Abstract
Mycobacterium tuberculosis, the pathogen that causes tuberculosis, has evolved sophisticated mechanisms for evading assault by the human host. This review focuses on M. tuberculosis regulatory metalloproteins that are sensitive to exogenous stresses attributed to changes in the levels of gaseous molecules (i.e., molecular oxygen, carbon monoxide and nitric oxide) to elicit an intracellular response. In particular, we highlight recent developments on the subfamily of Whi proteins, redox sensing WhiB-like proteins that contain iron-sulfur clusters, sigma factors and their cognate anti-sigma factors of which some are zinc-regulated, and the dormany survival regulon DosS/DosT-DosR heme sensory system. Mounting experimental evidence suggests that these systems contribute to a highly complex and interrelated regulatory network that controls M. tuberculosis biology. This review concludes with a discussion of strategies M. tuberculosis has developed to maintain redox homeostasis, including mechanisms to regulate endogenous nitric oxide and carbon monoxide levels.
Keywords: Mycobacterium tuberculosis, redox sensing, molecular gas sensing, metalloproteins, hypoxia
1. Introduction
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), is one of the world’s most successful human pathogens. To date, TB kills over 1.4 million people per year with approximately one third of the world’s population infected with the latent form of the disease [1]. For a bacterial pathogen such as Mtb to be successful, it must respond to and overcome the human host defense system of toxic compounds such as hydrogen peroxide (H2O2), nitric oxide (NO) and carbon monoxide (CO), as well as lowered levels of environmental pH (excess H+) and molecular oxygen (O2). In order to survive the host-produced reactive oxygen and nitrogen species, ROS and RNS, respectively, Mtb has evolved many strategies to evade assault, including utilizing superoxide dismutase, catalase peroxidase, alkyl hydroperoxidase and peroxiredoxins, which convert these compounds to non-reactive products. These classes of enzymes have previously been widely reviewed [2]. Moreover, the transition from actively growing Mtb to its latent state is triggered by redox sensing of lowered host O2 levels [3], elevated host NO [4] and CO [5, 6] levels as well as high concentrations of H+ [7] and nutrient starvation [8]. Latent Mtb may be undetected for several years and reactivation occurs when the host becomes immunocompromised, as commonly observed in AIDS patients [9, 10].
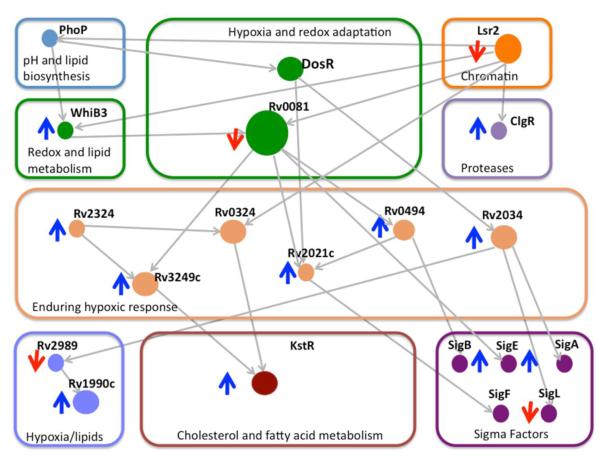
A recent tour-de-force in Mtb systems biology revealed a complicated regulatory network during Mtb adaptation to hypoxia and reactivation due to increased oxygen availability comprising extensive interactions amongst many transcription factors, several of which contain metal cofactors [11], Figure 1. The authors also show through gene expression profiles and lipid analyses that Mtb’s response to low oxygen levels greatly alters lipid content, metabolite utilization, protein degradation and iron/sulfur metabolism. Despite an ever-increasing wealth of pertinent information, it is not the intention of this review to provide a comprehensive description of the complex Mtb systems that react to redox potential modulations and gaseous molecule stresses produced by the host. For a more detailed description, readers are referred to the literature cited in the references. Instead, we focus on Mtb regulatory proteins with metal-ion centers; specifically, the iron-sulfur cluster containing transcription regulatory WhiB-like proteins from the Whi family of proteins, several sigma factors that are regulated by zinc-dependent anti-sigma factors, and the heme-containing sensory domains of the dormancy survival (Dos) regulon.
Figure 1.
Transcriptional factor regulatory subnetwork linking hypoxia, lipid metablosim and protein degradation. Grey arrows pointing down or up indicate gene down- or up-regulation, respectively, under hypoxic conditions. Grey lines represent correlated activation or repression between genes.
2. WhiB-like (Wbl) family of proteins
The Wbl family of proteins has been shown to exhibit important pleitropic roles. The Wbl protein family is unique to actinomycetes and was first described in Streptomyces coelicolor to be necessary for sporulation whereby white colonies were inhibited from forming grey spores [12]. In Mtb, seven Wbl proteins (WhiB1-7) have been identified [13, 14] and implicated in cell division, nutrient starvation, virulence and pathogenesis, antibiotic resistance, and stress sensing (reviewed in [15]). Notably, the genes encoding for WhiB1 and WhiB2 were shown to be essential [16-18] while virulence in Mtb whiB3 and whiB5 deletion mutants was attenuated in infected mice compared to wild-type Mtb [19, 20]. Furthermore, WhiB3 has been shown to regulate fatty acid metabolism by controlling the production of lipids such as poly- and diacyltrehaloses, sulfolipids and phthiocerol dimycocerosates [21].
Mtb Wbl proteins are relatively small (~10-15 kDa) proteins with four conserved cysteine residues, C-X18-35-C-X2-3-C-X5-7-C (where X is any residue), predicted to coordinate Fe to form [4Fe-4S] iron-sulfur clusters [14]. Although there is a lack of Wbl protein structural information, amino acid sequence analysis reveals two additional motifs. First, the second and third cysteine residues from the conserved [4Fe-4S] cluster motif (with the exception of WhiB5 – CLRRC) constitute a CXXC motif that is thought to function as a redox rheostat through the maintenance of thiol:disulfide homeostasis [22]. A second conserved GXWXG motif, which is found several residues after the last cysteine in the [4Fe-4S] cluster motif, has a predicted β-turn and is presumed to be the start of the C-terminal DNA-binding domain of Wbl proteins followed by a putative DNA-recognition helix. Furthermore, circular dichroism spectra showed different profiles for Mtb Wbl proteins, suggesting overall dissimilar structures despite sharing 49–66% sequence homology [23].
Comprehensive investigations have reported gene expression, structural and functional differences amongst the seven Mtb Wbl proteins [23, 24]. Real-time reverse transcription-PCR analyses revealed differential levels of Wbl transcripts in distinct growth phases (from early exponential to late stationary phase) as well as in response to anti-mycobacterial agents and a variety of stress conditions such as hypoxia, NO, low-iron, acidic pH and nutrient deficiency [24, 25]. These distinct response differences most likely govern the respective roles of the Mtb Wbl proteins and, more importantly, demonstrate their sensitivity to redox environments. In a separate study, Mtb Wbl proteins were shown to harbor iron-sulfur clusters that further exhibited differing susceptibilities to analytes such as molecular oxygen, oxidized glutathione, dithiothreitol, and iron chelators (e.g., EDTA), suggesting that accessibility to the clusters differs within Mtb Wbl proteins [23]. Biochemical characterization of Mtb apo-Wbl (iron free) proteins revealed that most of them have significantly different rates of disulfide reductase activity [23]. Recently, mounting evidence suggest that Mtb Wbl proteins can bind DNA [18, 21, 26-28] and may function as transcriptional regulators [20, 26]. Based on these observations and the redox sensitivity of [4Fe-4S] clusters, Mtb Wbl proteins are predicted to be susceptible to environmental cues to elicit an intracellular response. Thus, it appears that Mtb Wbl proteins are at least bifunctional in which they are disulfide reductases in their apo forms and transcriptional regulators dictated by degradation of their [4Fe-4S] clusters.
2a. Wbls are protein disulfide reductases
Thioredoxin-like proteins contain CXXC motifs and have long been established as important for maintaining cellular redox states (reviewed in [22]). From previous reports, in the absence of a [4Fe-4S] cluster, the conserved cysteines in all seven of the Mtb Wbl proteins form two intramolecular disulfide bonds: one between the CXXC motif and the other between the other two conserved cysteines [29-31]. Additionally, the Wbl proteins were characterized as protein disulfide reductases whereby the reduction of an intermolecular bond between α- and β-chains of insulin was dependent upon the CXXC motif [23, 29-31]. Notably, insulin reduction was not observed for WhiB2 [23]. Distinct from the other Mtb Wbl proteins, WhiB2 displayed chaperone-like activity by suppressing the aggregation of several protein substrates, including citrate synthase, rhodanese, and luciferase [32]. Interestingly, this ability is independent of the redox state of the conserved cysteines but instead relies on a non-conserved serine (Ser42) which when mutated to alanine abolishes activity. The authors speculate that phosphorylation of Ser42 may play a key role in WhiB2’s chaperone-like property [32].
