SUMMARY
The hexosamine biosynthetic pathway (HBP) generates UDP-GlcNAc (uridine diphosphate N-acetylglucosamine) for glycan synthesis and O-linked GlcNAc (O-GlcNAc) protein modifications. Despite the established role of the HBP in metabolism and multiple diseases, regulation of the HBP remains largely undefined. Here, we show that spliced X-box binding protein 1 (Xbp1s), the most conserved signal transducer of the unfolded protein response (UPR), is a direct transcriptional activator of the HBP. We demonstrate that the UPR triggers HBP activation via Xbp1s-dependent transcription of genes coding for key, rate-limiting enzymes. We further establish that this previously unrecognized UPR-HBP axis is triggered in a variety of stress conditions. Finally, we demonstrate a physiologic role for the UPR-HBP axis, by showing that acute stimulation of Xbp1s in heart by ischemia/reperfusion confers robust cardioprotection in part through induction of the HBP. Collectively, these studies reveal that Xbp1s couples the UPR to the HBP to protect cells under stress.
INTRODUCTION
Post-translational modification of proteins by O-linked coupling of GlcNAc (N-acetylglucosamine) is a dynamic process that governs the function of numerous proteins, both cytosolic and nuclear. O-GlcNAc modifications have been implicated in physiological and pathological responses to nutrient availability and cellular stress (Hanover et al., 2010; Zachara, 2012). Sustained increases in O-GlcNAc protein modification have been suggested to contribute to the pathogenesis of cancer, diabetes, and neurodegenerative diseases (Lazarus et al., 2009; Slawson and Hart, 2011). That said, acute up-regulation of O-GlcNAc modification promotes cell survival in the setting of various stresses (Darley-Usmar et al., 2012; Zachara, 2012).
O-GlcNAc modifications are mediated by O-GlcNAc transferase (OGT), the sole and highly conserved enzyme that conjugates O-GlcNAc groups to appropriate targets; its actions are dynamically counteracted by O-GlcNAcase (Hart et al., 2011; Slawson and Hart, 2011). The substrate of OGT is UDP-GlcNAc (uridine diphosphate N-acetylglucosamine), a nucleotide sugar which is the final product of the hexosamine biosynthetic pathway (HBP). Generation of UDP-GlcNAc by the HBP provides a substrate critical to multiple biological processes, including O-GlcNAc modification, N-glycan synthesis, and proteoglycan production. However, mechanisms governing activation of the HBP are unclear.
The UPR (unfolded protein response) is an evolutionarily conserved cellular process to cope with protein folding stress (Schroder and Kaufman, 2005; Walter and Ron, 2011). Accumulation of misfolded proteins in the ER lumen activates three major signal transducers, viz. PERK, ATF6 and IRE1. The resulting ER stress response retards protein translation, increases ER chaperone production, and enhances ER-associated protein degradation (ERAD), which together serve to restore cellular homeostasis. The IRE1 pathway is the most ancient branch of the UPR, being conserved from yeast to mammals (Hetz et al., 2011). IRE1, when activated by phosphorylation, manifests endoribonuclease activity, which cleaves a cryptic exon of 26 bp from the downstream target gene X-box binding protein 1 (Xbp1). The resulting spliced Xbp1 (Xbp1s) is a highly active transcription factor, which promotes gene expression of ER chaperones and molecules involved in ERAD. Accumulating evidence suggests that Xbp1s exerts strong pro-survival effects under various conditions, including cancer cell proliferation (Romero-Ramirez et al., 2004), plasma cell differentiation (Iwakoshi et al., 2003), inflammatory bowel disease (Kaser et al., 2008), Alzheimer disease (Casas-Tinto et al., 2011), and pancreatic acinar cell differentiation (Hess et al., 2011).
Myocardial infarction is a leading cause of mortality worldwide (Go et al., 2013). Restoration of blood flow to the infarct-related artery provokes a second wave of cell death, as the cardiomyocyte shifts to an oxygen-rich environment. Recent reports show that ischemia/reperfusion (I/R) is associated with potent increases in O-GlcNAc modification (Ngoh et al., 2011). We therefore set out to investigate the regulation of the HBP and O-GlcNAc modification under these conditions. Pathological events occurring with I/R, including Ca2+ mishandling and ROS accumulation, are potent inducers of the UPR (Murphy and Steenbergen, 2008; Turer and Hill, 2010). Here, we report that the HBP, O-GlcNAc protein modification, and the UPR are each robustly activated in heart by I/R. We demonstrate that the rate-limiting enzyme of the HBP, GFAT1 (glutamine fructose-6-phosphate aminotransferase 1), is a direct target of the UPR protein Xbp1s. Xbp1s overexpression in vivo significantly enhanced HBP flux and O-GlcNAc modification. Moreover, we report that Xbp1s is sufficient and necessary to protect heart from I/R injury, and GFAT1 is required for this cardioprotective response. Collectively, our results provide the first evidence for mechanistic coupling of the UPR and HBP, as well as uncovering a previously unrecognized role of Xbps1 in conferring robust cardioprotection from I/R injury.
RESULTS
O-GlcNAc protein modification and HBP are induced by cardiac I/R
The catalytic activity of OGT is highly sensitive to changes in UDP-GlcNAc levels, and as such, increased flux through the HBP can drive increases in O-GlcNAc protein modification (Boehmelt et al., 2000; Kreppel and Hart, 1999). Numerous studies have shown that acute induction of the HBP and O-GlcNAc protein modification protects cells from a variety of stresses, including heart disease (Darley-Usmar et al., 2012; Zachara, 2012). I/R stress results in an increase in O-GlcNAc protein modification in the ischemic region of the myocardium (Ngoh et al., 2011). Using a murine model of cardiac I/R injury, we triggered this I/R-induced increase in O-GlcNAc protein modification and assessed whether induction of HBP enzymes is a driving force underlying increased HBP flux.
Wild type (WT) male mice were subjected to cardiac ischemia for 45 min followed by reperfusion overnight (24 hrs). Hearts were separated into 3 regions: ischemic, border, and remote, according to previous triphenyltetrazolium chloride (TTC) staining patterns. Examination of the ischemic region confirmed that O-GlcNAc protein modification was increased two-fold as compared with the border zone, remote region, and sham-operated hearts (Figure 1A). This increase in O-GlcNAc modification was apparent as early as 4 hrs after reperfusion (Figure S1A). The specificity of the O-GlcNAc antibody was verified by antigen competition (Figure S1B). These data are consistent with previous reports demonstrating augmentation of O-GlcNAc modification by ischemic stress in vitro and in vivo (Champattanachai et al., 2007, 2008; Ngoh et al., 2011), and establish persistence of this modification in I/R-stressed mouse heart for at least 24 hrs.
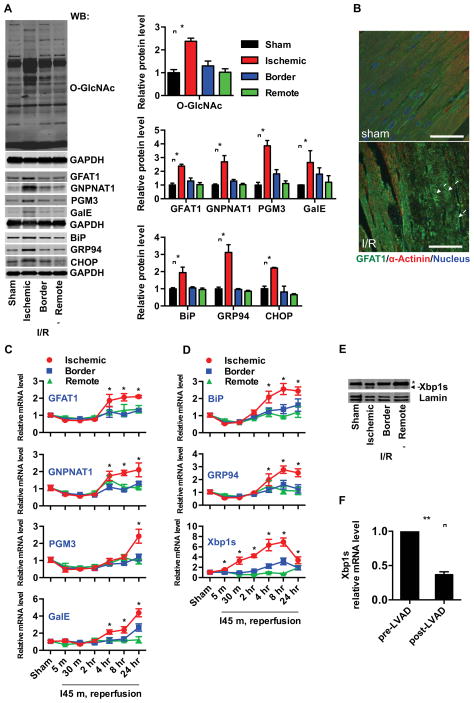
Figure 1. Induction of HBP, O-GlcNAc protein modification, and the UPR in heart by ischemia/reperfusion (I/R).
(A) Protein O-GlcNAc modification was increased in the infarct zone of I/R-stressed heart. Protein levels of the hexosamine biosynthetic pathway (HBP) and UPR markers were elevated in the same region. GAPDH was used as loading control. N = 3 for each group.
(B) Protein expression and localization of GFAT1 were visualized by fluorescence immunostaining in heart tissue from sham-operated and I/R animals. Arrows indicate cardiomyocytes in the ischemic region. Scale bars: 50 μm.
(C) Transcription of the HBP genes, GFAT1, GNPNAT1, and PGM3, as well as GalE, was induced in the infarct region of heart during I/R as assessed by qRT-PCR. Sham samples of 4 hrs and 24 hrs were pooled and used as control. N = 3–9.
(D) Transcription of UPR genes was induced in the infarct region of heart during I/R as assessed by qRT-PCR. N = 3–9.
(E) Xbp1s was increased in the infarct region of I/R-stressed heart. Lamin was used as loading control for nuclear extracts. * denotes a non-specific signal across all samples.
