Abstract
Arbuscular mycorrhizae are ancient symbioses that are thought to have originated >400 million years ago in the roots of plants, pioneering the colonization of terrestrial habitats. In these associations, a key process is the transfer of phosphorus as inorganic phosphate to the host plant across the fungus–plant interface. Mycorrhiza-specific phosphate transporter genes and their regulation are conserved in phylogenetically distant plant species, and they are activated selectively by fungal species from the phylum Glomeromycota. The potato phosphate transporter gene StPT3 is expressed in a temporally defined manner in root cells harboring various mycorrhizal structures, including thick-coiled hyphae. The results highlight the role of different symbiotic structures in phosphorus transfer, and they indicate that cell–cell contact between the symbiotic partners is required to induce phosphate transport.
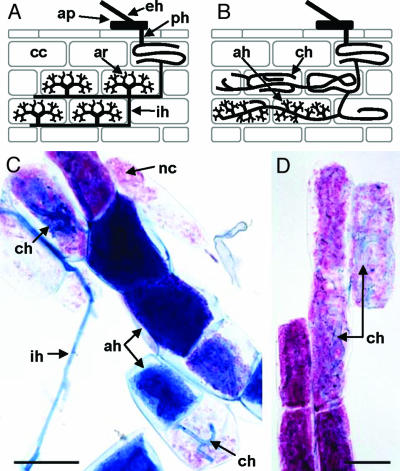
Arbuscular mycorrhizal fungi (AMF) are important biotrophic organisms, which live in symbiosis with ≈80% of land plants, forming a mycorrhiza (i.e., a root colonized by a symbiotic fungus). AMF affect plant biodiversity, as well as the variability and productivity of ecosystems (1, 2). As few as ≈150 fungal species are known to form arbuscular mycorrhizae (AM) in the roots of a vast number of plant species. No evidence for recombination has been found in the fungi, suggesting that they reproduce clonally and have been asexual for the entire period of their association with plants (3). Recently, AMF were placed into a new monophyletic group, the phylum Glomeromycota, which probably originated from the same ancestral group as the Ascomycota and Basidiomycota (4) ≈1,400–1,200 million years ago and is much older than the earliest land plants, which appeared ≈800 million years ago and whose primitive root systems were associated with ancestral AMF. AMF may, thus, have played a crucial role in facilitating the colonization of land by plants (5–8). It is also assumed that the ancient signaling pathways evolved in AM symbiosis were recruited subsequently for the establishment of the evolutionary younger legume–Rhizobia nodulation symbiosis (9). Today, despite the large number of plant species forming AM associations worldwide, only two major morphological types have been defined: the Arum and Paris types, respectively. In Arum-type mycorrhizae, fungal hyphae spread between cortical cells and form short-lived heavily branched symbiotic structures (arbuscules) within cells. In Paris-type mycorrhizae, cortical cells are colonized by intracellularly growing thick coiled hyphae, which occasionally form fine arbuscule-like ramifications (10).
Materials and Methods
Plant and Fungal Material. The plant material used was Daucus carota (carrot), Lotus japonicus cv. Gifu, Medicago truncatula cv. Jemalong, Petunia × hybrida (petunia), Plantago lanceolata (plantain), and Solanum tuberosum cv. Désirée (potato). AMF isolates used were Acaulospora delicata (JJ1094), Archaeospora nicolsonii (W4147), Gigaspora margarita (BEG34), Glomus caledonium (BEG15 and BEG20), Glomus etunicatum (CZ), Glomus geosporum (BEG11), Glomus intraradices (BEG75), Glomus mosseae (BEG76), Glomus spurcum (W4217), and Paraglomus brasilianum (W4121). Pathogenic fungi used were Rhizopus stolonifer var. stolonifer (MUCL20416), Nectria ventricosa (MUCL33), Rhizoctonia solani (MUCL30282), and the oomycete Phytophthora infestans (CRA208). The dark septate root endophyte Phialocephala fortinii, the binucleate Rhizoctonia sp., which is a root parasite that does not cause root-disease symptoms, and the plant growth stimulating root endophyte Piriformospora indica (DSM11827) (11, 12) were used as well.
G. caledonium (BEG15) was maintained in monoxenic culture on carrot hairy roots (13), and all other AMF isolates were maintained on P. lanceolata grown in a soil–quartz sand mixture (1:10) fertilized with quarter-strength Hoagland solution (14) containing 5 μM NH4H2PO4 by using drop irrigation. Sterile cultures of fungi other than AMF were maintained on potato dextrose agar (binucleate Rhizoctonia sp., N. ventricosa, P. indica, P. infestans, and R. solani), malt 2% yeast extract agar (R. stolonifer), and Melin–Norkrans nutrient media (P. fortinii).