The functional assignment of Mtb Wbl proteins as disulfide reductases raises two central questions, one regarding potential protein partners that maintain the CXXC motif redox state and the other regarding their cognate substrates. While Wbl proteins’ redox partners have not been fully investigated, the first substrate has been identified through a yeast two-hybrid screen. To that end, alpha (1,4)-glucan branching enzyme (GlgB), which is implicated in trehalose biosynthesis [33] and essential for optimal in vitro Mtb growth [34], was found to interact with WhiB1 [35]. Furthermore, WhiB1 reduces an intramolecular disulfide bond that covalently links two of the four GlgB domains; however the physiological significance of interdomain linkage remains unclear. Further characterization of this WhiB1-GlgB interaction, as well as the identification of other substrates of WhiB1 and the other Wbl proteins, is required to provide a better understanding of Mtb’s thiol-disulfide oxidoreductase systems (reviewed in [22]), and thus Mtb’s ability to cope with oxidative stress.
2b. Wbls contain [4Fe-4S] clusters sensitive to O2 and NO
[4Fe-4S] cluster assembly
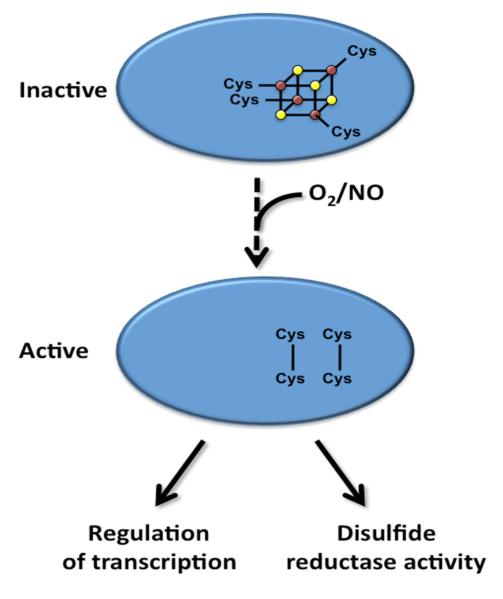
For the Wbl protein family, one of the early challenges was to obtain proteins with fully complexed [4Fe-4S] clusters. To date, there are at least three in vivo bacterial iron-sulfur cluster assembly systems that have been reported, namely Nif, Isc, and Suf [36, 37]; in Mtb, the suf operon has been identified [38] as well as a gene usually contained in a nif or isc gene cluster, nifS (iscS); however this gene does not appear to be within a gene cluster in Mtb [39]. Mtb IscS, an L-cysteine desulfurase, under anaerobic conditions in the presence of L-cysteine (sulfur donor), an iron source and a reducing agent can produce fully reconstituted [4Fe-4S]-WhiB3 [39]. Moreover, it has been shown for both WhiB1 and WhiB3 that all four cysteine residues are crucial for cluster acquisition [39, 40], providing each Fe within the cubane type [4Fe-4S] cluster a tetragonal S coordination (Figure 2).
Figure 2.
The proposed mechanism of Wbl activation. Under the current model, the [4Fe-4S] cluster bound to the Wbl proteins degrade via a series of intermediates upon exposure to NO and O2. The resulting conserved cysteine residues oxidize to form disulfide bonds and the apo-Wbl proteins are then able to bind to their respective DNA promoter regions to regulate transcription or perform their disulfide reductase activity. The oligomeric states of the Wbl proteins remain to be elucidated.
[4Fe-4S]-WhiB3 responds to O2 and NO
Due to the highly reactive nature of [4Fe-4S] clusters, Mtb Wbl proteins are predicted to be sensitive to external environmental signals. In fact, many reports have shown that the [4Fe-4S] clusters within proteins degrade upon exposure to O2 and NO [41, 42]. Recently, WhiB4 was demonstrated to be highly sensitive to both gases [27], although other Mtb Wbl proteins appear to have a preference for a gas analyte. WhiB3, in particular, was shown to be sensitive to both O2 and NO although the O2-dependent degradation of the [4Fe-4S] cluster occurs approximately 12 times faster in fumarate nitrate reductase (FNR), suggesting that O2 may not be WhiB3’s cognate analyte [39]. Furthermore, WhiB1 is highly sensitive to NO, with degradation of its [4Fe-4S] cluster occurring four orders of magnitude faster in the presence of NO in comparison to O2 [18]. Taken together, Mtb Wbl proteins are cognate O2 and/or NO sensors albeit their analyte specificities remain to be investigated. A first step to elucidate substrate specificity is to probe the mechanism of oxidation and nitrosylation that leads to [4Fe-4S] cluster degradation. While oxidative degradation within Wbl proteins is still not very well understood, recent efforts have been made with regards to nitrosative degradation with the essential WhiB1 and a close S. coelicolor homolog, WhiD, which are described in the following section.
Mechanism of nitrosylation and [4Fe-4S] cluster degradation
The mechanism of [4Fe-4S] cluster degradation by NO has been unraveled in recent years through a series of elegant experiments with WhiB1 and S. coelicolor WhiD [18, 43, 44]. Titrating a NO-releasing compound under anaerobic conditions and monitoring the UV/vis absorption profiles revealed that eight equivalents of NO to WhiB1, or S. coelicolor WhiD, were required for complete nitrosylation of the [4Fe-4S] cluster [18, 44]. Furthermore, stopped-flow experiments showed that [4Fe-4S] cluster degradation followed a multiphasic mechanism with distinct intermediates [44]. Additionally, nitrosylation of the [4Fe-4S] cluster oxidizes 3S2− to yield 3S0 [44]. Based on the final UV/vis spectrum, the predominant degradation product was proposed to be Roussin’s red ester (RRE) instead of the previously suggested tetrahedral dinitrosyl iron complex (DNIC) [39]. Moreover, the final degradation product was found to be EPR silent, which is in agreement with RRE as the product, as opposed to DNIC (spin = ½) that exhibits an EPR signal [44]. Taken together, nitrosylation of Wbl proteins was proposed to follow reaction (I).
| (I) |
Interestingly, a recent report on FNR, which is also NO sensitive, revealed a common reaction mechanism by which nitrosylated [4Fe-4S] clusters degrade despite possessing different cluster environments [28]. NO-initiated degradation was also followed using EPR to provide further insights to the distinct intermediates, which ultimately yielded RRE as the major degradation product with DNIC as a minor species.
Physiological relevance of [4Fe-4S] nitrosylation and degradation
The NO-mediated degradation of [4Fe-4S] clusters results in Mtb apo-Wbl proteins, which can function as transcription regulators. To that end, oxidized apo-WhiB1, in response to NO, has been shown to bind DNA whereas [4Fe-4S]-WhiB1 does not [18]. Additionally, footprinting experiments revealed that apo-WhiB1 bound to its own promoter to repress whiB1 transcription. Significantly, nitrosylation results in an oligomeric transition from an active heterodimeric [4Fe-4S]-FNR capable of binding DNA to control transcription to an inactive monomeric apo-FNR [28]; therefore, it is predicted that a switch in oligomeric state occurs in all Mtb Wbl proteins although it remains unknown what the oligomeric states of active (apo) and inactive ([4Fe-4S] cluster-bound) forms are. Accumulating evidence reinforces the hypothesis that Wbl proteins can bind to DNA. Apo-WhiB2 and a mycobacteriophage homolog, apo-WhiBTM4, were able to specifically bind to the promoter region upstream of the whiB2 gene [26]. Interestingly, mutating conserved cysteine residues of WhiBTM4, as well as truncating its C-terminal to remove the proposed DNA-binding domain, resulted in the loss of WhiBTM4’s inhibitory effect [26]. In a separate study, Mtb whiB3 knockout mutant displayed depleted levels of lipids; indeed, this phenotype was determined to be a direct result of WhiB3 binding to the promoters of polyketide synthesis genes responsible for lipid biosynthesis [21]. Moreover, WhiB4 can bind specifically to the promoter regions of ahpC (encoding a protein required to suppress oxidative assault) and whiB4, as well as non-specifically to GC-rich DNA sequences [27] and its interaction with DNA is dependent on the presence and oxidation of the four conserved cysteine residues (Figure 2).