(F) Xbp1s mRNA levels were reduced in human heart following LVAD mechanical support. N = 8. Data are represented as mean ± SEM. *, p < 0.05, **, p < 0.01.
We next measured the abundance of HBP enzymes in these hearts. Levels of GFAT1, the rate-limiting enzyme of the HBP (Figure S1C), were significantly increased in the ischemic region, as shown by immunoblotting and immunofluorescence staining (Figures 1A, 1B, and S1D). Furthermore, the increased steady-state levels of GFAT1 protein mirrored the increased O-GlcNAc modification within the time points examined (Figures 1A and S1A). Examination of transcript levels confirmed the increase in GFAT1 specifically in the ischemic region (Figure 1C). By examining all members of the HBP pathway, we found that two additional key enzymes, were also up-regulated, viz. GNPNAT1 (glucosamine-phosphate N-acetyltransferase) and PGM3 (phosphoglucomutase 3) (Figures 1A, C, and S1A). In addition, transcript and protein levels of UDP-glucose 4-epimease (GalE), the enzyme that drives conversion between UDP-GalNAc and UDP-GlcNAc and thereby contributes to the pool of UDP-GlcNAc, were also significantly increased (Figures 1A, 1C, and S1A). Together, these data suggest that transcription of genes coding for key enzymes of the HBP is significantly up-regulated in the ischemic region of heart in a time course similar to O-GlcNAc protein modification. Further, they suggest that the O-GlcNAc protein modification is driven by increased HBP flux during I/R stress in the heart.
GFAT1 is a direct target of Xbp1s
UDP-GlcNAc, the end product of the HBP, serves not only as the substrate for O-GlcNAc modification, but also as a critical donor for protein glycosylation, which is necessary for proper protein folding. These facts, combined with the realization that previous studies reported UPR activation during cardiac ischemia (Qi et al., 2007; Thuerauf et al., 2006), led us to investigate a possible link between the HBP and the UPR. Immunofluorescence staining for the ER stress chaperone BiP revealed significant increases in the ischemic zone (Figure S1D). Examination of protein levels by immunoblotting revealed an increase in BiP, and additionally showed that other UPR target proteins, GRP94 and CHOP, were also significantly up-regulated (Figures 1A and S1A). Increases in protein levels were accompanied by significant increases in their transcripts (Figure 1D). Interestingly, the pattern of transcriptional response of the UPR target genes matched that observed for the HBP genes in both location and timing. Additionally, the transcription factor Xbp1s, a driver of UPR gene expression, was significantly induced as early as 5 min post reperfusion and increased approximately 6-fold by 4 hrs (Figure 1D). An increase in Xbp1s protein levels in the ischemic zone was also observed by immunoblotting (Figure 1E). Together, these results confirm that the UPR is activated in the ischemic zone of myocardium during I/R in vivo and suggest a link between the transcriptional control of the HBP pathway and UPR target genes.
To examine whether the increased expression of Xbp1s was relevant to human disease, we examined levels of Xbp1s in myocardial samples from patients with end-stage heart failure. Samples were obtained from patients prior to left ventricular assist device implantation, and a second sample from the same patient was obtained after the device was removed for transplantation. qRT-PCR analysis revealed significant expression of Xbp1s in the stressed hearts, which was uniformly decreased in all samples after mechanical unloading with assist device support (Figure 1F).
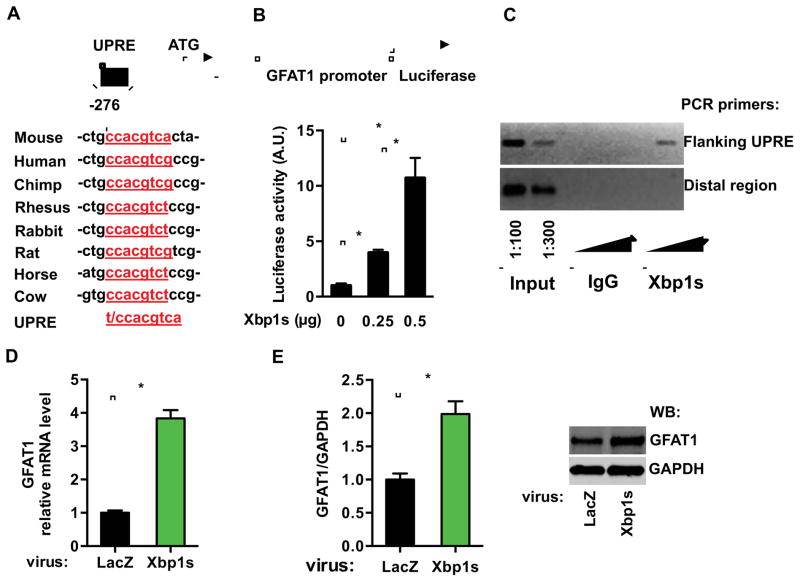
Having demonstrated that both the UPR and HBP are increased during I/R, we next tested for a link between these two processes. We first focused on GFAT1, the rate-limiting enzyme of the HBP. Sequence analysis of the GFAT1 promoter uncovered a region at −270 bp, which was highly conserved and is similar to the consensus sequence of the UPRE (Unfolded Protein Response Element), an established Xbp1s binding site (Figure 2A) (Yamamoto et al., 2004). To test whether the GFAT1 promoter could be activated by Xbp1s, we subcloned the promoter of mouse GFAT1 into a luciferase reporter vector. Co-expression of Xbp1s dramatically increased luciferase activity in HEK293T cells in a dose-dependent fashion, suggesting that Xbp1s directly stimulates the transcription of GFAT1 (Figure 2B). To corroborate this finding, we expressed Xbp1s in C2C12 cells and performed ChIP analysis (Figure S2A). Semi-quantitative PCR showed enrichment of the GFAT promoter region in the Xbp1s precipitate (Figure 2C). These results establish occupancy by Xbp1s on the endogenous GFAT1 promoter.
Figure 2. GFAT1 is a direct target of Xbp1s.
(A) A conserved DNA motif, similar to the UPRE, was identified in the GFAT1 promoter from different species.
(B) GFAT1 promoter was stimulated by Xbp1s overexpression. The GFAT1 promoter activity was measured by a luciferase assay upon Xbp1s co-transfection in HEK293T cells. N = 3 for each group.
(C) Xbp1s was associated with A ChIP assay was conducted in C2C12 cells after Xbp1s overexpression. PCR amplification was performed using primers spanning the UPRE or from a distal region in the GFAT1 promoter. The triangles indicate increasing amounts of immunoprecipitated DNA for PCR reaction.
(D) GFAT1 transcription was stimulated by Xbp1s in vitro. qPCR was conducted to quantify relative mRNA levels of GFAT1 after Xbp1s overexpression in NRVM. N = 6.
(E) GFAT1 protein levels were elevated by Xbp1s induction. N = 3–4. Data are represented as mean ± SEM. *, p < 0.05.
See also Figure S2.
To verify this relationship in cardiomyocytes, we infected neonatal rat ventricular myocytes (NRVM) in culture with lentivirus expressing either LacZ or Xbp1s. Expression of Xbp1s triggered robust up-regulation of GFAT1 at both mRNA and protein levels (Figures 2D and 2E). Collectively, these results reveal that GFAT1 is a direct transcriptional target of Xbp1s.
Xbp1s is an upstream activator of the HBP
Transcript and protein levels of the HBP-related enzymes, GNPNAT1, PGM3 and GalE, were also increased in the infarct region of myocardium following I/R (Figures 1A and 1C). Moreover, expression of Xbp1s in NRVM led to a significant up-regulation of these transcripts (Figure S2B). The induction of a number of HBP genes by I/R and Xbp1s overexpression suggests a common mechanism. Examination of the promoters of the GNPNAT1, PGM3 and GalE genes uncovered a conserved DNA motif consistent with a UPRE (Figure S2C) and consistent with our previous findings (Deng et al., 2013). Moreover, lentivirus-mediated overexpression of Xbp1s in NRVM triggered increases in O-GlcNAc protein modification (Figure S2D). Additional analysis of other enzymes from the HBP pathway, including GLUL1, GPI1, and UAP1, did not reveal consistent induction in the ischemic region of I/R hearts or in hearts from Xbp1s transgenic mice (vide infra); further, a conserved UPRE was not found in the promoter or intron regions (Figures S2E and S2F). Likewise, O-GlcNAcase (OGA), the enzyme which catalyzes removal of O-GlcNAc from proteins, was not altered by Xbp1s overexpression (Figure S2F). Collectively, these results suggest that Xbp1s is a key regulator of HBP flux by activating transcription of multiple enzyme-encoding genes in the pathway.