Plant Growth Conditions. Stock cultures of wild-type and transgenic potato plants were maintained in tissue culture by using MS medium (15) supplemented with 2% sucrose. In pot culture, inoculation of potato plants with all AMF isolates except G. margarita was performed as described (16). For G. margarita, potato plantlets were cocultivated with sunflower, which served as nurse plants. Each pot, containing the potato–sunflower combination, was inoculated with 15–20 spores of G. margarita. Composite plants of L. japonicus and M. truncatula (see below) were introduced to already-established mycorrhizae of G. mosseae on P. lanceolata, growing in a soil–quartz sand mixture. Plants were harvested 2–6 weeks after inoculation. Freshly collected potato roots were frozen in liquid N2 and stored at -80°C for subsequent RNA isolation, or they were immediately stained for β-glucuronidase (GUS) activity and for visualization of intraradical fungal mycelium (17).
RT-PCR. RT-PCR was performed as described (18) by using gene-specific primers, as described in Supporting Methods, which is published as supporting information on the PNAS web site. GenBank accession nos. for StPT3, StPT1, MtPT4, MtPT1, and PHT2;1 are AJ318822, AF156695, AY116210, AF000354, and AF533081, respectively.
Plant Hairy-Root Cultures. Agrobacterium rhizogenes were transformed with pBin19 carrying StPT3 promoter-GUS (16), MtPT4 promoter-GUS, OsPT11 promoter-GUS, and StPT3 promoter-Fluorescent Timer chimeric genes, respectively, by means of electroporation. To initiate hairy-root formation, plant tissues were inoculated with overnight cultures of A. rhizogenes grown on solid YEB medium. Potato and carrot hairy roots were generated as described (19) by using tubers and tap roots, respectively. Sterile potato plantlets were also used for the generation of hairy roots. The apical part of the plantlet was excised and the sectioned stem surface was inoculated with A. rhizogenes. Inoculation of L. japonicus and M. truncatula sectioned seedling radicles with A. rhizogenes as described (20) allowed generation of vigorous composite plants consisting of wild-type shoots and hairy roots harboring appropriate constructs, which developed from the inoculation site. All inoculated plant material was kept in a growth chamber at low light at 23°C until hairy roots appeared. Formed hairy roots were transferred aseptically to nutrient medium, containing 100 mg/liter ampicillin. We selected 10–15 independently transformed hairy roots of each plant and inoculated them with AMF (see below). Mycorrhizal hairy roots exhibiting strong reporter gene expression were selected, amplified, and used for further experiments.
Monoxenic Cultures. Monoxenic cultures of various fungal isolates on plant hairy roots were established as described for AMF (13, 21). Medium for cultivation of potato hairy roots was supplied with 3.6 mg/liter KH2PO4 and 7.5 g/liter sucrose, whereas carrot hairy roots were grown on 4.8 mg/liter KH2PO4 and 10 g/liter sucrose. Media were solidified with 0.4% agarose (Fluka) containing very low levels of contaminant elements, especially phosphorus (22), and pH was adjusted to 5.5. Hairy roots were inoculated with either 5–10 sterile AMF spores or small pieces of nutrient medium containing hyphae of all other species, respectively. Plates were incubated at RT in the dark. Formation of plant–fungus associations was tracked by using light microscopy. Colonized roots were extracted from media and stained for GUS activity and for intraradical fungal mycelium, or they were used directly for fluorescence analysis.
Histochemical Analysis. For GUS assays, fresh root material was incubated in 0.1% Magenta-β-d-glucuronide cyclohexylammonium salt (Biosynth, Basel) and 0.1% Triton X-100 in 0.05 M sodium phosphate buffer (pH 7.2) either at room temperature for 30 min to 1 h for plant roots colonized by AMF or at 37°C overnight for all other root–fungus associations after vacuum infiltration. To visualize fungal hyphae, the stained material was transferred directly to 10% KOH (wt/vol) at 90°C for 1 h to clear the tissues, acidified with 1% HCl (vol/vol), and then counter-stained for 1–3 h at 90°C with 0.05 or 0.3% trypan blue, respectively, dissolved in lactoglycerol (lactic acid/glycerol/water, 1:1:1 vol/vol/vol). Excess staining was removed by immersing the samples in 50% glycerol. Colonized cortex cells were separated carefully from roots. Stained material was analyzed by light microscopy.
Confocal Microscopy. Fluorescence in mycorrhizal plant roots was visualized by using two confocal laser scanning microscopes: DM IRBE and TCS SP (Leica, Deerfield, IL) or an LSM 510 META (Zeiss) under oil with a ×20 or ×40 objective, respectively, at excitation and emission wavelengths of 488 and 568, 488–530, and 575–620 nm (for DM IRBE and TCS SP) or 488 and 543, 505–530, and 560–615 nm for LSM 510 META for green and red fluorescence, respectively. The data were processed by using the nt (Leica) or the lsm 510 meta (Zeiss) software, respectively. Images were assembled by using photoshop 7.0 (Adobe Systems, Mountain View, CA).