3. Regulation of σ Factors in Response to Redox Stress
Sigma (σ) factors are protein subunits responsible for much of the specificity in cellular gene expression profiles, whereby they recruit RNA polymerase to specific genes for transcription and expression [45, 46]. Of the thirteen Mtb σ factors [46], SigH, SigE, SigL, SigF, as well as the general stress response factor SigB, have been shown to be involved in redox stress response [47-51]. Notably, these σ factors are unable to directly sense and react to the extracellular environment, relying on careful regulation by anti-σ factors (or by other σ factors for SigB), which are capable of sensing stress signals. Upon activation, these σ factors will recognize and bind their respective promoter regions to recruit RNA polymerase to induce transcription of redox homeostasis genes including thioredoxins, peroxiredoxins, and other transcription factors [52]. While the function, interactions and gene regulation patterns of Mtb redox σ factors have been well characterized, thus far, there has been little success with structural elucidation.
3a. Role of zinc ions in regulation: the thiol-disulfide redox switch
Redox σ factors are tightly regulated to ensure cell survival when exposed to redox stress. This regulation occurs via the inhibitory anti-σ factors, which typically possess a conserved HXXXCXXC peptide motif capable of coordinating Zn2+, earning them the title ‘zinc-associated anti-σ factors’, or ZAS proteins [53]. Under non-stress or reducing conditions, ZAS proteins are tightly bound to the -35 promoter-binding region of their respective σ-factors [50]. Coordination of a structural Zn2+ ion both stabilizes the ZAS protein and increases the reactivity of the two cysteine thiol groups [54]. By acting as a Lewis acid, this Zn2+ ion can lower the pKa of the cysteine thiols, increasing their nucleophilicity and ability to interact with electrophilic host-induced ROS. This metal coordination also prevents the two conserved cysteines from forming a disulfide bond in the absence of redox stress. Upon exposure to oxidizing conditions (such as peroxides and hydroxyl radicals), the coordinated Zn2+ ion is released and the cysteines form a disulfide bond, causing conformational changes in the ZAS protein which results in σ-factor dissociation to regulate gene expression [55]. Due to the importance of the conserved cysteine residues in complex formation, even a single cysteine mutation results in loss of inhibitory activity of the ZAS protein [53]. This mechanism of protein complex regulation has been extensively characterized for the S. coelicolor RsrA/SigR complex [53, 56], which shows significant homology to σ factor/anti-σ factor complexes in other actinomycetes, including Mtb, and functions in a similar manner. Specifically, this thioldisulfide redox switch motif is observed in several Mtb extracytoplasmic function (ECF) anti-σ factors involved in redox stress sensing and response.
3b. SigH is released from its anti-σ factor RshA in an oxidative stress-dependent manner
One of the most important σ factors in Mtb stress response and survival is SigH, a global regulator which has been shown to upregulate nearly 40 genes involved in general stress response [57]. SigH is activated upon Mtb entry into host macrophages, implicating its role in pathogenesis and survival within a host. The role of SigH in redox stress response was first seen when a Mycobacterium smegmatis sigH deletion mutant displayed significantly increased sensitivity to hyperoxides such as diamide [48]; however sigH deletion in Mtb shows an increase in expression of chemokines involved in granuloma formation [58], suggesting a probable immunomodulatory role. SigH confers protection from oxidative stress by upregulating the expression of several genes, such as the thioredoxins trxA, trxB1, and trxC, along with thioredoxin reductase trxB2, and two other σ factors sigE and sigB [57, 59]. SigH is regulated by the anti-σ factor RshA, a ZAS protein. Despite a lack of functional evidence, RshA was thought to regulate SigH via Zn2+ coordination due to the presence of a ZAS motif [60]. Further support for this hypothesis is that a close homolog S. coelicolor RsrA has been experimentally shown to utilize a Zn2+ binding redox switch [55]. In contrast, recent research revealed that an [4Fe-4S] cluster can displace Zn2+ from RshA, suggesting a higher binding affinity although neither metal cofactor has been co-purified with RshA [61]. In fact, it appears that Zn2+ and the [4Fe-4S] cluster, as well as the conserved cysteines, are dispensable for RshA/SigH complex formation [61], indicating that the method of redox-sensing may be distinct from other Mtb redox-regulated σ factors. Interestingly, further mutational analyses showed that RshA His49Ala and Glu168Ala mutants hindered RshA/SigH complex formation, implying that the interaction may not be mediated by a metal ion-based redox switch but by a series of salt bridges that may be disrupted in the presence of hyperoxides.
While Mtb RshA and SigH structures have not been solved, close homologs from S. coelicolor have [56], providing insight into the interactions between these two proteins in Mtb. Based on the S. coelicolor SigH and RshA structures and hydrogen/deuterium exchange mass spectrometry experiments, a model has been generated for the formation of the Mtb SigH/RshA complex [61]. Like S. coelicolor, Mtb SigH and RshA are predicted to be completely α-helical, globular proteins, interacting via a series of ion pairs to form a stable inactive complex. An essential serine/threonine protein kinase, PknB, was shown to be involved in SigH regulation by preventing SigH/RshA complex formation through phosphorylation of RshA, possibly disrupting the electrostatic potential at the complex interface [62]. PknB cannot directly sense redox stress, therefore this phosphorylation event may serve to prolong the SigH signal thereby inhibiting expression of stress-response genes and ensuring cell-survival.
3c. SigE is activated upon exposure to redox and membrane stress
Among the targets upregulated by SigH under redox stress is SigE, another alternative σ factor involved in stress response. Similar to SigH, SigE has been shown to be vital for Mtb survival within activated macrophages, implicating it as an important virulence factor [63-65]. SigE is regulated by binding its cognate anti-σ factor RseA, a ZAS protein that utilizes the Zn2+-mediated thiol-disulfide redox switch to sense the presence of ROS and disulfide stress [47]. Upon activation, SigE upregulates clgR, a transcriptional activator that in turn upregulates protein expression of ClpC1P2 protease, SigB, cell-wall maintenance proteins, as well as itself in a positive feedback loop. Much like SigH, SigE is also regulated by phosphorylation via PknB. However, while SigH and its cognate anti-σ factor are both phosphorylated via PknB to maintain SigH-upregulated genes, phosphorylation of RseA at a conserved threonine residue (Thr39) targets it for phosphorylation-dependent degradation by the ClpC1P2 protease, allowing for extended upregulation of SigE-activated stress response genes [47]. Of note, RseA proteolysis only occurs under membrane/cell-wall stress (whereby its disruption is induced by sodium dodecyl sulfate or an antibiotic such as vancomycin), while oxidative or heat stress results in no change in RseA levels. Due to the use of this protease to aid in propagation of the SigE signal, disrupting the function of the ClpC1P2 protease machinery may potentially sensitize Mtb to the harsh environment within host macrophages, attenuating Mtb virulence, particularly when administered along with vancomycin. SigE-deficient Mtb shows significantly decreased survival during host infection [63-65], thus blocking the function of SigE would likely serve as an effective method for Mtb cell arrest.
3d. Alternate σ factor SigB aids in propogation of SigH and SigE signals
Similarly to SigH, SigB plays a major role in several forms of stress response in Mtb, including heat stress, cell envelope stress, and redox stress [49, 66]. Unlike the four other ECF σ factors, SigB is unable to sense extracytoplasmic signals in the same manner. As opposed to relying on an inhibitory anti-σ factor that can sense stress, SigB relies on upregulation via SigH and SigE when stimulated by redox stress. An Mtb sigB deletion mutant exposed to either cell-wall stress (via sodium dodecyl sulfate) or oxidative stress (via diamide) yields a significant decrease in cell growth viability. Complementation of this mutant with SigB restores near wild type growth in vitro. These results suggest that SigB targets similar genes to SigH/SigE due to their similar activation responses, however experimental evidence shows that SigB shares very few gene targets with either ECF σ factor, implicating SigB as an important downstream signal in the stress response activities of both SigE and SigH. Among SigB unique targets activated by oxidative stress are furA, an iron uptake regulator protein, katG, a catalase-peroxidase, and whiB2 [66]. While a sigB deletion mutant seems to play an important role in Mtb growth under in vitro hypoxic conditions, when grown within THP-1 macrophage cells or guinea pig lung tissues it does not show any noticeable change in viability compared to wild type. The disparity between the importance of SigB in vitro versus in vivo may be explained by the poor representation of current animal models, as a similar effect is observed for a dosR deletion mutant [67], which shows varying levels of significance to Mtb growth under various conditions.