Increases in O-GlcNAc modifications correlate with UPR activation in a wide range of stress conditions
O-GlcNAc modification is known to play an important role in the cellular response to stress, above and beyond cardiac I/R. Our data suggest that activation of the UPR may be a universal link between O-GlcNAc modification and the cellular stress response. To test this, we examined a number of stress conditions in which O-GlcNAc protein modification has been reported to be increased. COS-7 cells were serum-starved overnight and then treated with NaCl (100 mM), CoCl2 (50 μM), or sodium arsenite (75 μM) for 8 hrs. Consistent with previous findings (Zachara et al., 2004), we observed an increase in O-GlcNAc modification (Figure S3A). Also, we found expression levels of BiP, the classical marker of the UPR, were significantly elevated. Thus, activation of O-GlcNAc protein modification correlates with the induction of the UPR. To extend these observations, we first determined that Xbp1s was indeed induced by these stress treatments. Next, we tested the requirement for Xbp1s in this process by siRNA-induced silencing. Knockdown of Xbp1s led to a significant reduction in stress-mediated augmentation of O-GlcNAc modification (Figure S3B). These data provide strong support for a universal link between UPR activation and increased cellular O-GlcNAc protein modification.
Recent studies demonstrate that glucose deprivation in NRVM leads to robust up-regulation of O-GlcNAc modification (Zou et al., 2012). We subjected NRVM to glucose starvation for 18 hrs. Protein O-GlcNAc modification was increased, which correlated with BiP induction (Figure S3C). Further, knockdown of Xbp1s significantly diminished starvation-induced increases in O-GlcNAc modification (Figure S3D). These data lend additional support to the notion that the UPR is a generalizeable, upstream trigger of the HBP and O-GlcNAc modifications.
We also examined the effects of known ER stress inducers on O-GlcNAc protein modification. Consistent with a model in which Xbp1s is an upstream activator of the HBP and O-GlcNAc modification, thapsigargin (Tg), tunicamycin (TM), or DTT each significantly augmented O-GlcNAc levels (Figure S3E). Knockdown of Xbp1s prior to administration of each ER stress inducer significantly attenuated GFAT1 induction and O-GlcNAc modification (Figure S3F). Thus induction of ER stress is itself a bona fide trigger, through Xbp1s, of cellular O-GlcNAc protein modification.
While our in vitro data strongly support a general model linking the UPR and HBP, we set up to determine whether Xbp1s induction is correlated with HBP up-regulation and increases in O-GlcNAc modification in models beyond I/R in vivo. TM is a potent inhibitor of protein N-Glycan synthesis and a well-established inducer of ER stress. We injected TM into adult male mice and harvested hearts 24 hrs later, noting robust up-regulation of genes involved in the UPR (Figure S3G). Importantly, we found enzymes within the HBP pathway and O-GlcNAc protein modification were also significantly increased. These results indicate that induction of Xbp1s, elicited by conditions other than I/R, is correlated with augmentation of the HBP and O-GlcNAc modification in heart. Collectively, these data, then, point to a direct link between activation of the UPR elicited by a variety of cellular stresses and increases in O-GlcNAc protein modification.
Cardiomyocyte-specific Xbp1s expression drives HBP flux and O-GlcNAc protein modification in vivo
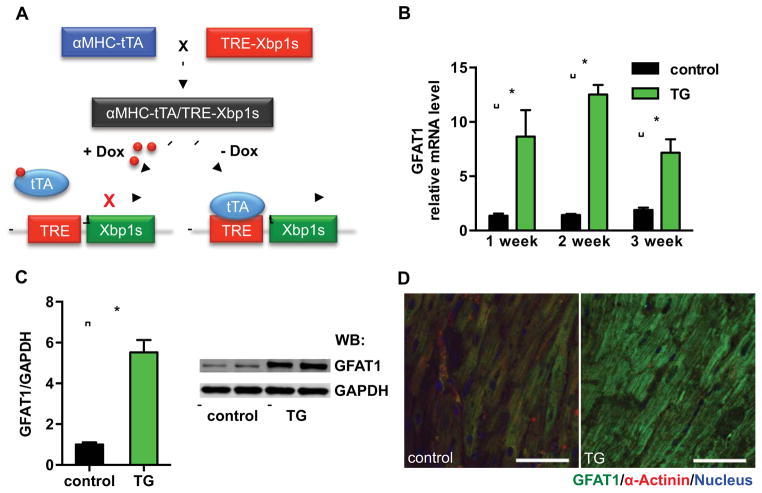
To test the link between the UPR and HBP flux in vivo, we engineered mice to induce Xbp1s expression specifically in cardiomyocytes. These animals harbored the Xbp1s coding sequence, with expression driven by seven tetracycline responsive elements. By breeding with cardiomyocyte-specific αMHC-tTA transgenic mice, Xbp1s is expressed only in cardiomyocytes and suppressed in the presence of doxycycline (Dox) (Figure 3A). These animals (including breeding pairs) were maintained on Dox-containing water to prevent transgene expression, and induction of Xbp1s was accomplished by removing Dox from the water supply.
Figure 3. Xbp1s drives GFAT1 expression in vivo.
(A) Inducible overexpression of Xbp1s in cardiomyocytes in vivo. Xbp1s expression was suppressed by doxycycline (Dox).
(B) GFAT1 transcription was significantly induced by Xbp1s overexpression. Control (αMHC-tTA only) and TG (αMHC-tTA, TRE-Xbp1s double transgenic) mice were placed on regular water (1–3 weeks) to stimulate Xbp1s expression. Cardiac GFAT1 mRNA levels were quantified by qRT-PCR. N = 3.
(C) GFAT1 protein expression was compared between control and TG mouse hearts after 2-week induction of Xbp1s. GAPDH was used as loading control. N = 3. Data are represented as mean ± SEM. *, p < 0.05.
(D) GFAT1 protein levels and expression patterns were assessed by immunostaining. Scale bars: 50 μm.
See also Figure S4.
To test the fidelity of this system, we evaluated all possible combinations of transgene and Dox. Only the double transgenic mice in the absence of Dox manifested induction of Xbp1s (Figure S4A), confirming the efficiency, tightness, and specificity of the system. Xbp1s protein abundance in these animals was approximately 3-fold higher than that induced by I/R (Figures 1E and S4B). Consistent with functional expression of the transgene, the downstream target of Xbp1s, BiP, was significantly induced (Figure S4C). We induced Xbp1s for 1–3 weeks and found that GFAT1 manifested 10-fold induction in mRNA levels and 5-fold increases in protein levels (Figures 3B and 3C). Immunofluorescence staining for GFAT1 protein confirmed the gene expression changes (Figures 3D and S4D). In contrast, GFAT2, the other member of the GFAT family, was not up-regulated by Xbp1s, but rather displayed a trend toward decreased abundance, possibly due to compensation for GFAT1 induction (Figure S4E).
Xbp1s induction leads to increases in nucleotide sugars and O-GlcNAc modification
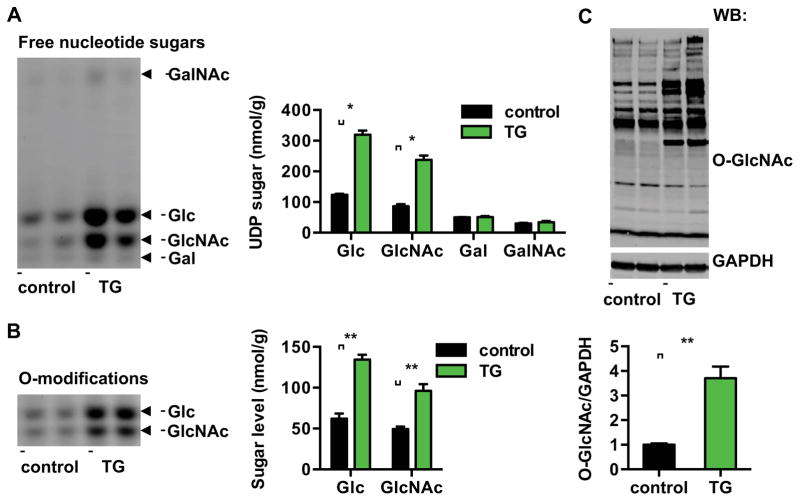
We next set out to determine whether Xbp1s-dependent activation of the HBP leads to increases in the nucleotide sugar end products of the pathway. We isolated free UDP-sugars from hearts and analyzed them by FACE (Fluorophore-Assisted Carbohydrate Electrophoresis) (Gao and Lehrman, 2006). Free nucleotide sugars of UDP-Glucose (UDP-Glc) and UDP-GlcNAc were significantly increased in Xbp1s transgenics relative to control mice (Figures 4A and S4F).
Figure 4. Xbp1s induction leads to increases in nucleotide sugars and O-GlcNAc modification.
(A) Induction of Xbp1s in cardiomyocytes led to significant increases in free nucleotide sugars in heart. Control and TG mice were placed on regular water (2 weeks) to stimulate Xbp1s expression. Free nucleotide sugars were analyzed by FACE. Samples corresponding to equal amount of total cellular proteins were loaded for each mouse strain. N = 6.
(B) Induction of Xbp1s in cardiomyocytes led to augmentation of O-Glc and O-GlcNAc protein modifications. Total cellular O-linked monosaccharides were separated by FACE gel. N = 6 for each group.