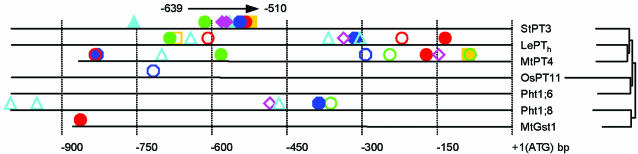
Phylogenetic Footprinting. The phylogenetic relationship between the coding sequence of the Pht1 family genes StPT3 (AJ318822), LePTh (tomato; Lycopersicon esculentum; Avraham A. Levy, Weizmann Institute of Sciences, Rehovot, Israel, personal communication), MtPT4 (GenBank accession no. AY116211), OsPT11 (GenBank accession no. AF536971), ARAth;Pht1;6 (At5g43340), and ARAth;Pht1;8 (GenBank accession no. At1g20860) (Arabidopsis thaliana; ref. 23), and the AMF-inducible glutathione S-transferase MtGst1 (M. truncatula; GenBank accession nos. AY134608 and AY134609; ref. 24) was determined by alignment with ClustalW. The corresponding promoter regions up to 1 kb upstream of the respective initiator ATGs were analyzed for conserved motifs by using footprinter 2.0 Web serve (available at http://bio.cs.washington.edu/software.htm) (25), a computer algorithm designed to identify conserved motifs in noncoding regions of orthologous genes from many species. Motifs were identified by using a motif size of 8–10 with a maximal parsimony score of 0–2 in a subregion of 1,000, allowing for motif losses. The promoter sequences were then scanned manually for variations of the six resulting motifs.
See Supporting Methods for further description of the methods used in this study.
Results and Discussion
Several hundred-million years of coevolution of plants and AMF, the worldwide distribution of AMF, and the absence of host specificity in plant–AMF interactions suggest that the cascade of events leading to the establishment of the functional AM symbiosis is conserved between modern, evolutionary distant plant species. To test this possibility, promoter sequences from mycorrhiza-specific phosphate transporter genes from the two eudicot species potato (S. tuberosum, StPT3) (16) and the model legume M. truncatula (MtPT4) (26), as well as from the monocot rice (Oryza sativa, OsPT11) (27) were fused to the GUS reporter gene and the constructs were introduced into hairy roots of five different plant species via A. rhizogenes-mediated root transformation. Subsequently, the roots were challenged with three different species of AMF, and GUS expression was monitored (Table 1). In summary, promoters of eudicot species directed GUS expression in a mycorrhiza-specific manner in the eudicot host plants, whereas the promoter from the monocot rice was not regulated correspondingly in the eudicot plants (Table 1). Microscopic inspection revealed that GUS expression invariably colocalized to intraradical AMF mycelium (data not shown). These data suggest that signal perception and the intracellular signal transduction pathway mediating the mycorrhiza-specific regulation of phosphate transport are conserved in at least three orders of the plant kingdom, i.e., the Solanales, Apiales, and Fabales, which are distributed between the two subclasses Rosidae and Asteridae within the eudicots. The evolutionary distance between rice and potato apparently did not allow conservation of the respective regulatory mechanisms, although both species exhibit mycorrhiza-specific phosphate transport (16, 27). Promoter-dissection studies indicated the presence of a regulatory domain within a 129-bp region of the StPT3 promoter. Truncated versions of the StPT3 promoter lacking this region failed to direct GUS reporter gene expression on AM fungal colonization of potato roots with G. mosseae, whereas the presence of this region, irrespective of the length of the upstream sequence, was necessary to activate GUS expression (data not shown). Phylogenetic footprinting was performed to identify conserved candidate regulatory elements in nonconserved promoter sequences of different phosphate transporter genes and one AMF-inducible gene of unrelated function. From six motifs found within the 129-bp regulatory region of StPT3, the CTTC motif was abundant in the promoters of mycorrhiza-specific phosphate transporters from eudicot tomato (LePTh; R. N. and M. B., unpublished data), and M. truncatula (MtPT4), and an AMF-up-regulated glutathione S-transferase (MtGst1) from M. truncatula, whereas it was absent in the OsPT11 promoter from monocot rice, the promoters of the flower-specific ARAth; Pht1;6 gene, and the promoters of the root-specific and phosphate starvation-inducible ARAth;Pht1;8 gene, respectively, from A. thaliana (23) (Fig. 1 and see Table 3, which is published as supporting information on the PNAS web site). Of all promoters analyzed, the TAAT motif was present exclusively in the AMF-up-regulated Pht1 family genes from eudicot potato, M. truncatula, and tomato, respectively. In the MtGst1 promoter, only one of six motifs was conserved, whereas four elements were conserved in the ARAth;Pht1;8 promoter. None of the motifs was present in the flower-specific ARAth;Pht1;6 gene from A. thaliana. Ultimately, it must be elucidated whether these motifs alone or in combination specify root and/or mycorrhiza specificity to StPT3 expression and have functions in other mycorrhizal plant species that are similar to functions in potato.