3e. SigF/UsfX complex is regulated in a Zn2+-independent manner
Mtb SigF is an extracytoplasmic σ factor which is implicated in redox stress response as an M. smegmatis SigF knockout mutant shows increased sensitivity to hyperoxides [51]. Also, the sigF gene is upregulated when Mtb is exposed to antibiotics or cumene hyperoxide [68]. Interestingly, genomic analysis of the SigF regulon shows that SigF may play a role in upregulating the expression of genes whiB1 and whiB4 [69] as well as whiB7 [70]. Despite a similar stress-response function to other redox-σ factors, SigF has several differences from SigH and SigE. SigF is co-transcribed with its anti-σ factor, UsfX, since both genes lie adjacent to each other on the genome. Additionally, UsfX does not appear to contain the HXXXCXXC motif and thus displays a unique method of interaction as well as redox sensing to regulate SigF activity [71]. Intriguingly, this system also has two anti-anti-σ factors, RsfA and RsfB. RsfA and RsfB were shown to bind the UsfX/SigF complex through UsfX interactions under reducing conditions and in an ATP-dependent manner, respectively [71]. In particular, RsfA, which contains four cysteine residues, is stable and complexed to UsfX/SigF under reducing conditions but rapidly degrades under oxidizing conditions when Cys73 and Cys109 form a disulfide bond. Interestingly, both SigF and UsfX display equivalent tight DNA binding affinities, thus RsfA (and potentially RsfB) may serve as a redox switch similar to those observed in other anti-σ factors. While UsfX shows some homology to anti-σ factors, based on its unique DNA binding capabilities it may aid in SigF activation as opposed to its inhibition. If this were the case, one may speculate that RsfA binding under reducing conditions may in fact inactivate SigF, and only under oxidizing conditions, when the RsfA Cys73-Cys109 disulfide bond is formed, can UsfX and SigF function in gene regulation.
3f. Insights into the membrane associated SigL/RslA complex interface
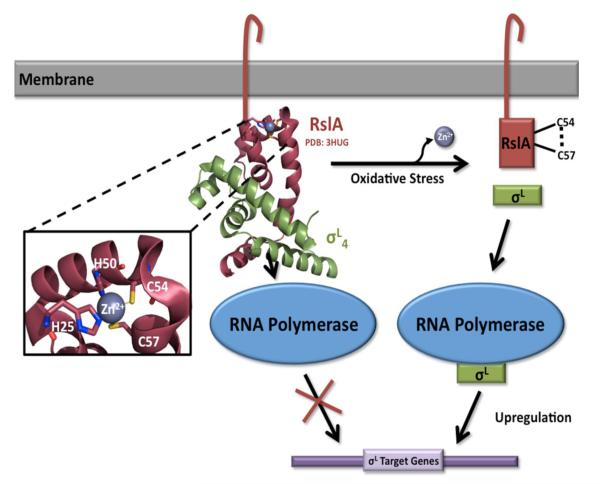
SigL has been shown to be vital for Mtb virulence [72] and is regulated by a redox-sensing anti-σ factor, the membrane-associated protein RslA. Similar to SigF, SigL and RslA are co-transcribed and thus the two proteins are tightly associated. Much like the anti-σ factor of SigE, RslA is a ZAS protein with a cytosolic domain containing the HXXXCXXC motif to allow for the coordination of a Zn2+ ion under reducing conditions and an extracellular C-terminal domain (Figure 3). Under extracellular redox stress, the two cysteines of the CXXC motif (Cys54 and Cys57) form a disulfide bond, releasing the bound Zn2+ ion and thus RslA dissociates from SigL, allowing SigL to bind its target promoters and regulate gene expression [50]. SigL is also unique from the other redox σ factors as it upregulates genes involved in maintenance of the cell-wall envelope and secreted protein modifications, implicating it with SigE and SigH as important in virulence. The crystal structure of full-length SigL protein has still not been solved, however the structure of the -35 binding domain of SigL complexed with RslA has been solved by Thakur et al in 2010 [50], Figure 3. As expected, RslA binds to the predicted -35 promoter binding region of SigL (based on SigE homology) to prevent its association with target genes and inhibit its function. This interaction shows a large buried surface area of 1332 Å2, whereby this interaction prevents DNA binding. The cytosolic C-terminal ZAS domain also shows a tight association to a Zn2+ ion at the CXXC motif, confirming previous biochemical results for RslA and other mycobacterial anti-σ factors (Figure 3). Despite extensive evidence of this mechanism of SigL regulation and release, Dainese et al. were unable to see an increase in SigL activity within an oxidative environment [73], indicating that this redox-switch may play another unexpected role in Mtb biology.
Figure 3.
The mechanism of SigL regulation. The membrane-associated anti-σ factor, RslA, contains a conserved cytosolic ZAS (HXXXCXXC) motif, which coordinates a structural Zn2+ ion under reducing conditions. SigL function is inhibited through complex formation occluding SigL’s predicted -35 promoter binding domain (PDB code 3HUG). Upon exposure to oxidative stress, the two cysteines of the CXXC motif (Cys54 and Cys57) form a disulfide bond, releasing the Zn2+ ion and leading to a conformational change in RslA. This conformational change results in the dissociation of the RslA/SigL complex, allowing SigL to interact with RNA polymerase and upregulate target genes.
3g. Role of serine/threonine kinases (STKs) in redox stress response
The serine/threonine kinase (STK) PknB has previously been shown to play a role in the regulation of ECF σ factors SigH and SigE [47, 62]. Additionally, three other STKs, PknA, PknE, and PknG have also been shown to play roles in redox stress response and survival [74-77]. PknA has been characterized as phosphorylating several proteins including PknB, FtsZ (a tubulin homolog), and FipA (a FtsZ interacting protein) when exposed to environmental stress [75]. Experimental data have shown that phosphorylation of these targets is required for cell survival when exposed to oxidative stress, signifying an important role for PknA in both redox sensing as well as stress survival. PknA, PknE, and PknG show significant differences from human STKs and have important functions in cell survival; this makes Mtb STKs attractive drug targets and thus deserving of extensive functional and regulatory studies. While PknA and PknB are not known to have a direct redox sensing ability, PknE displays a thioredoxin fold typical of redox sensing proteins [74], and PknG has been shown to have a CXXCG motif and inhibits macrophage lysosome fusion with Mtb-containing phagosomes [76, 77]. Together, these studies demonstrate that PknE and PknG are redox-sensing kinases are important for Mtb to colonize its host, making them potent targets along with the ECF σ factors for potential new drugs to fight Mtb infection.
4. The Sensory Domains for the Dos two-component system
The Mtb two/three-component DosS/DosT-DosR system comprises two membrane-tethered histidine kinases, DosS and DosT, which mediate the response to host gaseous small molecules such as O2, NO and CO [5, 6, 78, 79] as well as proton concentrations [78]. The Dos response regulatory system is postulated to signal the bacterium to enter into a persistent state [80, 81]. When activated, DosS and DosT undergo autophosphorylation and transfer their signaling phosphate group to the response regulator, DosR [81-83]. DosR is a transcriptional regulator that regulates a subset of genes (known as the dormancy regulon) required to enter dormancy [84], resulting in the redirection of the electron transport chain due to lack of O2. The consequence of this change includes upregulation of alternate electron acceptors such as nitrate/fumarate and succinate systems [85, 86], downregulation of basal metabolism [87, 88], upregulation of triacylglycerol production [89, 90], and thickening of the mycobacterial cell-wall [91].
DosS and DosT have similar four-domain structures, both consisting of two extracytoplasmic tandem cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA (GAF) domains followed by a histidine kinase-like and ATPase domains [83, 92, 93]. The first N-terminal extracellular GAF domain (GAF-A) of both DosS and DosT contains a heme cofactor whereas the second GAF domain (GAF-B) does not. DosS and DosT sense small gaseous host molecules and changes in external free proton concentrations via the heme cofactor of each GAF-A domain. Both kinases are inactive in their ferrous O2-bound states but active in other ferrous states, for example as in their nitrosyl form [78, 93, 94]. Comprehensive studies have been carried out on the single heme-containing GAF-A domains [95-97] although little is known regarding the mechanism of intra-domain communication from the GAF-A heme-binding pocket through to the kinase domain. Interestingly, DosS and DosT lacking heme, either by truncation of GAF-A domains or production of full-length apo proteins, retain their autophosphorylation activities suggesting that the isolated kinase domains alone are fully functional [80, 81, 83]. Further, these results imply that the heme cofactor is the on-off switch for both DosS and DosT kinase activity since in its absence the kinase domains are always activated.
Thus far, studies suggest that DosT senses O2, CO and NO [78, 93] while DosS is proposed to sense the redox state and/or gases [78, 93, 94, 98, 99]. Furthermore, structural and spectroscopic analyses of DosT and DosS GAF-A domains have been carried out to shed light on their sensory functions in diatomic ligand binding and have been recently reviewed in depth by Ortiz de Montellano [97] and others [95, 96].