(C) Cardiac induction of Xbp1s led to significant increases in O-GlcNAc protein modification, as evaluated by immunoblotting for O-GlcNAc. GAPDH was used as loading control. N = 6 for control and n = 8 for TG mice. Data are represented as mean ± SEM. *, p < 0.05, **, p < 0.01.
See also Figure S4.
We next evaluated the O-linked monosaccharide composition of covalently modified proteins. Total proteins were isolated from control and transgenic mouse hearts and processed for β-elimination to cleave modified sugars. After separation by FACE, we detected significant increases in O-Glc and O-GlcNAc monosaccharide levels (Figures 4B and S4G).
The obligate substrate UDP-GlcNAc is transferred to accepting residues by OGT, and OGT enzymatic activity and O-GlcNAc protein modification are largely dependent on intracellular free UDP-GlcNAc levels. Consistent with the elevated levels of free UDP-GlcNAc in transgenic hearts, O-GlcNAc protein modification was significantly increased (Figure 4C). These data demonstrate that increased Xbp1s expression is sufficient to drive HBP flux and O-GlcNAc protein modification in heart.
UDP-GlcNAc is an important precursor of N-Glycan and O-Glycan sugars. FACE analysis showed a moderate but significant increase in total neutral N-Glycan in transgenic mice (Figure S4H). These findings are consistent with up-regulation of the HBP and GalE, as the latter is the epimerase that orchestrates the balance between UDP-Glc and UDP-Gal and between UDP-GlcNAc and UDP-GalNAc. No difference in negatively charged N-Glycan or O-Glycan was detected (data not shown). Together, these results reveal strong induction of O-GlcNAc protein modification, and significant increases in neutral N-Glycan by Xbp1s overexpression in heart, thereby supporting a model in which increases in the HBP pathway during cell stress are driven by Xbp1s. Further, the orchestrated up-regulation of multiple enzymes involved in the synthesis and inter-conversion of UDP-sugars provides a satisfying explanation for the increases in N-Glycan species, which may contribute to enhanced protein folding and relief of ER stress.
Xbp1 is required for induction of HBP and O-GlcNAc protein modification
Our data point to a model in which activation of the UPR by I/R triggers Xbp1s-dependent activation of the HBP and increased O-GlcNAc protein modification. To investigate this further in vivo, we engineered a mouse line harboring a cardiomyocyte-specific silencing construct of Xbp1 by crossing mice in which the Xbp1 locus is floxed with αMHC-Cre transgenic mice (cardiomyocyte-specific knockout, cKO). DNA from isolated cardiomyocytes manifested approximately 90% recombination efficiency in cKO cardiomyocytes, as measured by semi-quantitative PCR analysis. No detectable excision was identified in F/F myocytes or in non-cardiomyocytes (Figures S5A and S5B).
Xbp1 silencing caused no significant changes in basal levels of GFAT1, GNPNAT1, PGM3, GalE, or a number of UPR markers (Figure S5C). This is consistent with our findings where Xbp1 was silenced by siRNA in vitro (Figure S5D). Moreover, when we subjected the heart samples to FACE, no significant differences were found in either free nucleotide sugars or O-linked modifications (Figure S5E). Consistently, O-GlcNAc protein modification did not differ (Figure S5F).
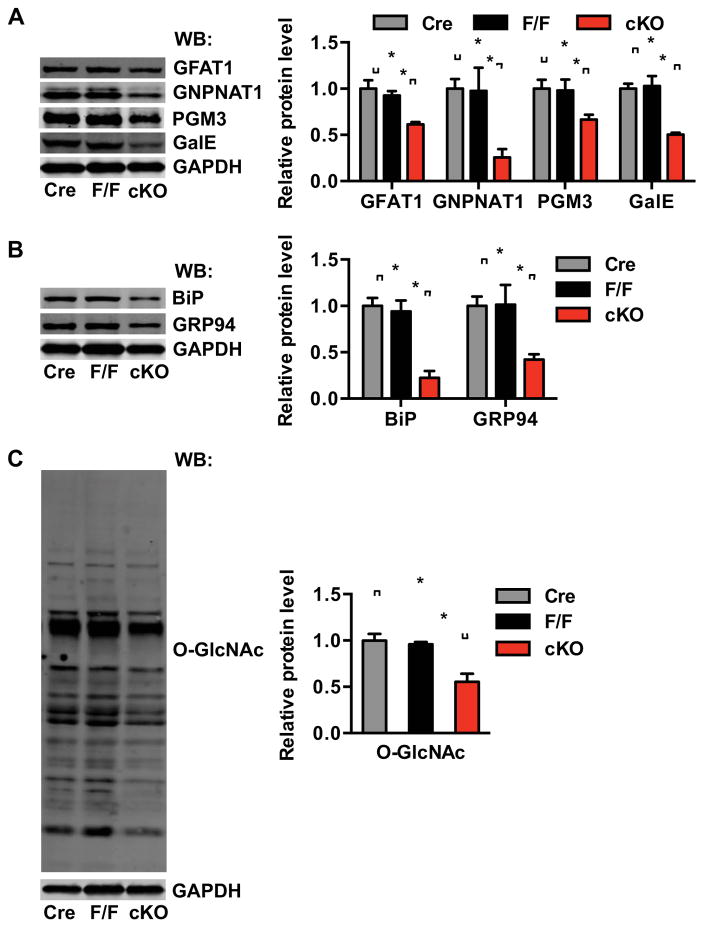
These cKO animals and littermate controls were then subjected to I/R surgery. I/R-induced GFAT1 protein expression was significantly attenuated in the cKO (Figure 5A). I/R induction of GNPNAT1, PGM3 and GalE was also significantly reduced by Xbp1 silencing. Likewise, ER stress markers were less abundant (Figure 5B). Consistent with less induction of HBP genes in cKO hearts, protein O-GlcNAc levels after I/R were significantly reduced (Figure 5C). These data demonstrate a requirement for Xbp1 in the induction of HBP and O-GlcNAc protein modification after I/R.
Figure 5. Xbp1s is required for induction of the UPR, the HBP, and O-GlcNAc modification in heart after I/R.
(A) Induction of the HBP genes, GFAT1, GNPNAT1, and PGM3, as well as GalE was analyzed in αMHC-Cre, F/F and cKO hearts 24 hrs after I/R. N = 3–4 per group.
(B) Expression of the UPR markers BiP and GRP94 was analyzed 24 hrs after I/R. N = 3–4.
(C) O-GlcNAc modification was reduced in cKO hearts compared with controls. N = 3–4. Data are represented as mean ± SEM. *, p < 0.05.
See also Figure S3, S5.
The requirement of Xbp1 in TM-induced up-regulation of the HBP and O-GlcNAc modification was also examined. TM triggered potent up-regulation of UPR markers, enzymes within the HBP pathway, and protein O-GlcNAc modification in F/F hearts (Figure S3H). As anticipated, this response was significantly blunted by loss of Xbp1 in cKO hearts. These data, then, highlight the importance of Xbp1 in ER stress-induced activation of the HBP and resulting increases in O-GlcNAc protein modification.
Xbp1s induction is sufficient and necessary to protect heart from I/R injury in vivo
We have shown that the UPR is robustly induced in heart by I/R, consistent with previous findings (Thuerauf et al., 2006). While a number of in vivo studies have focused on the ATF6 pathway in response to ischemia (Doroudgar et al., 2009; Martindale et al., 2006), a specific role for the IRE1/Xbp1s pathway in heart during I/R has not been reported.
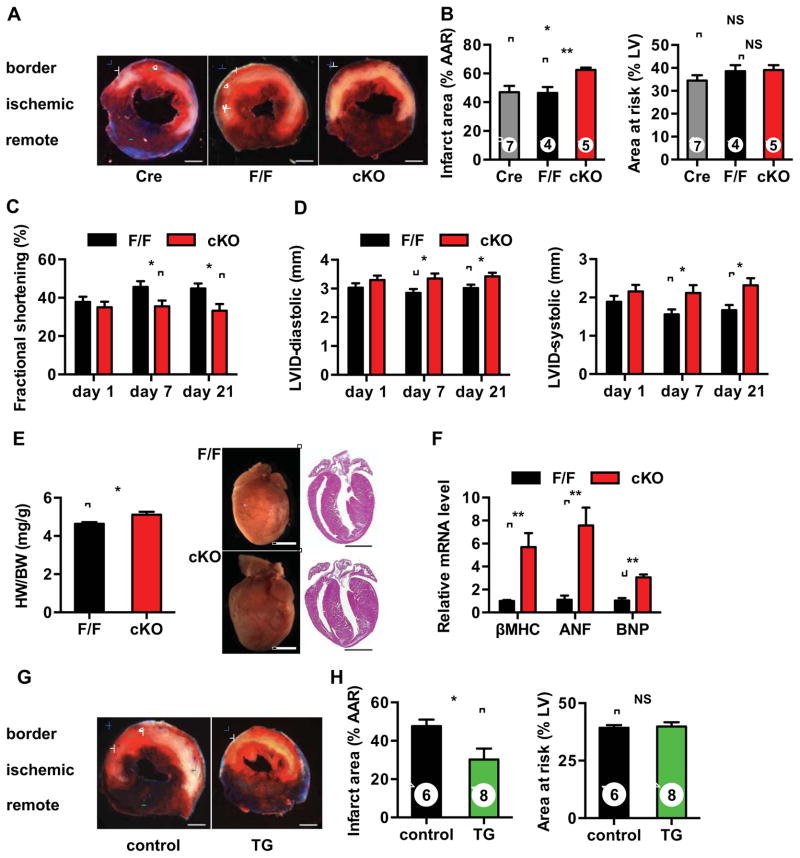
To investigate this question, we subjected cKO animals to cardiac ischemia for 45 min followed by reperfusion overnight. TTC staining was performed to measure infarct size and area at risk. These experiments revealed a significant increase in myocyte death in cKO mice compared with either F/F or αMHC-Cre controls; greater than 30% increases in infarct size were seen (Figures 6A and 6B). Surgical injury was similar in each line as evidenced by similar areas at risk of infarction.