Table 1. Induction of mycorrhiza-specific phosphate transporters in plant species of different taxonomic positions.
| Promoter region fused to GUS
|
|||
|---|---|---|---|
| Plants studied | StPT3 | MtPT4 | OsPT11 |
| Eudicots | |||
| Asteridae | |||
| Solanales | |||
| Solanum tuberosum | +, G.m., P | +, G.m., P | —, G.m., P |
| Petunia hybrida | +, G.m.,I | ||
| Apiales | |||
| Daucus carota | +, G.L., P | ||
| Rosidae | |||
| Fabales | |||
| Lotus japonicus | +, G. mos., I | ||
| Medicago truncatula | +, G.mos., I | +, G.mos., I | —, G.mos., I |
Potato (S. tuberosum) and carrot (D. carota) transgenic hairy roots. L. japonicus, and M. truncatula composite plants with transgenic hairy roots and transgenic petunia (Petunia × hybrida) were generated by means of agrobacterial transformation with constructs carrying promoter regions of mycorrhiza-specific phosphate transporters from potato (StPTs; 1.72 Kb), M. truncatula (MtPT4; 0.87 Kb), and rice (O. sativa) (OsPT11; 0.8, 1.54, and 3.16 Kb) fused to the GUS reporter gene. Roots were inoculated with one of three AMF species: G.m., Gigaspora margarita; G.i., Glomus intraradices; and G. mos., Glomus mosseae. Mycorrhizal roots were stained for GUS activity. +, Positive GUS staining in root zones colonized by AMF; —, absence of GUS staining. P and I indicate formation of either Paris- or intermediate-type AM, respectively.
Fig. 1.
Putative cis-regulatory elements identified by phylogenetic footprinting. Putative cis-regulatory elements identified by footprinter 2.0 in the 129-bp regulatory region of the StPT3 promoter (arrow) and in promoter regions of ≈1 kb, as shown at bottom relative to the start ATG, of other AMF-inducible genes, i.e., phosphate transporters from tomato (LePTh), M. truncatula (MtPT4), and rice (Ospt11), as well as glutathione S-transferase MtGst1 from M. truncatula, are shown. The promoters of flower-specific phosphate transporter ARAth;Pht1;6 and root-specific phosphate starvation-inducible phosphate transporter ARAth;Pht1;8 from A. thaliana were used as controls. Highly conserved and less-conserved motifs are shown as filled and open symbols, respectively (see Table 3). Sequences are ordered according to the phylogenetic relatedness of the corresponding coding regions, as indicated on the right.
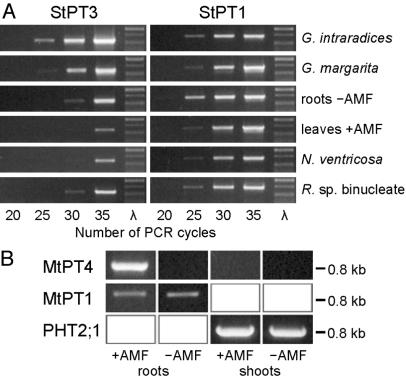
To learn more about the specificity of StPT3 expression in various plant–fungus associations, wild-type and transgenic potato plants, as well as hairy roots harboring a StPT3 promoter-GUS chimeric gene, were inoculated with 17 species from the fungal phyla Glomeromycota (10 species), Zygomycota (1 species), Basidiomycota (3 species), Ascomycota (2 species), and the nonfungal phylum Oomycota (1 species), all of which are known to enter either mutualistic symbiotic, pathogenic, plant-growth promoting, or neutral associations, respectively, with plants (Table 2). Histochemical staining for GUS activity revealed that StPT3 promoter activity colocalized with root zones colonized by AMF (Fig. 2 C and D), as shown (16). No GUS activity was observed in noninoculated control roots, root zones of mycorrhizal plants that were not colonized by AMF, or in roots that were colonized by fungal pathogens or root endophytes other than AMF (Table 2). Interestingly, low levels of StPT3 expression were detected by RT-PCR using StPT3 gene-specific primers and RNA from noninoculated roots and leaves of potato (Fig. 3A). These low transcript levels had not been detected (16) by RNA gel blot analysis. StPT3 transcript abundance was increased exclusively in potato roots inoculated with AMF, whereas it remained low in roots of noninoculated control plants and plants inoculated with species from phyla other than the Glomeromycota (Fig. 3A). In comparison, mRNA abundance of the phosphate transporter StPT1 (16) was comparable in all associations (Fig. 3A). Gene-specific RT-PCR to detect MtPT4 gene transcripts in M. truncatula confirmed mycorrhiza-specific expression of MtPT4 in roots colonized by AMF (Fig. 3B) (26). In contrast to basal StPT3 expression in nonmycorrhized potato, no MtPT4 transcripts were detectable in roots of noncolonized M. truncatula and shoots, thus indicating differential regulation of StPT3 and MtPT4 expression in the absence of AMF in the respective plant species. Overall, the expression of an AM-associated phosphate transporter attributes a functional parameter with predictive value to the new taxonomy of AMF and the phylum Glomeromycota with respect to a key parameter in the biology of these biotrophic fungi, i.e., the delivery of large amounts of phosphorus to the host plant.