4a. Structures of GAF-A domains of DosS and DosT
The crystal structures of heme-bound GAF-A domains of DosS [100] and DosT [101] have been determined, and are structurally similar. Each GAF-A domain comprises a five-stranded antiparallel β-sheet decorated with four α-helices, two long helices lie diagonally across one side of the β-sheet and on its other side, two shorter helices run almost parallel. Additionally, two elaborate loops near the shorter helices form the heme-binding pocket. The heme-binding cavity is a hydrophobic pocket, which tightly binds heme in an orientation perpendicular to the β-sheet. Although DosS/DosT GAF-A domains are structurally similar, there are subtle differences in the accessibility of small molecules to the heme-binding pocket, which are described below. Of note, the structure of the second GAF-B domain of M. smegmatis DosS has been solved [102] and despite sharing a similar fold as the DosS/DosT GAF-A domains, the GAF-B domain does not contain heme and has a six-stranded β-sheet that is curved, almost forming a β-barrel, whereas the GAF-A domains have almost flat β-sheets.
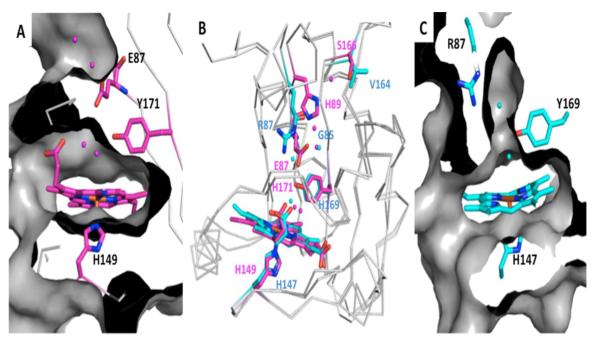
Within the DosS GAF-A heme-binding pocket, heme-iron is ligated in its proximal position through His149, whereas at the distal position heme-iron is ligated to the oxygen of water (Figure 4). The water molecule hydrogen bonds to Tyr171, which in turn interacts with His89 via Glu87, which further interacts with Ser166 via a water molecule [100]. Along this hydrogen-bonding network, there is no clear channel where water or small gaseous molecules can traverse from protein surface to heme-iron (Figure 4). Within the DosT GAF-A binding pocket, the corresponding proximal ligand to heme-iron is also histidine, His147, and the distal water ligand also hydrogen-bonds to a tyrosine, Tyr169 that interacts with Arg87 via Gly85 rather than a bulky glutamate as seen for DosS GAF-A, Figure 4 [101].
Figure 4.
Structures of DosS and DosT. Molecular surface representation for A & C, with a grey ribbon model and important residues in stick representation where oxygen and nitrogen are colored red and blue, respectively, and DosS and DosT carbons are pink and cyan, respectively. (A) Active site of DosS (PDB code 2W3E) with a H-bonding network from heme to protein surface that does not allow access of gases to the heme-iron distal site due in part to Glu87. (B) Superimposition of DosS and DosT where important residues and heme molecules are in stick representation. (C) Active site of DosT (PDB code 2VZW) where Gly85, which is in the equivalent position of DosS Glu87, opens a channel for gases to access the distal position of heme-iron.
4b. Exposure of GAF-A domains of DosS and DosT to Oxygen
After reduction followed by exposure to O2, DosT forms a stable ferrous-dioxygen heme complex (oxy-DosT) [103], and thus acts in a similar manner as the PAS-domain sensor FixL [104] and E. coli Dos [105], implying that the heme domain is an effective O2 trap. In contrast, when DosS is reduced and exposed to oxygen, ferric protein (Met-DosS) was observed. It was suggested that auto-oxidation of DosS occurred rather than forming a stable complex with O2 and thus, it appears that dissociation of O2 is faster for DosS compared to DosT [103]. Contrary to these observations, equilibrium dissociation constants of O2 are reportedly similar for both deoxy ferrous DosT and DosS, 26 μM and 3 μM, respectively [93]. It should be noted that for both DosS and DosT, NO and CO bind to heme-iron in a similar manner [97].
The difference in oxygen binding of reduced DosT and DosS could be the consequence of two separate events. First, DosT GAF-A has an apparent tunnel from its surface to the heme-iron (Figure 4) whereas Glu87 in DosS GAF-A blocks access for small gaseous molecules to heme-iron (Figure 4). Substitution of Glu87 with a glycine in the DosS GAF-A (to mimic DosT Gly85) results in a channel from the domain’s surface to the heme cofactor and consequently allows small molecules to reach the heme-iron as observed by the formation of a stable ferrous dioxygen complex [101]. Conversely, when reduced DosT GAF-A Gly85Glu mutant is exposed to air, there is no formation of the ferrous-dioxygen heme species [101]. Second, DosT has distal residues keeping oxygen in close proximity to the heme molecule after dissociation (i.e., Try169 H-bonds to O2). This is supported by molecular dynamic simulation that demonstrates that H-bonding of O2 to the conserved tyrosine (Tyr169 in DosT and Tyr171 in DosS) keeps the dissociated molecule closer to the heme-bound configuration in DosT compared to DosS [103]. However, it remains unclear why there is a difference in O2 dissociation and rebinding since DosS also has a conserved tyrosine (Tyr171). Interestingly, the DosT Tyr169Phe mutant allows O2 to escape more easily whereas CO cannot [103]. One potential mechanism may be that Tyr is locked in an alternate conformation when the ligand is not O2, thus mutating Tyr unlocks this proposed alternate conformation and allows the conformation for O2 dissociation which hinders CO escape. These results suggest that there is a Tyr H-bonding trigger in DosT signal transmission, and plays an important role in ligand discrimination as it does in DosS [106]. Additionally, the conserved Tyr within the DosS and DosT heme binding sites have been proposed to modulate the on-off switch of kinase activites for both proteins, demonstrated by the DosS Tyr171Phe mutant being an inactive kinase in all heme-iron states.
4c. Is DosS a redox sensor?
In vitro biochemical experiments of the DosS GAF-A domain suggest that it could indeed be a redox sensor. Over the last several years there has been more biological evidence supporting this hypothesis. Kumar, et al. demonstrated that ferrous DosS and deoxy DosT show increased autokinase activity compared with Met-DosS and oxy-DosT [78]. Furthermore, DosS and DosT respond to different decreased oxygen concentrations, where DosT is more sensitive [98]. Finally, Mtb DosS responds to a reduced electron transport system. The Dos regulon was induced by DosS and not DosT under aerobic conditions by ascorbate, a powerful cytochrome c reductant, changing the redox signal [107], thus confirming that DosS is primarily a redox sensor with a probable secondary function in sensing molecular gases.
5. Additional systems that maintain Mtb redox balance
Apart from the metal-containing regulatory proteins discussed within this review, there are other important systems that mycobacteria employ to maintain its redox homeostasis. The Mtb thiol-disulfide oxidoreductase system encompasses the thioredoxin system and mycothiol (reviewed in [2, 22]). The thioredoxin system, which comprises of thioredoxin as well as members from the Ahp and Tpx peroxiredoxin systems, has been shown to maintain intracellular thiols and peroxide levels. Interestingly, the Mtb disulfide-bond forming (Dsb) proteins, which include DsbA [108-110], DsbE [111] and DsbF [112], are responsible for disulfide formation of secreted and often virulent proteins through canonical thioredoxin-folds and have been implicated in the correct folding of a putative peroxiredoxin [112], suggesting their roles in redox maintenance. Additionally, in the absence of glutathione, Mtb produces millimolar concentrations of mycothiol, a major actinobacteria-specific low molecular weight thiol that functions as a redox buffer [113]. While the role of mycothiol has been extensively investigated, much less is known about a second low molecular weight thiol, ergothioneine, although initial characterization in M. smegmatis reveals its role in protection against oxidative stress [114]. Finally, Mtb’s ability to cope with oxidative stress also relies on catalase peroxidase, superoxide dismutase and methionine sulfoxide reductases (reviewed in [2]).
Host-derived gaseous molecules that activate hypoxia are also produced within mycobacteria during metabolic processes. To that end, Mtb has evolved mechanisms to circumvent toxic buildup of endogenous levels of NO and CO. As such, truncated Mtb hemoglobins, GlbO and GlbN, were discovered to possess nitric oxide dioxygenase-like activity, which capture NO molecules at the entrance of the active site channel [115, 116]. However, this finding is rather controversial and requires experimental clarification. Moreover, Mtb flavohemoglobins may also function in the conversion of NO and O2 to nitrate; however this has not been tested. Of note, the assault of CO upon Mtb in host macrophages is due to the upregulation of host heme oxygenase, which breaks down heme to release CO. Mtb has a heme uptake system [117, 118], which requires a heme-degrading protein to release iron for cellular utilization. A cytosolic Mtb heme degrading protein, MhuD, has been identified [119] where heme is degraded to produce iron without the formation of CO [120]. Significantly, Mtb MhuD is the first heme-degrading protein shown not to produce CO presumably to avoid signaling to itself to transition into a latent state. On the contrary, Mtb can tolerate low levels of CO, suggesting a potential CO resistance pathway [6]. It has also been suggested that Mtb may utilize CO as a carbon and electron source [121]. In support of this hypothesis, mycobacteria possess a homolog of the metalloprotein complex [122], carbon monoxide dehydrogenase (CODH), which hydrolyzes CO to carbon dioxide, electrons and protons. Furthermore, mycobacteria have been observed to grow on CO and methanol [123]. However, Mtb utilization of CO as a carbon source still requires experimental validation.