Figure 6. Xbp1s induction protects heart from I/R injury in vivo.
(A) Xbp1 silencing led to increased injury from I/R. Male mice were subjected I/R. Cardiac injury was assayed by TTC staining. Blue, unaffected, viable tissue; red, area at risk; white, infarct area. Scale bars: 1 mm.
(B) Infarct area (relative to area at risk) and area at risk (relative to left ventricle) were quantified. Number of animals used is indicated.
(C) Ventricular function in cKO mice manifested significant deterioration as measured by % fractional shortening 7 days and 21 days post I/R. N = 5 for F/F and n = 7 for cKO.
(D) Left ventricular internal diameters (LVID) in diastole (left) and systole (right) were compared. N = 5–7.
(E) cKO mice developed more severe hypertrophy following I/R. N = 3–5. Scale bars: 2 mm.
(F) cKO mice manifested more robust fetal gene reactivation after I/R. N = 3–5.
(G) Xbp1s overexpression protected hearts from I/R injury. Control and TG mice were placed on normal water for 2 weeks to induce transgene expression. I/R surgery was performed and cardiac injury was assessed by TTC staining. Scale bars: 1 mm.
(H) Infarct area (relative to area at risk) and area at risk (relative to left ventricle) were quantified. Data are represented as mean ± SEM. *, p < 0.05, **, p < 0.01. NS, not significant.
See also Figure S5, S6.
To test for functional relevance, we examined cardiac function post I/R. At baseline, myocardial function and morphology of cKO hearts did not differ from control (Figures S5G and S5H). Significant differences, however, did become apparent after these mice were subjected to I/R. Echocardiographic analysis revealed significant deteriorations in systolic performance, quantified as percent fractional shortening, in cKO mice 7 days post I/R (Figure 6C). Left ventricular internal dimensions were also greater in cKO hearts (Figure 6D). Gross analysis revealed more profound cardiac hypertrophy in cKO mice, which was corroborated by robust fetal gene reactivation (Figures 6E and 6F). These data, then, support a model in which induction of Xbp1s during I/R is cardioprotective.
We next set out to address whether Xbp1s expression is sufficient to drive cardioprotection from I/R injury, taking advantage of our inducible transgenic mouse model. Xbp1s expression was induced by removal of Dox for two weeks. Echocardiographic analysis revealed no differences in baseline cardiac function (Figure S6A). After I/R, we observed dramatic protection against ischemic injury by Xbp1s induction, with the infarct area of the transgenic group reduced by nearly 50% (Figures 6G, 6H, and S6B). No difference in area at risk was identified between groups, confirming that the ischemic insult was similar. Functionally, echocardiographic analysis showed that the transgenic animals manifested significantly improved heart function 1 week after I/R (Figure S6C). This result is anticipated by the observed reduction in infarct size in the transgenic group. Collectively, these data demonstrate that Xbp1s expression is necessary and sufficient to protect the heart from I/R injury.
GFAT1 is required for Xbp1s-dependent cardioprotection in I/R
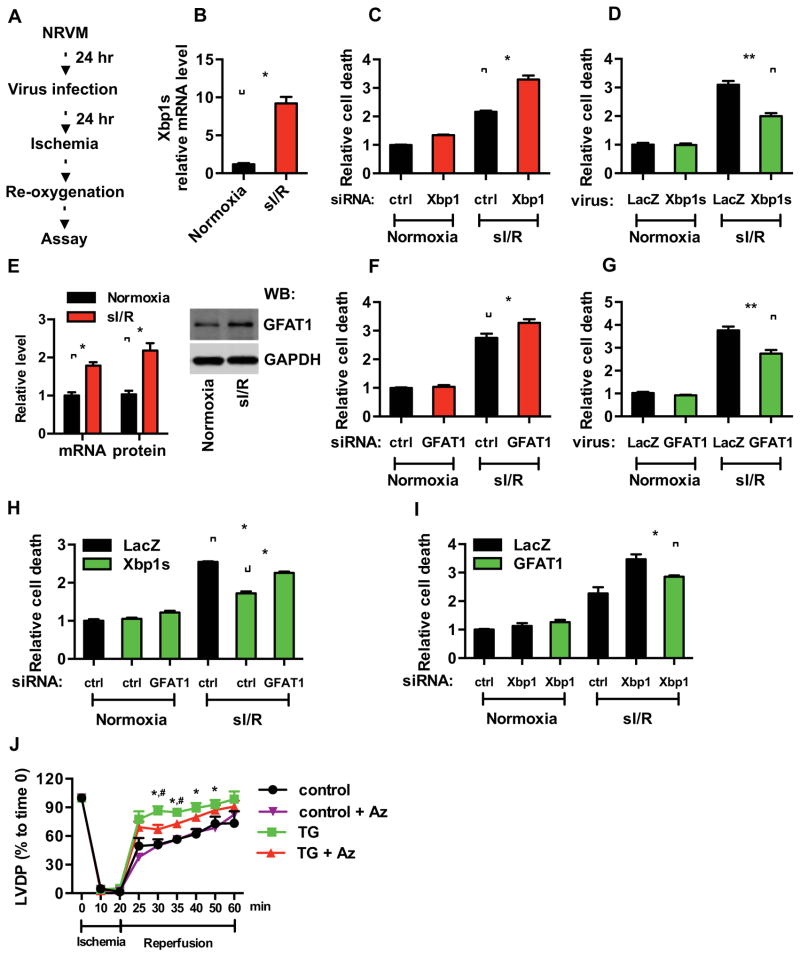
We hypothesized that Xbp1s-driven augmentation of O-GlcNAc protein modification contributes to the cardioprotective response. To examine whether GFAT1 stimulation is required in this process, we first turned to an in vitro model of I/R using NRVM (Figure 7A). Simulated I/R (sI/R) was accomplished where NRVM were exposed to ischemic conditions followed by overnight reperfusion. We found sI/R effectively induced Xbp1s expression (Figure 7B). To confirm that this model accurately mimics the protective effects of Xbp1s observed in vivo, we reduced Xbp1s levels by siRNA. Xbp1s knockdown led to significantly increased cell death as assessed by LDH release (Figures 7C and S7A). Additionally, overexpression of Xbp1s by lentiviral infection conferred significant protection from sI/R injury (Figures 7D and S7B). These data were corroborated by propidium iodide staining and ATP measurements (Figures S7C–F). Together, these in vitro results support our in vivo findings of the protective role of Xbp1s against I/R injury and afforded us a tractable model with which to test the role of the HBP pathway.
Figure 7. GFAT1 is required for Xbp1s-dependent cardioprotection during I/R.
(A) Experimental procedures for NRVM in vitro.
(B) Simulated I/R (sI/R) induced Xbp1s expression as assessed by qRT-PCR. N = 3.
(C) Knockdown of Xbp1 led to enhanced cell death in response to sI/R. Xbp1s expression was reduced by siRNA. Cell death was measured by LDH release. N = 3 for each group.
(D) Xbp1s overexpression protected cardiomyocytes from sI/R injury. NRVM were infected with lentivirus expressing either LacZ or Xbp1s. Cell death from sI/R was quantified by LDH measurements. N = 6.
(E) GFAT1 mRNA and protein levels were significantly induced in NRVM after sI/R. N = 3.
(F) Knockdown of GFAT1 exacerbated sI/R injury. GFAT1 expression was reduced by siRNA. NRVM were then subjected to sI/R, and cell death was assessed by LDH release. N = 3.
(G) GFAT1 overexpression protected cardiomyocytes from sI/R injury. NRVM were infected with lentivirus expressing either LacZ or GFAT1. Cell death from sI/R was quantified by LDH measurements. N = 6 for each group.
(H) GFAT1 knockdown diminished Xbp1s cardioprotection in sI/R. NRVM were first infected with LacZ or Xbp1s lentivirus. GFAT1 was reduced by siRNA. After sI/R, LDH assays were conducted. N = 3.