Table 2. Induction of StPT3 expression in potato inoculated with fungi from different taxonomic positions.
| Taxonomic position (kingdom/phylum/order/family)† | Fungal species | Fungal status | GUS activity‡ | Mycorrhiza type (only for AMF)§ | |
|---|---|---|---|---|---|
| Oomycota | Phytophthora infestans | Pathogen | —, ms | ||
| Fungi | |||||
| Zygomycota | Rhizopus stolonifer | Pathogen | —, ms | ||
| Glomeromycota | |||||
| Paraglomerales | Paraglomus brasilianum | — | +, s | P; ah, ch | |
| Archaeosporales | Archaeospora nicolsonii | — | +, s | P; ch | |
| Diversisporales | Acaulosporaceae | Acaulospora delicata | — | +, s | P; ah, ch |
| Diversisporaceae | Glomus spurcum | — | +, s | P; ah, ch | |
| Gigasporaceae | Gigaspora margarita | — | +, ms | P; ch | |
| Glomerales | Glomeraceae (Glomus-group A) | Glomus caledonium | Mutualistic symbiont | +, ms | I; ah, ch, ih |
| Glomus geosporum | — | +, s | I; ah, ch, ih | ||
| Glomus intraradices | — | +, ms | I; a, ah, ch, ih | ||
| Glomus mosseae | — | +, ms | I; a, ah, ch, ih | ||
| Glomeraceae (Glomus-group B) | Glomus etunicatum | — | +, ms | P; ch | |
| Basidiomycota | |||||
| Piriformospora indica | Plant growth-promoting endophyte | —, m | — | ||
| Rhizoctonia solani | Pathogen | —, ms | — | ||
| Binucleate Rhizoctonia sp. | Neutral endophyte | —, s | — | ||
| Ascomycota | |||||
| Nectria ventricosa | Pathogen | —, ms | — | ||
| Phialocephala fortinii | Mutualistic symbiont to weak pathogen | —, m | — | ||
Potato plants and/or hairy roots harboring the StPT3 promoter—GUS reporter gene construct were inoculated with 17 strains of root-colonizing organisms. GUS activity in colonized tissues corresponds to the induction of StPT3 expression. +, Positive GUS staining in root zones colonized by AMF; —, absence of GUS staining.
* Systematics of AMF is given according to ref. 4
Order and family names are given only for AMF
GUS activity assays were performed in monoxenic (m), soil (s), or in both cultures (ms)
Details of AM morphology: P, Paris-type AM; I, intermediate-type AM; a, arbuscules; ah, arbusculate hyphae; ch, coiled hyphae; ih, intercellular hyphae
Fig. 2.
StPT3 promoter activity in AM symbioses. (A and B) Features of Arum and Paris types, respectively, of AM. Hyphae of extraradical mycelium (eh) make contact with the host root by forming appressoria (ap) on the root surface. Appressorium-borne hyphae penetrate (ph) the root and give rise to the intraradical mycelium, which in different plant–AMF associations forms two main morphological types, i.e., Arum and Paris. In Arum-type AM (A), intercellular hyphae (ih) grow between the cortical cells (cc) and form fine heavily branched arbuscules (ar) within cortex cells. In Paris-type AM (B), thick coiled hyphae (ch) spread intracellularly. In some cortical cells, lateral branches of coiled hyphae develop into fine branched arbuscule-like structures forming arbusculate hyphae (ah). Intermediate type AM combines features of Arum and Paris types. (C and D) Isolated cortex cells of potato hairy roots harboring the StPT3 promoter-GUS reporter chimeric gene colonized by G. caledonium (C) and G. margarita (D) histochemically stained for GUS activity and fungal mycelium. Magenta staining indicates GUS activity, and AMF hyphae are stained blue. The StPT3 promoter is expressed in cells colonized by both fine branched arbusculate hyphae (ah) and thick-coiled hyphae (ch). GUS products can diffuse into neighboring noncolonized cells (nc); ih, intercellular hypha. (Bars, 50 μm.)
Fig. 3.
Expression of mycorrhiza-inducible phosphate transporters in potato and M. truncatula. (A) Semiquantitative RT-PCR performed with 1 μg of total RNA extracted from potato roots (or leaves, where indicated) colonized by fungi of different status (see Table 2). cDNAs were amplified with StPT3 and StPT1 sequence-specific primers, respectively. (B) MtPT4 expression in M. truncatula colonized by the AMF fungus G. mosseae. RT-PCR was performed with 1 μg of total RNA extracted from M. truncatula tissues. cDNAs were amplified with MtPT1-, MtPT4-, and PHT2;1-sequence-specific primers, respectively, over 35 cycles. MtPT1 and PHT2;1 served as marker genes for expression in roots and shoots, respectively. As size marker, PstI-restricted λ DNA was used.