6. Conclusion
Redox stress response is a vital process for Mtb, as bacterial strains deficient in this process tend to have reduced or loss of virulence and/or the inability to successfully colonize a host [2]. Intriguingly, the Wbl proteins, the DosS/DosT-DosR system, and the σ factors appear to be interrelated in response to stress [11]. In fact, direct links between Wbl proteins and σ factors have been observed in Corynebacterium galactum and Mtb. C. galactum SigH was shown to control expression of whcE, a Wbl homolog, as well other genes involved in redox sensing [124] whereas Mtb SigB and SigF have been implicated in upregulating the expression of genes whiB1, whiB2, whiB4 and whiB7 [66, 69, 70].
Understanding the intricacies of Mtb’s complex systems for redox maintenance and stress sensing, as well as its transition into a latent state is crucial to combat TB, especially with the recent emergence of multiple drug-resistant Mtb strains. To that end, significant strides have been made especially for Mtb Wbl proteins, sigma factors and the Dos regulon, with the eventual hope that these data can be exploited for the development of anti-TB therapeutics.
Highlights.
Mycobacterium tuberculosis (Mtb) has evolved strategies to evade human host assault
Mtb redox stress response utilizes metalloproteins to regulate gene expression
The Wbl protein family contains iron-sulfur clusters that are sensitive to NO and O2
The heme cofactor in DosS/DosT senses NO, CO and O2 and regulates DosR activity
Zn-dependent anti-σ factors regulate σ factor activity in the presence of ROS
Acknowledgements
Authors of this review were supported by the National Institutes of Health, PO1-AI095208 (co-PI C.W.G) and R01-AI081161 (C.W.G). We thank Heidi Contreras and Robert Morse for critical reading of this article.
7. Abbreviations
- Mtb
Mycobacterium tuberculosis
- TB
tuberculosis
- Fe-S
iron-sulfur
- ZAS
zinc-associated anti-σ
- ECF
extracytoplasmic function
- WhiB
Subfamily of Whi proteins found in actinomycetes and originally described in Streptomyces coelicolor whereby mutations in their genes turn gray colonies white
- Wbl
WhiB-like
- ROS
reactive oxygen species
- RNS
reactive nitrogen species
- Dos
dormancy survival
- GAF
cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].W.H. Organization 2012.
- [2].Kumar A, Farhana A, Guidry L, Saini V, Hondalus M, Steyn AJ. Expert Rev Mol Med. 2011;13:e39. doi: 10.1017/S1462399411002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wayne LG, Sohaskey CD. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- [4].Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kumar A, Deshane JS, Crossman DK, Bolisetty S, Yan BS, Kramnik I, Agarwal A, Steyn AJ. J Biol Chem. 2008;283:18032–18039. doi: 10.1074/jbc.M802274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shiloh MU, Manzanillo P, Cox JS. Cell Host Microbe. 2008;3:323–330. doi: 10.1016/j.chom.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- [9].Guelar A, Gatell JM, Verdejo J, Podzamczer D, Lozano L, Aznar E, Miro JM, Mallolas J, Zamora L, Gonzalez J, et al. AIDS. 1993;7:1345–1349. doi: 10.1097/00002030-199310000-00007. [DOI] [PubMed] [Google Scholar]
- [10].Linas BP, Wong AY, Freedberg KA, Horsburgh CR., Jr. Am J Respir Crit Care Med. 2011;184:590–601. doi: 10.1164/rccm.201101-0181OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, Gomes A, Rustad T, Dolganov G, Glotova I, Abeel T, Mahwinney C, Kennedy AD, Allard R, Brabant W, Krueger A, Jaini S, Honda B, Yu WH, Hickey MJ, Zucker J, Garay C, Weiner B, Sisk P, Stolte C, Winkler JK, Van de Peer Y, Iazzetti P, Camacho D, Dreyfuss J, Liu Y, Dorhoi A, Mollenkopf HJ, Drogaris P, Lamontagne J, Zhou Y, Piquenot J, Park ST, Raman S, Kaufmann SH, Mohney RP, Chelsky D, Moody DB, Sherman DR, Schoolnik GK. Nature. 2013;499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Davis NK, Chater KF. Mol Gen Genet. 1992;232:351–358. doi: 10.1007/BF00266237. [DOI] [PubMed] [Google Scholar]
- [13].Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- [14].Soliveri JA, Gomez J, Bishai WR, Chater KF. Microbiology. 2000;146(Pt 2):333–343. doi: 10.1099/00221287-146-2-333. [DOI] [PubMed] [Google Scholar]
- [15].Zheng F, Long Q, Xie J. Cell Biochem Biophys. 2012;63:103–108. doi: 10.1007/s12013-012-9348-z. [DOI] [PubMed] [Google Scholar]
- [16].Raghunand TR, Bishai WR. J Bacteriol. 2006;188:6966–6976. doi: 10.1128/JB.00384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Raghunand TR, Bishai WR. Microbiology. 2006;152:2735–2747. doi: 10.1099/mic.0.28911-0. [DOI] [PubMed] [Google Scholar]
- [18].Smith LJ, Stapleton MR, Fullstone GJ, Crack JC, Thomson AJ, Le Brun NE, Hunt DM, Harvey E, Adinolfi S, Buxton RS, Green J. Biochem J. 2010;432:417–427. doi: 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Steyn AJ, Collins DM, Hondalus MK, Jacobs WR, Jr., Kawakami RP, Bloom BR. Proc Natl Acad Sci U S A. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Casonato S, Cervantes Sanchez A, Haruki H, Rengifo Gonzalez M, Provvedi R, Dainese E, Jaouen T, Gola S, Bini E, Vicente M, Johnsson K, Ghisotti D, Palu G, Hernandez-Pando R, Manganelli R. Infect Immun. 2012;80:3132–3144. doi: 10.1128/IAI.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJ. PLoS Pathog. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].den Hengst CD, Buttner MJ. Biochim Biophys Acta. 2008;1780:1201–1216. doi: 10.1016/j.bbagen.2008.01.008. [DOI] [PubMed] [Google Scholar]
- [23].Alam MS, Garg SK, Agrawal P. FEBS J. 2009;276:76–93. doi: 10.1111/j.1742-4658.2008.06755.x. [DOI] [PubMed] [Google Scholar]
- [24].Geiman DE, Raghunand TR, Agarwal N, Bishai WR. Antimicrob Agents Chemother. 2006;50:2836–2841. doi: 10.1128/AAC.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Larsson C, Luna B, Ammerman NC, Maiga M, Agarwal N, Bishai WR. PLoS One. 2012;7:e37516. doi: 10.1371/journal.pone.0037516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rybniker J, Nowag A, van Gumpel E, Nissen N, Robinson N, Plum G, Hartmann P. Mol Microbiol. 2010;77:642–657. doi: 10.1111/j.1365-2958.2010.07235.x. [DOI] [PubMed] [Google Scholar]
- [27].Chawla M, Parikh P, Saxena A, Munshi M, Mehta M, Mai D, Srivastava AK, Narasimhulu KV, Redding KE, Vashi N, Kumar D, Steyn AJ, Singh A. Mol Microbiol. 2012;85:1148–1165. doi: 10.1111/j.1365-2958.2012.08165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Crack JC, Stapleton MR, Green J, Thomson AJ, Le Brun NE. J Biol Chem. 2013;288:11492–11502. doi: 10.1074/jbc.M112.439901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Alam MS, Agrawal P. Protein Expr Purif. 2008;61 doi: 10.1016/j.pep.2008.04.010. [DOI] [PubMed] [Google Scholar]
- [30].Alam MS, Garg SK, Agrawal P. Mol Microbiol. 2007;63:1414–1431. doi: 10.1111/j.1365-2958.2007.05589.x. [DOI] [PubMed] [Google Scholar]
- [31].Garg SK, Alam MS, Soni V, Radha Krishnan KV, Agrawal P. Protein Expr Purif. 2007;52:422–432. doi: 10.1016/j.pep.2006.10.015. [DOI] [PubMed] [Google Scholar]
- [32].Konar M, Alam MS, Arora C, Agrawal P. FEBS J. 2012;279:2781–2792. doi: 10.1111/j.1742-4658.2012.08662.x. [DOI] [PubMed] [Google Scholar]
- [33].De Smet KA, Weston A, Brown IN, Young DB, Robertson BD. Microbiology. 2000;146(Pt 1):199–208. doi: 10.1099/00221287-146-1-199. [DOI] [PubMed] [Google Scholar]