(I) Overexpression of GFAT1 significantly rescued cell death by Xbp1 silencing. NRVM were first infected with LacZ or GFAT1 lentivirus. Xbp1 was reduced by siRNA. After sI/R, cell death was quantified by LDH assay. N = 6 for each group. Data are represented as mean ± SEM. *, p < 0.05, **, p < 0.01.
(J) Inhibition of GFAT1 diminished cardioprotection by Xbp1s. Control and TG mouse hearts were processed for Langendorff analysis (ischemia 20 min; reperfusion 40 min). Cardiac function was assessed as left ventricular developed pressure (LVDP). N = 7 for control, n = 4 for TG, n = 4 for control + Azaserine (Az), and n = 4 for TG + Az. Data are represented as mean ± SEM. *, TG vs control, p < 0.05. #, TG vs TG + Az, p < 0.05.
See also Figure S7.
As GFAT1 is the rate-limiting enzyme in the HBP, we targeted GFAT1 as a means of modulating the HBP. Consistent with a model in which GFAT1 is a direct target of Xbp1s, sI/R stimulated GFAT1 expression at both mRNA and protein levels (Figures 7E). GFAT1 depletion by RNAi significantly increased NRVM death (Figures 7F and S7G). Overexpression of GFAT1 by lentivirus infection enhanced cell survival against sI/R (Figures 7G and S7H). In parallel experiments, we infected NRVM with lentivirus expressing Xbp1s and reduced GFAT1 expression by siRNA. Upon exposure to sI/R, we found that GFAT1 knockdown significantly diminished the protective effects of Xbp1s (Figure 7H). Conversely, overexpression of GFAT1 rescued the cell death observed under conditions of Xbp1s knockdown (Figure 7I). These data support the role of the HBP in Xbp1s-dependent cardioprotection.
UDP-GlcNAc is a final product of the HBP. We therefore asked whether supplementation of GlcNAc itself, thereby bypassing the need for HBP induction, would rescue the loss of Xbp1s in I/R-induced cell death. We first silenced Xbp1s by siRNA transfection and then subjected the cells to sI/R. We supplemented GlcNAc during reperfusion. Here, we found that GlcNAc significantly reduced Xbp1s knockdown-induced cell death, highlighting the importance of GlcNAc in the cardioprotection afforded by Xbp1s (Figure S7I).
OGT is the enzyme that catalyzes O-GlcNAc conjugation. To further define the role of O-GlcNAc modification from other possible protective effects of Xbp1s induction, we targeted OGT in NRVM by siRNA. We found that silencing of OGT significantly inhibited Xbp1s-mediated protection against sI/R damage (Figure S7J). Collectively, these results indicate that O-GlcNAc protein modification contributes significantly to the cardioprotective actions of Xbp1s in I/R-stressed cardiomyocytes.
To further evaluate the dependence on GFAT1 of Xbp1s-mediated cardioprotection, we next turned to the Langendorff model in which the heart is isolated and undergoes controlled I/R. Cardiac function is assessed by measuring contractile strength by left ventricular developed pressure (LVDP). Cardiomyocyte-specific Xbp1s overexpression conferred significant protection during I/R as measured by LVDP recovery (Figure 7J). Azaserine, an enzymatic inhibitor of GFAT1, was used at a concentration that does not affect basal cardiac function (Liu et al., 2007a). Azaserine treatment reduced the protective effect of Xbp1s in the transgenic hearts. Importantly, azaserine did not completely abolish the protective effect of Xbp1s expression in this assay, just as GFAT1 knockdown did not abolish the cardioprotection observed in the sI/R experiments (Figure 7H). Likewise, 6-diazo-5-oxo-l-norleucine (DON), another inhibitor of GFAT1, showed similar suppression of Xbp1s-mediated rescue of cardiac function (Figure S7K). These data, then, suggest that additional pathways of protection are triggered by the UPR during I/R (Glembotski, 2007; Groenendyk et al., 2010; Minamino et al., 2010). Collectively, these results point to a critical role of GFAT1 in the beneficial effects of Xbp1s, highlighting Xbp1s and activation of its transcriptional targets in the HBP, and consequent O-GlcNAc protein modification as pivotal factors in the cellular response to I/R stress (Figure S7L).
DISCUSSION
O-GlcNAc protein modification is a prevalent post-translational modification of numerous proteins and can modulate protein stability and function. The enzyme responsible for this conjugation, OGT, is largely dependent on cellular free UDP-GlcNAc levels for enzymatic activity. Thus, synthesis of UDP-GlcNAc through the HBP is pivotal in regulating O-GlcNAc modification. Here, we report that Xbp1s, a highly active transcription factor of the UPR, directly promotes transcription of the gene coding for GFAT1, the rate-limiting enzyme of the HBP. We also provide evidence that Xbp1s activates transcription of two additional enzyme-encoding genes within the HBP, viz. GNPNAT1 and PGM3, and the related gene GalE. Xbp1s strongly induces HBP flux, as revealed by increases in free UDP-GlcNAc and O-GlcNAc protein modifications. Further, we show that Xbp1s, the most highly conserved arm of the UPR, is induced during I/R in heart. Using a combination of gain- and loss-of-function strategies, we demonstrate that this response confers robust cardioprotection, serving to preserve myocyte viability and contractile function. Finally, we show that Xbp1s-dependent cardioprotection is largely dependent on GFAT1. Together, these studies uncover for the first time a mechanistic axis directly linking the UPR, HBP, O-GlcNAc protein modification, and associated cell survival under stress conditions.
Approximately 5% of glucose entering cells may be metabolized through the HBP; under certain conditions, the HBP contribution can be significantly higher (Darley-Usmar et al., 2012; Slawson and Hart, 2011). After uptake, glucose is fixed by phosphorylation and metabolized by various pathways. In the HBP, GFAT transfers the amine moiety from glutamine to fructose-6-phosphate. Subsequently, glucosamine-6-phosphate is modified by GNPNAT to generate N-acetylglucosamine-6-phosphate, which is rapidly converted to the ultimate product of the HBP, UDP-GlcNAc (Wells and Hart, 2003).
UDP-GlcNAc is the obligate substrate for O-GlcNAc protein modification, which has attracted considerable attention due to its immediate association with developmental and disease conditions (Slawson and Hart, 2011; Wells and Hart, 2003). Indeed, O-GlcNAc modifications can modulate protein stability and function, and are implicated in various diseases, including diabetes, neurodegeneration and cardiovascular disease (Darley-Usmar et al., 2012; Lazarus et al., 2009). Multiple lines of evidence suggest that O-GlcNAc modification is increased in various cardiac diseases, and that this increase may be protective (Champattanachai et al., 2008; Jones et al., 2008; Laczy et al., 2010; Liu et al., 2007b; Ngoh et al., 2009a; Watson et al., 2010). However, prior to this report, mechanisms triggering the upstream HBP synthetic cascade have remained elusive.
OGT manifests significantly higher affinity for UDP-GlcNAc compared to several other enzymes of UDP-GlcNAc-consuming reactions (Darley-Usmar et al., 2012). Moreover, deficiency of GNPNAT1 leads to substantial reductions in UDP-GlcNAc levels, which translate into decreased O-GlcNAc modification (Boehmelt et al., 2000). OGT activity and O-GlcNAc levels are largely dependent on free UDP-GlcNAc levels (Kreppel and Hart, 1999). Data reported here provide direct evidence that Xbp1s stimulates the HBP pathway, which leads to increases in UDP-GlcNAc production and O-GlcNAc modification. Interestingly, O-GlcNAc modification has been shown to reduce ER stress-induced cell death in cardiomyocytes (Ngoh et al., 2009b). Likewise, GlcNAc treatment can significantly attenuate cell death elicited by glucose deprivation and UPR activation, suggesting a possible feedback role for HBP flux in antagonizing the detrimental effects of ER stress (Palorini et al., 2013).
The UPR is a ubiquitous response to a variety of cellular insults, many of which provoke protein folding stress (Schroder and Kaufman, 2005). Even under physiological conditions, the UPR toggles on and off to cope with the normal protein folding demands of cell growth and proliferation. The UPR is an elegant and complex response to transduce signaling from the affected organelles (e.g. ER for ER stress) to the nucleus for restoration of homeostasis. At least three UPR branches have been described in the ER. The IRE1/Xbp1s branch in mammals functions to resolve protein-folding stress and restore ER homeostasis in multiple cell types under various conditions (Casas-Tinto et al., 2011; Hess et al., 2011; Iwakoshi et al., 2003; Kaser et al., 2008; Romero-Ramirez et al., 2004; Thuerauf et al., 2006).
In heart, restoration of coronary blood flow is critical in limiting damage due to acute myocardial infarction and improving clinical outcomes. A large body of literature, however, reveals that reperfusion paradoxically triggers lethal damage to cardiomyocytes (Hausenloy and Yellon, 2013; Murphy and Steenbergen, 2008; Yellon and Hausenloy, 2007). Despite this fundamental fact, no therapies targeting reperfusion injury are available for clinical use. Calcium mishandling in both the cytosol and mitochondria and mitochondrial ROS over-production are among the most critical contributors to the pathogenesis of I/R injury. Importantly, both of these events are well-established inducers of protein misfolding and the UPR. Indeed, Glembotski and colleagues have reported up-regulation of ER chaperones upon I/R in both in vitro and ex vivo models, highlighting the possible involvement of the UPR in I/R pathogenesis (Doroudgar et al., 2009; Martindale et al., 2006; Thuerauf et al., 2006). Here, we employed an in vivo approach, finding that UPR chaperones are stimulated in the ischemic zone of I/R-stressed tissue. Additionally, we provide evidence that the UPR is activated in human hearts under stress.