Despite the large number of plant species in AM associations worldwide, only two major morphological types have been defined in AM, i.e., the Arum and Paris types, respectively (Fig. 2 A and B) (28, 34). It is generally assumed that the arbuscule is a key structure in AM symbiosis and that the interface between arbuscules and the invaginated plant cell membrane, the so-called periarbuscular membrane, is the site of phosphate, and possibly carbon, exchange between the symbionts (26, 29, 30). Much less is known about nutrient-transfer mechanisms at interfaces in Paris-type mycorrhiza, although it reportedly is the most common morphological type of AM. Moreover, conflicting physiological data on plant growth and phosphorus uptake in response to the formation of the Paris-type AM have been published (31, 32). Because AMF formed either Paris or intermediate types of AM both in roots and hairy roots of potato (Fig. 2 and Table 2), this association was studied subsequently in more detail. Potato hairy roots harboring a StPT3 promoter-GUS chimeric gene were inoculated in vitro with either G. margarita or G. caledonium. These two species of AMF allowed examination of the StPT3 expression pattern in cells colonized by either only coiled hyphae (G. margarita) or coiled and arbusculate hyphae (G. caledonium). Colocalization of stained mycelium and GUS activity in mycorrhizal potato roots clearly showed that the StPT3 promoter was active not only in cortical cells colonized by heavily branched hyphae (i.e., arbuscules and arbusculate hyphae; ref. 16 and Fig. 2C) but also in cells containing only thick coiled hyphae (Fig. 2 C and D), indicating that expression of the StPT3 gene is induced in root cells hosting diverse AM structures including types other than arbuscules. This molecular evidence suggests possible phosphate transfer at the fungus–root interface of the Paris-type mycorrhiza.
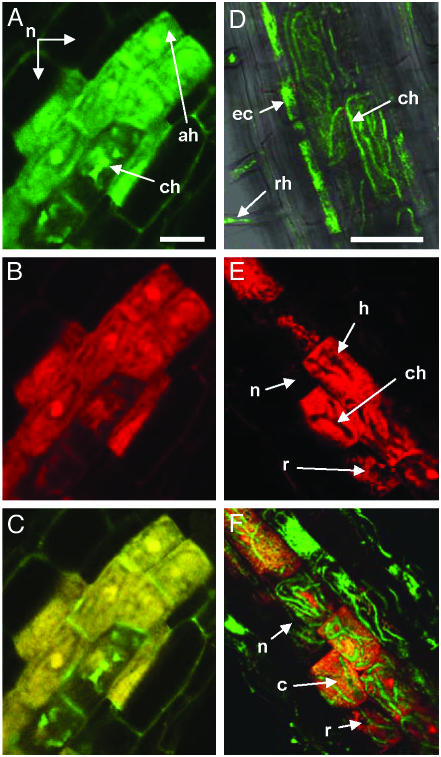
It is, however, generally known that the product of the GUS reaction may diffuse and, thus, obscure the precise localization of GUS expression. To confine reporter gene expression precisely to the site of promoter activity and simultaneously obtain information on the temporal profile of StPT3 expression, a reporter system based on the Fluorescent Timer, a mutant form of the DsRed fluorescent protein from the coral Discosoma, was used. The Fluorescent Timer protein shifts fluorescence color from green to red over time because of slow fluorophore maturation (33). The rate of color conversion has been reported to be independent of protein concentration and, therefore, can be used to trace time-dependent expression. To trace time-dependent expression, potato hairy roots harboring a StPT3 promoter-Fluorescent Timer chimeric gene colonized with G. margarita and G. caledonium, respectively, were inspected by laser scanning confocal microscopy. Within a single colonization unit, differentially fluorescing cells were present. Generally, background green autofluorescence in cortical cells originated mainly from fungal hyphae (Fig. 4D), whereas strong red fluorescence was detectable exclusively in cells colonized by AMF hyphae (Fig. 4 B and E). Neither noncolonized cortical cells nor fungal hyphae emitted strong red fluorescence. Colonized cells usually exhibited homogeneously distributed green and red fluorescence independent of the type of colonization, i.e., coiled hyphae or arbusculate hyphae (Fig. 4 A, B, E, and F). Interestingly, cells emitting strong red fluorescence were bordered by colonized cells without red fluorescence (Fig. 4E). An overlay of both red and green fluorescence of the same section allowed us to see not only where in a colonization unit the StPT3 promoter is active but also when it becomes inactive (Fig. 4F). Green fluorescence originating from the Fluorescent Timer was accompanied by red fluorescence, giving rise to yellow-to-orange cells signifying continuous promoter activity. Red fluorescent cells denoted cells in which promoter activity had ceased after an extended “on” period. Nonsynchronous pattern of promoter activity was apparent among individual cells within a single mycorrhizal unit, i.e., in morphologically similar potato cortex cells colonized by thick-coiled hyphae from G. margarita, and cells emitted green and red, red only, or no fluorescence (Fig. 4F), whereas in potato most cortical cells containing arbusculate hyphae from G. caledonium emitted green and red fluorescence, indicating that the StPT3 promoter was simultaneously active in most colonized cells (Fig. 4 A–C). Somewhat weaker and green fluorescence in two cells of the same colonization unit was not related to fungal biomass (data not shown) and, therefore, suggested that in these cells StPT3 expression has just been turned on (Fig. 4C). This dynamic event within single colonization units was previously thought to be impossible to visualize in mycorrhizal roots (16, 26), and it could reflect a different functional status of individual colonized cells within mycorrhizal units. We observed that in old monoxenic cultures of G. margarita and potato hairy roots, StPT3 promoter activity ceased at the time of fungal spore formation (data not shown). Overall, these data indicate that induction of StPT3 expression is a temporally controlled cell-autonomous process with no signaling occurring to neighboring noncolonized cells. Thus, StPT3 expression is most likely induced during cell-to-cell contact between the two symbionts and depends on the functional state of the fungus–plant association within a single cell. A combination of different approaches, including the Fluorescent Timer technique, could shed more light on structure–function relationships within symbiotic units of a mycorrhiza.