- [34].Sassetti CM, Boyd DH, Rubin EJ. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- [35].Garg S, Alam MS, Bajpai R, Kishan KR, Agrawal P. BMC Biochem. 2009;10:1. doi: 10.1186/1471-2091-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Johnson DC, Dean DR, Smith AD, Johnson MK. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- [37].Roche B, Aussel L, Ezraty B, Mandin P, Py B, Barras F. Biochim Biophys Acta. 2013;1827:455–469. doi: 10.1016/j.bbabio.2012.12.010. [DOI] [PubMed] [Google Scholar]
- [38].Huet G, Daffe M, Saves I. J Bacteriol. 2005;187:6137–6146. doi: 10.1128/JB.187.17.6137-6146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Singh A, Guidry L, Narasimhulu KV, Mai D, Trombley J, Redding KE, Giles GI, Lancaster JR, Jr., Steyn AJ. Proc Natl Acad Sci U S A. 2007;104:11562–11567. doi: 10.1073/pnas.0700490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Smith LJ, Stapleton MR, Buxton RS, Green J. PLoS One. 2012;7:e40407. doi: 10.1371/journal.pone.0040407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Crack JC, Green J, Thomson AJ, Le Brun NE. Curr Opin Chem Biol. 2012;16:35–44. doi: 10.1016/j.cbpa.2012.02.009. [DOI] [PubMed] [Google Scholar]
- [42].Green J, Paget MS. Nat Rev Microbiol. 2004;2:954–966. doi: 10.1038/nrmicro1022. [DOI] [PubMed] [Google Scholar]
- [43].Crack JC, den Hengst CD, Jakimowicz P, Subramanian S, Johnson MK, Buttner MJ, Thomson AJ, Le Brun NE. Biochemistry. 2009;48:12252–12264. doi: 10.1021/bi901498v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Crack JC, Smith LJ, Stapleton MR, Peck J, Watmough NJ, Buttner MJ, Buxton RS, Green J, Oganesyan VS, Thomson AJ, Le Brun NE. J Am Chem Soc. 2011;133:1112–1121. doi: 10.1021/ja109581t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Errington J. Microbiol. Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bashyam MD, Hasnain SE. Infection, Genetics, and Evolution. 2004;4:301–308. doi: 10.1016/j.meegid.2004.04.003. [DOI] [PubMed] [Google Scholar]
- [47].Barik S, Sureka K, Mukherjee P, Basu J, Kundu M. Mol Microbiol. 2010;75:592–606. doi: 10.1111/j.1365-2958.2009.07008.x. [DOI] [PubMed] [Google Scholar]
- [48].Fernandes ND, Wu Q, Kong D, Puyang X, Garg SK, Husson RN. Journal of Bacteriology. 1999;181:4266–4274. doi: 10.1128/jb.181.14.4266-4274.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Fontan PA, Voskuil MI, Gomez M, Tan D, Pardini M, Manganelli R, Fattorini L, Schoolnik GK, Smith I. J Bacteriol. 2009;191:5628–5633. doi: 10.1128/JB.00510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Thakur KG, Praveena T, Gopal B. J Mol Biol. 2010;397:1199–1208. doi: 10.1016/j.jmb.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wu H, Xiao J, Zhang J, Tao J, H. H., M. K. Wei Sheng Wu Xue Bao. 2012;52:1352–1359. [PubMed] [Google Scholar]
- [52].Bhat SA, Singh N, Trivedi A, Kansal P, Gupta P, Kumar A. Free Radic Biol Med. 2012;53:1625–1641. doi: 10.1016/j.freeradbiomed.2012.08.008. [DOI] [PubMed] [Google Scholar]
- [53].Paget MSB, Bae J, Hahn M, L. W., Kleanthous C, R. J., Buttner MJ. Molecular Microbiology. 2001;39:1036–1047. doi: 10.1046/j.1365-2958.2001.02298.x. [DOI] [PubMed] [Google Scholar]
- [54].Ilbert M, Graf PCF, Jakob U. Antioxidants and Redox Signaling. 2006;8:835–846. doi: 10.1089/ars.2006.8.835. [DOI] [PubMed] [Google Scholar]
- [55].Li W, Bottrill AR, Bibb MJ, Buttner MJ, Paget MSB, Kleanthous C. Journal of Molecular Biology. 2003;333:461–472. doi: 10.1016/j.jmb.2003.08.038. [DOI] [PubMed] [Google Scholar]
- [56].Li W, Stevenson CE, Burton N, Jakimowicz P, Paget MS, Buttner MJ, Lawson DM, Kleanthous C. J Mol Biol. 2002;323:225–236. doi: 10.1016/s0022-2836(02)00948-8. [DOI] [PubMed] [Google Scholar]
- [57].Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith I. Molecular Microbiology. 2002;45:365–374. doi: 10.1046/j.1365-2958.2002.03005.x. [DOI] [PubMed] [Google Scholar]
- [58].Dutta NK, Mehra S, Martinez AN, Alvarez X, Renner NA, Morici LA, Pahar B, Maclean AG, Lackner AA, Kaushal D. PLoS One. 2012;7:e28958. doi: 10.1371/journal.pone.0028958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Raman S, Song T, Puyang X, Bardarov S, Jacobs WR, Jr., Husson RN. J Bacteriol. 2001;183:6119–6125. doi: 10.1128/JB.183.20.6119-6125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Song T, Dove SL, Lee KH, Husson RN. Molecular Microbiology. 2003;50:949–959. doi: 10.1046/j.1365-2958.2003.03739.x. [DOI] [PubMed] [Google Scholar]
- [61].Kumar S, Badireddy S, Pal K, Sharma S, Arora C, Garg SK, Alam MS, Agrawal P, Anand GS, Swaminathan K. PLoS One. 2012;7:e43676. doi: 10.1371/journal.pone.0043676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Park ST, Kang CM, Husson RN. Proc Natl Acad Sci U S A. 2008;105:13105–13110. doi: 10.1073/pnas.0801143105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ando M, Yoshimatsu T, Ko C, Converse PJ, Bishai WR. Infect Immun. 2003;71:7170–7172. doi: 10.1128/IAI.71.12.7170-7172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Manganelli R, Fattorini L, Tan D, Iona E, Orefici G, Altavilla G, Cusatelli P, Smith I. Infect Immun. 2004;72:3038–3041. doi: 10.1128/IAI.72.5.3038-3041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Manganelli R, Voskuil MI, Schoolnik GK, Smith I. Mol Microbiol. 2001;41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- [66].Lee JH, Karakousis PC, Bishai WR. J Bacteriol. 2008;190:699–707. doi: 10.1128/JB.01273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Rustad TR, Harrell MI, Liao R, Sherman DR. PLoS One. 2008;1502 doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Michele TM, Ko C, Bishai WR. Antimicrobial Agents and Chemotherapy. 1999;43:218–225. doi: 10.1128/aac.43.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Humpel A, Gebhard S, Cook GM, Berney M. J Bacteriol. 2010;192:2491–2502. doi: 10.1128/JB.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Hartkoorn RC, Sala C, Uplekar S, Busso P, Rougemont J, Cole ST. J Bacteriol. 2012;194:2001–2009. doi: 10.1128/JB.06692-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Malik SS, Luthra A, Ramachandran R. Biochim Biophys Acta. 2009;1794:541–553. doi: 10.1016/j.bbapap.2008.11.007. [DOI] [PubMed] [Google Scholar]
- [72].Hahn MY, Raman S, Anaya M, Husson RN. J Bacteriol. 2005;187:7062–7071. doi: 10.1128/JB.187.20.7062-7071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Dainese E, Rodrigue S, Delogu G, Provvedi R, Laflamme L, Brzezinski R, Fadda G, Smith I, Gaudreau L, Palu G, Manganelli R. Infect Immun. 2006;74:2457–2461. doi: 10.1128/IAI.74.4.2457-2461.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Jayakumar D, Jacobs WR, Jr., Narayanan S. Cell Microbiol. 2008;10:365–374. doi: 10.1111/j.1462-5822.2007.01049.x. [DOI] [PubMed] [Google Scholar]
- [75].Sureka K, Hossain T, Mukherjee P, Chatterjee P, Datta P, Kundu M, Basu J. PLoS One. 2010;5:e8590. doi: 10.1371/journal.pone.0008590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [76].Tiwari D, Singh RK, Goswami K, Verma SK, Prakash B, Nandicoori VK. J Biol Chem. 2009;284:27467–27479. doi: 10.1074/jbc.M109.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, Klebl B, Thompson C, Bacher G, Pieters J. Science. 2004;304:1800–1804. doi: 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- [78].Kumar A, Toledo JC, Patel RP, Lancaster JR, Jr., Steyn AJ. Proc Natl Acad Sci U S A. 2007;104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Proc Natl Acad Sci U S A. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Saini DK, Malhotra V, Dey D, Pant N, Das TK, Tyagi JS. Microbiology. 2004;150:865–875. doi: 10.1099/mic.0.26218-0. [DOI] [PubMed] [Google Scholar]
