekevitz, P. (1985) Proc. Natl. Acad. Sci. USA 82, 3494-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, K., Aoki, C., Elste, A., Rogalski-Wilk, A. A. & Siekevitz, P. (1997) Proc. Natl. Acad. Sci. USA 94, 13273-13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friel, D. D. (2000) Cell Calcium 28, 307-316. [DOI] [PubMed] [Google Scholar]
- 42.Rizzuto, R. (2001) Curr. Opin. Neurobiol. 11, 306-311. [DOI] [PubMed] [Google Scholar]
- 43.Gorman, A. M., Ceccatelli, S. & Orrenius, S. (2000) Dev. Neurosci. 22, 348-358. [DOI] [PubMed] [Google Scholar]
- 44.Mattson, M. P. (2000) Nat. Rev. Mol. Cell. Biol. 1, 120-129. [DOI] [PubMed] [Google Scholar]
- 45.Soltys, B. J. & Gupta, R. S. (1999) Trends Biochem. Sci. 24, 174-177. [DOI] [PubMed] [Google Scholar]
- 46.Soltys, B. J. & Gupta, R. S. (2000) Int. Rev. Cytol. 194, 133-196. [DOI] [PubMed] [Google Scholar]
- 47.Huang, Y., Lu, W., Ali, D. W., Pelkey, K. A., Pitcher, G. M., Lu, Y. M., Aoto, H., Roder, J. C., Sasaki, T., Salter, M. W., et al. (2001) Neuron 29, 485-496. [DOI] [PubMed] [Google Scholar]
- 48.Malenka, R. C. & Nicoll, R. A. (1999) Science 285, 1870-1874. [DOI] [PubMed] [Google Scholar]
- 49.Sanna, P. P., Berton, F., Cammalleri, M., Tallent, M. K., Siggins, G. R., Bloom, F. E. & Francesconi, W. (2000) Proc. Natl. Acad. Sci. USA 97, 8653-8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo, W., Zou, S., Guan, Y., Ikeda, T., Tal, M., Dubner, R. & Ren, K. (2002) J. Neurosci. 22, 6208-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pei, L., Teves, R. L., Wallace, M. C. & Gurd, J. W. (2001) J. Cereb. Blood Flow Metab. 21, 955-963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.