Fig. 4.
Spatial and temporal pattern of StPT3 promoter activity in mycorrhizal potato hairy roots. Laser scanning confocal microscopy of potato hairy roots harboring the StPT3 promoter-Fluorescent Timer chimeric gene. Roots are colonized by G. caledonium (A–C) or G. margarita (D–F). (A) Green fluorescence of a transgenic root originating mainly from the Fluorescent Timer in cells colonized by arbusculate hyphae (ah) or fungal coiled hyphae (ch) and not from noncolonized cells (n). (B) Red fluorescence from the Fluorescent Timer. (C) Yellow fluorescence originating from merged images in A and B. (D) Merged images showing transmitted light, and red and green fluorescence, respectively, of a control root harboring the StPT3 promoter-GUS gene. Green autofluorescence originates from root hairs (rh), epidermal cells (ec), and fungal coiled hyphae (ch). (E) Red fluorescence from the Fluorescent Timer. StPT3 promoter activity is localized to cortical cells colonized by fungal coiled hyphae (ch). Colonized cells exhibit high (h), residual (r), or no (n) Fluorescent Timer fluorescence. (F) Merged images showing both red and green fluorescence. Colonized cells exhibit different levels of StPT3 promoter activity: c, continuous StPT3 promoter activity leading to yellow color because of overlay of green and red fluorescence as in C; r, Fluorescent Timer expression has ceased, and residual Fluorescent Timer protein fluoresces red; n, colonized cell with no Fluorescent Timer expression. Images in A–C and D–F were captured with laser scanning confocal microscopes obtained from Zeiss and Leica, respectively. (Bars, 50 μm.)
The functional AM fungus–root interface extends far beyond the periarbuscular space and includes Paris-type hyphae and hyphal coils colonizing root cells. Our results support the hypothesis that phosphate transport in mutualistic beneficial symbioses between eudicot host plants and evolutionary ancient monophyletic AM fungi exploits conserved signal perception and transduction mechanisms. Secondly, the corresponding signal(s) is released and/or perceived at root cortex cells in a temporally defined manner in the presence of intracellular AM fungal hyphae. In our model, either an AMF-specific signal molecule or a fungal component modified by a fungal or a root cortex cell-specific secretory enzyme elicits the response to AM fungal colonization and eventually the induction of StPT3 (and probably other symbiosis-related plant genes) (see Fig. 5, which is published as supporting information on the PNAS web site). It may be important to elucidate signal perception and intracellular transduction mechanisms involved in StPT3 gene regulation.
Supplementary Material
Acknowledgments
We thank many colleagues for supplying us with the microorganisms used in this study; Sigrid Unseld (Institute of Biology, University of Stuttgart, Stuttgart, Germany) for her Fluorescent Timer plasmid; Anna Mezzacasa and Dr. Ari Helenius (Institute of Biochemistry, ETH Zurich) for support with confocal laser scanning microscopy; and Dr. Christine Rausch (Institute of Plant Sciences, ETH Zurich) for RNA samples. The work was supported by Roche Research Foundation Grant 2001-146 (to M.B. and N.A.), Research Commission of ETH Zurich Grants TH-3/00-3 and TH-5/02-3 (to M.B. and N.A.), and Swiss National Science Foundation Grant 3100-066745 (to M.B.). Some of the monoxenic cultures had been initiated at the K. A. Timiryazev Institute of Plant Physiology of the Russian Academy of Sciences (Moscow) by V.K. and were supported by Russian Fund for Basic Research Grant 01-04-48320 to Dr. Inna Kuzovkina. All other work was performed at ETH Zurich.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AM, arbuscular mycorrhiza; AMF, arbuscular mycorrhizal fungus/fungi; GUS, β-glucuronidase.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AJ318822 (StPT3), AF156695 (StPT1), AY116211 and AY116210 (MtPT4), AF000354 (MtPT1), AF533081 (PHT2;1), AY134608 and AY134609 (MtGst1), At5g43340 (ARAth;Pht1;6), and At1g20860 (ARAth;Pht1;8)].