- [81].Roberts DM, Liao RP, Wisedchaisri G, Hol WG, Sherman DR. J Biol Chem. 2004;279:23082–23087. doi: 10.1074/jbc.M401230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, Schoolnik GK, Sherman DR. Mol Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Saini DK, Malhotra V, Tyagi JS. FEBS Lett. 2004;565:75–80. doi: 10.1016/j.febslet.2004.02.092. [DOI] [PubMed] [Google Scholar]
- [84].Boon C, Dick T. J Bacteriol. 2002;184:6760–6767. doi: 10.1128/JB.184.24.6760-6767.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Sohaskey CD. J Bacteriol. 2008;190:2981–2986. doi: 10.1128/JB.01857-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Watanabe S, Zimmermann M, Goodwin MB, Sauer U, Barry CE, 3rd, Boshoff HI. PLoS Pathog. 2011;7:e1002287. doi: 10.1371/journal.ppat.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Baek SH, Li AH, Sassetti CM. PLoS Biol. 2011;9:e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Leistikow RL, Morton RA, Bartek IL, Frimpong I, Wagner K, Voskuil MI. J Bacteriol. 2010;192:1662–1670. doi: 10.1128/JB.00926-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. PLoS Pathog. 2011;7:e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Low KL, Rao PS, Shui G, Bendt AK, Pethe K, Dick T, Wenk MR. J Bacteriol. 2009;191:5037–5043. doi: 10.1128/JB.00530-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kruh NA, Troudt J, Izzo A, Prenni J, Dobos KM. PLoS One. 2010;5:e13938. doi: 10.1371/journal.pone.0013938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Sardiwal S, Kendall SL, Movahedzadeh F, Rison SC, Stoker NG, Djordjevic S. J Mol Biol. 2005;353:929–936. doi: 10.1016/j.jmb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- [93].Sousa EH, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. Protein Sci. 2007;16:1708–1719. doi: 10.1110/ps.072897707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Ioanoviciu A, Yukl ET, Moenne-Loccoz P, de Montellano PR. Biochemistry. 2007;46:4250–4260. doi: 10.1021/bi602422p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Aono S. Antioxid Redox Signal. 2012;16:678–686. doi: 10.1089/ars.2011.4248. [DOI] [PubMed] [Google Scholar]
- [96].Farhana A, Saini V, Kumar A, Lancaster JR, Jr., Steyn AJ. Antioxid Redox Signal. 2012;17:1232–1245. doi: 10.1089/ars.2012.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Sivaramakrishnan S, Ortiz de Montellano PR. Biosensors. 2013;3:3390–3282. doi: 10.3390/bios3030259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kim MJ, Park KJ, Ko IJ, Kim YM, Oh JI. J Bacteriol. 2010;192:4868–4875. doi: 10.1128/JB.00550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Ioanoviciu A, Meharenna YT, Poulos TL, Ortiz de Montellano PR. Biochemistry. 2009;48:5839–5848. doi: 10.1021/bi802309y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Cho HY, Cho HJ, Kim YM, Oh JI, Kang BS. J Biol Chem. 2009;284:13057–13067. doi: 10.1074/jbc.M808905200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Podust LM, Ioanoviciu A, Ortiz de Montellano PR. Biochemistry. 2008;47:12523–12531. doi: 10.1021/bi8012356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Lee JM, Cho HY, Cho HJ, Ko IJ, Park SW, Baik HS, Oh JH, Eom CY, Kim YM, Kang BS, Oh JI. J Bacteriol. 2008;190:6795–6804. doi: 10.1128/JB.00401-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Vos MH, Bouzhir-Sima L, Lambry JC, Luo H, Eaton-Rye JJ, Ioanoviciu A, Ortiz de Montellano PR, Liebl U. Biochemistry. 2012;51:159–166. doi: 10.1021/bi201467c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Gilles-Gonzalez MA, Ditta GS, Helinski DR. Nature. 1991;350:170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- [105].Lechauve C, Bouzhir-Sima L, Yamashita T, Marden MC, Vos MH, Liebl U, Kiger L. J Biol Chem. 2009;284:36146–36159. doi: 10.1074/jbc.M109.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Yukl ET, Ioanoviciu A, Nakano MM, de Montellano PR, Moenne-Loccoz P. Biochemistry. 2008;47:12532–12539. doi: 10.1021/bi801234w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Honaker RW, Dhiman RK, Narayanasamy P, Crick DC, Voskuil MI. J Bacteriol. 2010;192:6447–6455. doi: 10.1128/JB.00978-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Chim N, Harmston CA, Guzman DJ, Goulding CW. BMC Struct Biol. 2013;13:23. doi: 10.1186/1472-6807-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Premkumar L, Heras B, Duprez W, Walden P, Halili M, Kurth F, Fairlie DP, Martin JL. Acta Crystallogr D Biol Crystallogr. 2013;69:1981–1994. doi: 10.1107/S0907444913017800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Wang L, Li J, Wang X, Liu W, Zhang XC, Li X, Rao Z. Protein Cell. 2013;4:628–640. doi: 10.1007/s13238-013-3033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Goulding CW, Apostol MI, Gleiter S, Parseghian A, Bardwell J, Gennaro M, Eisenberg D. J Biol Chem. 2004;279:3516–3524. doi: 10.1074/jbc.M311833200. [DOI] [PubMed] [Google Scholar]
- [112].Chim N, Riley R, The J, Im S, Segelke B, Lekin T, Yu M, Hung LW, Terwilliger T, Whitelegge JP, Goulding CW. J Mol Biol. 2010;396:1211–1226. doi: 10.1016/j.jmb.2009.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Newton GL, Fahey RC, Cohen G, Aharonowitz Y. J Bacteriol. 1993;175:2734–2742. doi: 10.1128/jb.175.9.2734-2742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Sao Emani C, Williams MJ, Wiid IJ, Hiten NF, Viljoen AJ, Pietersen RD, van Helden PD, Baker B. Antimicrob Agents Chemother. 2013;57:3202–3207. doi: 10.1128/AAC.02572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Ouellet H, Juszczak L, Dantsker D, Samuni U, Ouellet YH, Savard PY, Wittenberg JB, Wittenberg BA, Friedman JM, Guertin M. Biochemistry. 2003;42:5764–5774. doi: 10.1021/bi0270337. [DOI] [PubMed] [Google Scholar]
- [116].Savard PY, Daigle R, Morin S, Sebilo A, Meindre F, Lague P, Guertin M, Gagne SM. Biochemistry. 2011;50:11121–11130. doi: 10.1021/bi201059a. [DOI] [PubMed] [Google Scholar]
- [117].Jones CM, Niederweis M. J Bacteriol. 2011;193:1767–1770. doi: 10.1128/JB.01312-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Tullius MV, Harmston CA, Owens CP, Chim N, Morse RP, McMath LM, Iniguez A, Kimmey JM, Sawaya MR, Whitelegge JP, Horwitz MA, Goulding CW. Proc Natl Acad Sci U S A. 2011;108:5051–5056. doi: 10.1073/pnas.1009516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Chim N, Iniguez A, Nguyen TQ, Goulding CW. J Mol Biol. 2010;395:595–608. doi: 10.1016/j.jmb.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Nambu S, Matsui T, Goulding CW, Takahashi S, Ikeda-Saito M. J Biol Chem. 2013;288:10101–10109. doi: 10.1074/jbc.M112.448399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Zacharia VM, Shiloh MU. Med Gas Res. 2012;2:30. doi: 10.1186/2045-9912-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Santiago B, Schubel U, Egelseer C, Meyer O. Gene. 1999;236:115–124. doi: 10.1016/s0378-1119(99)00245-0. [DOI] [PubMed] [Google Scholar]
- [123].Park SW, Hwang EH, Park H, Kim JA, Heo J, Lee KH, Song T, Kim E, Ro YT, Kim SW, Kim YM. J Bacteriol. 2003;185:142–147. doi: 10.1128/JB.185.1.142-147.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Kim TH, Park JS, Kim HJ, Kim Y, Kim P, Lee HS. Biochem Biophys Res Commun. 2005;337:757–764. doi: 10.1016/j.bbrc.2005.09.115. [DOI] [PubMed] [Google Scholar]