Using both gain- and loss-of-function strategies, we show that Xbp1s induction protects the heart from I/R injury in vivo. Xbp1s is a powerful transcription factor, targeting an array of ER chaperones and a group of molecules involved in ERAD. Mechanistically, these proteins may serve as a link between Xbp1s and cellular protection. Overexpression of the ER chaperone BiP in cardiomyocytes in vitro reduces cell death triggered by proteasome inhibition (Fu et al., 2008). Similar findings have been reported for the chaperone PDI (Severino et al., 2007). Even brief pharmacological stimulation of ER stress using tunicamycin can improve heart function during I/R in vivo (Petrovski et al., 2011). All these reports indicate that ER chaperones may mediate, at least in part, the cardioprotective actions of Xbp1s. Involvement of other pathways was largely unknown.
Whereas protein chaperones and ERAD contribute importantly to protein homeostasis, correct protein folding also requires proper post-translational glycosylation. Nucleotide sugars are intermediate substrates of glycosylation, and their abundances and relative concentrations are significant determinants of protein folding. Among them, UDP-GlcNAc is synthesized de novo by the HBP. Moreover, the proper ratios of UDP sugars are governed by the epimerase GalE. We have shown here that GFAT1, the rate-limiting enzyme of the HBP, and GalE, are direct targets of Xbp1s. Thus, HBP activation and control of the relative abundances of nucleotide sugars may contribute significantly to the beneficial effects of Xbp1s during I/R.
Our data strongly suggest that O-GlcNAc protein modification contributes to Xbp1s-mediated cardioprotection against I/R. At least one major question remains: How does O-GlcNAc modification protect the cell? GlcNAc, a molecule containing a sugar backbone, an amine group, and an acetyl group, is a unique cellular element, potentially serving as a reservoir of basic nutrients. Moreover, O-GlcNAc targets Ser/Thr residues for modification, which may either suppress or enhance phosphorylation (Hart et al., 2011). The cardioprotection mediated by O-GlcNAc modification during I/R may stem, at least in part, from its modulation of phosphorylation of key elements in the cell death pathway.
In conclusion, Xbp1s is a direct inducer of the HBP and, as a consequence, O-GlcNAc modification. Xbp1s is induced in heart during I/R in vivo, and Xbp1s exerts robust cardioprotection against I/R injury. Thus, our findings uncover a direct mechanistic link between the UPR, the HBP, O-GlcNAc modification, and cardioprotection.
EXPERIMENTAL PROCEDURES
Animals
Wild-type male mice (8–12 weeks old) of C57/B6 background were used for ischemia/reperfusion surgery. The TRE-Xbp1s transgenic mouse was generated in FVB and then backcrossed into C57/B6 background for at least 9 generations (Deng et al., 2013). We crossed the TRE-Xbp1s mouse with the αMHC-tTA mouse model (Yu et al., 1996). Floxed Xbp1 mice (Kaser et al., 2008) were bred with αMHC-Cre mice to achieve cardiomyocyte-specific deletion of Xbp1.
Cardiomyocyte isolation and treatment
Neonatal rat ventricular cardiomyocytes (NRVM) were isolated from 1–2 day old Sprague-Dawley rat pups. For simulated I/R, cells were changed to I/R buffer and placed in a hypoxia chamber (Billups-Rothenberg) and flushed with 95% N2/5% CO2 for 30 min. The chamber was closed for an additional 3.5–4.5 hrs, followed by reperfusion with culture medium. NRVM incubated with control buffer were used as controls. LDH assays were conducted using the CytoTox96 cytotoxicity kit (Promega). Cell survival was also measured by the CellTiter-Glo viability assay kit (Promega), and propidium iodide staining (1 μg/mL).
RNA isolation and PCR analysis
Total RNA was isolated from heart tissue or NRVM using the Total RNA Fatty and Fibrous Tissue Kit (Bio-Rad). All primer sequences are provided (Table S1).
Immunoblotting
Heart tissue lysate was prepared in T-PER (Thermo) containing protease inhibitors and phospho-STOP (Roche). When preparing lysate for O-GlcNAc analysis, PUGNAc (40 μM) was included to inhibit OGA activity. To isolate nuclear extracts, the NE-PER kit was used (Thermo). Immunoblotting was done with an Odyssey scanner (LI-COR).
Langendorff experiments
Heart was quickly removed and the aorta was cannulated on a blunted 21 gauge needle. Ischemia was initiated by arresting perfusion for 20 min followed by reperfusion for 40 min. Left ventricular pressure was recorded with a pressure transducer using a ventricular balloon. Azaserine (80 μM) or 6-diazo-5-oxo-l-norleucine (DON) (50 μM) was employed to inhibit GFAT enzymatic activity.
Nucleotide sugar and protein glycosylation analysis in heart
Snap-frozen heart tissue was pulverized directly in methanol, dried, and extracted with chloroform:methanol, water, and chloroform:methanol:water. The water fraction was collected for free nucleotide sugar analysis. The remaining pellet was used to analyze sugar modification on proteins.
Patient samples
Human heart tissue samples were obtained from patients with advanced heart failure. Paired ventricular tissue samples were obtained from each patient at the time of left ventricular assist device implantation and then at the time of heart transplantation. Patient information is shown in Table S2.
Statistical analysis
All data were expressed as mean ± SEM. The Student t test was performed to compare 2 groups. One-way ANOVA was used to analyze multiple groups. Two-way ANOVA and subsequent Tukey tests were performed to analyze time course studies. A p value less than 0.05 was considered as significant.
Supplementary Material
HIGHLIGHTS.
Ischemia/reperfusion (I/R) in heart activates spliced X-box binding protein 1 (Xbp1s).
Xbp1s triggers expression of multiple enzymes of the hexosamine biosynthetic pathway.
Xbp1s increases O-GlcNAc modification on cardiac proteins.
Xbp1s protects heart from I/R injury.