References
- 1.van der Heijden, M. G. A., Klironomos, J. N., Ursic, M., Moutoglis, P., Streitwolf-Engel, R., Boller, T., Wiemken, A. & Sanders, I. R. (1998) Nature 396, 69-72. [Google Scholar]
- 2.Klironomos, J. N. (2002) Nature 417, 67-70. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn, G., Hijri, M. & Sanders, I. R. (2001) Nature 414, 745-748. [DOI] [PubMed] [Google Scholar]
- 4.Schussler, A., Schwarzott, D. & Walker, C. (2001) Mycol. Res. 105, 1413-1421. [DOI] [PubMed] [Google Scholar]
- 5.Simon, L., Bousquet, J., Levesque, R. C. & Lalonde, M. (1993) Nature 363, 67-69. [Google Scholar]
- 6.Remy, W., Taylor, T. N., Hass, H. & Kerp, H. (1994) Proc. Natl. Acad. Sci. USA 91, 11841-11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Redecker, D., Kodner, R. & Graham, L. E. (2000) Science 289, 1920-1921. [DOI] [PubMed] [Google Scholar]
- 8.Heckman, D. S., Geiser, D. M., Eidell, B. R., Stauffer, R. L., Kardos, N. L. & Hedges, S. B. (2001) Science 293, 1129-1133. [DOI] [PubMed] [Google Scholar]
- 9.Kistner, C. & Parniske, M. (2002) Trends Plant Sci. 7, 511-518. [DOI] [PubMed] [Google Scholar]
- 10.Smith, F. A. & Smith, S. E. (1997) New Phytol. 137, 373-388. [DOI] [PubMed] [Google Scholar]
- 11.Sahay, N. S. & Varma, A. (1999) FEMS Microbiol. Lett. 181, 297-302. [DOI] [PubMed] [Google Scholar]
- 12.Varma, A., Verma, S., Sudha, Sahay, N., Butehorn, B. & Franken, P. (1999) Appl. Environ. Microbiol. 65, 2741-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karandashov, V., Kuzovkina, I., Hawkins, H. J. & George, E. (2000) Mycorrhiza 10, 23-28. [Google Scholar]
- 14.Hoagland, D. & Broyer, T. (1936) Plant Physiol. 11, 471-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murashige, T. & Skoog, F. (1962) Physiol. Plant. 15, 473-497. [Google Scholar]
- 16.Rausch, C., Daram, P., Brunner, S., Jansa, J., Laloi, M., Leggewie, G., Amrhein, N. & Bucher, M. (2001) Nature 414, 462-466. [DOI] [PubMed] [Google Scholar]
- 17.Brundrett, M. C., Piche, Y. & Peterson, R. L. (1984) Can. J. Bot. 62, 2128-2134. [Google Scholar]
- 18.Bucher, M., Brunner, S., Zimmermann, P., Zardi, G. I., Amrhein, N., Willmitzer, L. & Riesmeier, J. W. (2002) Plant Physiol. 128, 911-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryder, M. H., Tate, M. E. & Kerr, A. (1985) Plant Physiol. 77, 215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boisson-Dernier, A., Chabaud, M., Garcia, F., Becard, G., Rosenberg, C. & Barker, D. G. (2001) Mol. Plant–Microbe Interact. 14, 695-700. [DOI] [PubMed] [Google Scholar]
- 21.Karandashov, V. E., Kuzovkina, I. N., George, E. & Marschner, H. (1999) Russ. J. Plant Physiol. 46, 87-92. [Google Scholar]
- 22.Calba, H., Jaillard, B., Fallavier, P. & Arvieu, J. C. (1996) Plant Soil 178, 67-74. [Google Scholar]
- 23.Mudge, S. R., Rae, A. L., Diatloff, E. & Smith, F. W. (2002) Plant J. 31, 341-353. [DOI] [PubMed] [Google Scholar]
- 24.Wulf, A., Manthey, K., Doll, J., Perlick, A. M., Linke, B., Bekel, T., Meyer, F., Franken, P., Kuster, H. & Krajinski, F. (2003) Mol. Plant–Microbe Interact. 16, 306-314. [DOI] [PubMed] [Google Scholar]
- 25.Blanchette, M. & Tompa, M. (2003) Nucleic Acids Res. 31, 3840-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison, M. J., Dewbre, G. R. & Liu, J. (2002) Plant Cell 14, 2413-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paszkowski, U., Kroken, S., Roux, C. & Briggs, S. P. (2002) Proc. Natl. Acad. Sci. USA 99, 13324-13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallaud, I. (1905) Revue Générale de Botanique 17, 5-48, 66-83, 123-136, 223-239, 313-325, 425-433. [Google Scholar]
- 29.Bago, B., Pfeffer, P. E. & Shachar-Hill, Y. (2000) Plant Physiol. 124, 949-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison, M. J. (1997) Trends Plant Sci. 2, 54-60. [Google Scholar]
- 31.Burleigh, S. H., Cavagnaro, T. & Jakobsen, I. (2002) J. Exp. Bot. 53, 1593-1601. [DOI] [PubMed] [Google Scholar]
- 32.Cavagnaro, T. R., Smith, F. A., Ayling, S. M. & Smith, S. E. (2003) New Phytol. 157, 127-134. [DOI] [PubMed] [Google Scholar]
- 33.Terskikh, A., Fradkov, A., Ermakova, G., Zaraisky, A., Tan, P., Kajava, A. V., Zhao, X. N., Lukyanov, S., Matz, M., Kim, S., et al. (2000) Science 290, 1585-1588. [DOI] [PubMed] [Google Scholar]
- 34.Gallaud, I. (1905) Revue Générale de Botanique 433, 479-500. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.