Acknowledgments
We thank Herman May and Yongli Kong for technical assistance. We thank Dr. Gary Wright (Department of Pharmacology, Quillen College of Medicine, East Tennessee State University) for kindly providing equipment for the Langendorff experiments. This work was supported by grants from the NIH (HL-080144, JAH: HL-0980842, JAH; HL-100401, JAH; DK-55758, PES; DK-088761, PES; GM-038545, MAL; HL-102478-02, PPAM), CPRIT (RP110486P3), the AHA DeHaan Foundation (0970518N), and the Fondation Leducq (11CVD04). Z.V.W. was supported by a postdoctoral fellowship from the AHA (10POST4320009). Y.D. was supported by a postdoctoral fellowship from the ADA (7-08-MN-53).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boehmelt G, Wakeham A, Elia A, Sasaki T, Plyte S, Potter J, Yang Y, Tsang E, Ruland J, Iscove NN, et al. Decreased UDP-GlcNAc levels abrogate proliferation control in EMeg32-deficient cells. EMBO J. 2000;19:5092–5104. doi: 10.1093/emboj/19.19.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Tinto S, Zhang Y, Sanchez-Garcia J, Gomez-Velazquez M, Rincon-Limas DE, Fernandez-Funez P. The ER stress factor XBP1s prevents amyloid-beta neurotoxicity. Hum Mol Genet. 2011;20:2144–2160. doi: 10.1093/hmg/ddr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champattanachai V, Marchase RB, Chatham JC. Glucosamine protects neonatal cardiomyocytes from ischemia-reperfusion injury via increased protein-associated O-GlcNAc. Am J Physiol Cell Physiol. 2007;292:C178–187. doi: 10.1152/ajpcell.00162.2006. [DOI] [PubMed] [Google Scholar]
- Champattanachai V, Marchase RB, Chatham JC. Glucosamine protects neonatal cardiomyocytes from ischemia-reperfusion injury via increased protein O-GlcNAc and increased mitochondrial Bcl-2. Am J Physiol Cell Physiol. 2008;294:C1509–1520. doi: 10.1152/ajpcell.00456.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darley-Usmar VM, Ball LE, Chatham JC. Protein O-linked beta-N-acetylglucosamine: a novel effector of cardiomyocyte metabolism and function. J Mol Cell Cardiol. 2012;52:538–549. doi: 10.1016/j.yjmcc.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Wang ZV, Tao C, Gao N, Holland WL, Ferdous A, Repa JJ, Liang G, Ye J, Lehrman MA, et al. The Xbp1s/GalE axis links ER stress to postprandial hepatic metabolism. J Clin Invest. 2013;123:455–468. doi: 10.1172/JCI62819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroudgar S, Thuerauf DJ, Marcinko MC, Belmont PJ, Glembotski CC. Ischemia activates the ATF6 branch of the endoplasmic reticulum stress response. J Biol Chem. 2009;284:29735–29745. doi: 10.1074/jbc.M109.018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu HY, Minamino T, Tsukamoto O, Sawada T, Asai M, Kato H, Asano Y, Fujita M, Takashima S, Hori M, et al. Overexpression of endoplasmic reticulum-resident chaperone attenuates cardiomyocyte death induced by proteasome inhibition. Cardiovasc Res. 2008;79:600–610. doi: 10.1093/cvr/cvn128. [DOI] [PubMed] [Google Scholar]
- Gao N, Lehrman MA. Non-radioactive analysis of lipid-linked oligosaccharide compositions by fluorophore-assisted carbohydrate electrophoresis. Methods Enzymol. 2006;415:3–20. doi: 10.1016/S0076-6879(06)15001-6. [DOI] [PubMed] [Google Scholar]
- Glembotski CC. Endoplasmic reticulum stress in the heart. Circ Res. 2007;101:975–984. doi: 10.1161/CIRCRESAHA.107.161273. [DOI] [PubMed] [Google Scholar]
- Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, et al. Heart disease and stroke statistics--2013 update: a report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenendyk J, Sreenivasaiah PK, Kim do H, Agellon LB, Michalak M. Biology of endoplasmic reticulum stress in the heart. Circ Res. 2010;107:1185–1197. doi: 10.1161/CIRCRESAHA.110.227033. [DOI] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta. 2010;1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J Clin Invest. 2013;123:92–100. doi: 10.1172/JCI62874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DA, Humphrey SE, Ishibashi J, Damsz B, Lee AH, Glimcher LH, Konieczny SF. Extensive pancreas regeneration following acinar-specific disruption of Xbp1 in mice. Gastroenterology. 2011;141:1463–1472. doi: 10.1053/j.gastro.2011.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol Rev. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- Jones SP, Zachara NE, Ngoh GA, Hill BG, Teshima Y, Bhatnagar A, Hart GW, Marban E. Cardioprotection by N-acetylglucosamine linkage to cellular proteins. Circulation. 2008;117:1172–1182. doi: 10.1161/CIRCULATIONAHA.107.730515. [DOI] [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem. 1999;274:32015–32022. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- Laczy B, Marsh SA, Brocks CA, Wittmann I, Chatham JC. Inhibition of O-GlcNAcase in perfused rat hearts by NAG-thiazolines at the time of reperfusion is cardioprotective in an O-GlcNAc-dependent manner. Am J Physiol Heart Circ Physiol. 2010;299:H1715–1727. doi: 10.1152/ajpheart.00337.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus BD, Love DC, Hanover JA. O-GlcNAc cycling: implications for neurodegenerative disorders. Int J Biochem Cell Biol. 2009;41:2134–2146. doi: 10.1016/j.biocel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Marchase RB, Chatham JC. Glutamine-induced protection of isolated rat heart from ischemia/reperfusion injury is mediated via the hexosamine biosynthesis pathway and increased protein O-GlcNAc levels. J Mol Cell Cardiol. 2007a;42:177–185. doi: 10.1016/j.yjmcc.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Marchase RB, Chatham JC. Increased O-GlcNAc levels during reperfusion lead to improved functional recovery and reduced calpain proteolysis. Am J Physiol Heart Circ Physiol. 2007b;293:H1391–1399. doi: 10.1152/ajpheart.00285.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale JJ, Fernandez R, Thuerauf D, Whittaker R, Gude N, Sussman MA, Glembotski CC. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res. 2006;98:1186–1193. doi: 10.1161/01.RES.0000220643.65941.8d. [DOI] [PubMed] [Google Scholar]
- Minamino T, Komuro I, Kitakaze M. Endoplasmic reticulum stress as a therapeutic target in cardiovascular disease. Circ Res. 2010;107:1071–1082. doi: 10.1161/CIRCRESAHA.110.227819. [DOI] [PubMed] [Google Scholar]
- Murphy E, Steenbergen C. Mechanisms underlying acute protection from cardiac ischemia-reperfusion injury. Physiol Rev. 2008;88:581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoh GA, Facundo HT, Hamid T, Dillmann W, Zachara NE, Jones SP. Unique hexosaminidase reduces metabolic survival signal and sensitizes cardiac myocytes to hypoxia/reoxygenation injury. Circ Res. 2009a;104:41–49. doi: 10.1161/CIRCRESAHA.108.189431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoh GA, Hamid T, Prabhu SD, Jones SP. O-GlcNAc signaling attenuates ER stress-induced cardiomyocyte death. Am J Physiol Heart Circ Physiol. 2009b;297:H1711–1719. doi: 10.1152/ajpheart.00553.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoh GA, Watson LJ, Facundo HT, Jones SP. Augmented O-GlcNAc signaling attenuates oxidative stress and calcium overload in cardiomyocytes. Amino Acids. 2011;40:895–911. doi: 10.1007/s00726-010-0728-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palorini R, Cammarata F, Balestrieri C, Monestiroli A, Vasso M, Gelfi C, Alberghina L, Chiaradonna F. Glucose starvation induces cell death in K-ras-transformed cells by interfering with the hexosamine biosynthesis pathway and activating the unfolded protein response. Cell Death Dis. 2013;4:e732. doi: 10.1038/cddis.2013.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovski G, Das S, Juhasz B, Kertesz A, Tosaki A, Das DK. Cardioprotection by endoplasmic reticulum stress-induced autophagy. Antioxid Redox Signal. 2011;14:2191–2200. doi: 10.1089/ars.2010.3486. [DOI] [PubMed] [Google Scholar]
- Qi X, Vallentin A, Churchill E, Mochly-Rosen D. deltaPKC participates in the endoplasmic reticulum stress-induced response in cultured cardiac myocytes and ischemic heart. J Mol Cell Cardiol. 2007;43:420–428. doi: 10.1016/j.yjmcc.2007.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Ramirez L, Cao H, Nelson D, Hammond E, Lee AH, Yoshida H, Mori K, Glimcher LH, Denko NC, Giaccia AJ, et al. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004;64:5943–5947. doi: 10.1158/0008-5472.CAN-04-1606. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Severino A, Campioni M, Straino S, Salloum FN, Schmidt N, Herbrand U, Frede S, Toietta G, Di Rocco G, Bussani R, et al. Identification of protein disulfide isomerase as a cardiomyocyte survival factor in ischemic cardiomyopathy. J Am Coll Cardiol. 2007;50:1029–1037. doi: 10.1016/j.jacc.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer. 2011;11:678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuerauf DJ, Marcinko M, Gude N, Rubio M, Sussman MA, Glembotski CC. Activation of the unfolded protein response in infarcted mouse heart and hypoxic cultured cardiac myocytes. Circ Res. 2006;99:275–282. doi: 10.1161/01.RES.0000233317.70421.03. [DOI] [PubMed] [Google Scholar]
- Turer AT, Hill JA. Pathogenesis of myocardial ischemia-reperfusion injury and rationale for therapy. Am J Cardiol. 2010;106:360–368. doi: 10.1016/j.amjcard.2010.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Watson LJ, Facundo HT, Ngoh GA, Ameen M, Brainard RE, Lemma KM, Long BW, Prabhu SD, Xuan YT, Jones SP. O-linked beta-N-acetylglucosamine transferase is indispensable in the failing heart. Proc Natl Acad Sci U S A. 2010;107:17797–17802. doi: 10.1073/pnas.1001907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L, Hart GW. O-GlcNAc turns twenty: functional implications for post-translational modification of nuclear and cytosolic proteins with a sugar. FEBS Lett. 2003;546:154–158. doi: 10.1016/s0014-5793(03)00641-0. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem. 2004;136:343–350. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- Yellon DM, Hausenloy DJ. Myocardial reperfusion injury. N Engl J Med. 2007;357:1121–1135. doi: 10.1056/NEJMra071667. [DOI] [PubMed] [Google Scholar]
- Yu Z, Redfern CS, Fishman GI. Conditional transgene expression in the heart. Circ Res. 1996;79:691–697. doi: 10.1161/01.res.79.4.691. [DOI] [PubMed] [Google Scholar]
- Zachara NE. The roles of O-linked beta-N-acetylglucosamine in cardiovascular physiology and disease. Am J Physiol Heart Circ Physiol. 2012;302:H1905–1918. doi: 10.1152/ajpheart.00445.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachara NE, O’Donnell N, Cheung WD, Mercer JJ, Marth JD, Hart GW. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J Biol Chem. 2004;279:30133–30142. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]
- Zou L, Zhu-Mauldin X, Marchase RB, Paterson AJ, Liu J, Yang Q, Chatham JC. Glucose Deprivation-induced Increase in Protein O-GlcNAcylation in Cardiomyocytes Is Calcium-dependent. J Biol Chem. 2012;287:34419–34431. doi: 10.1074/jbc.M112.393207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.